Abstract
Here we present the Coon OMSSA Proteomic Analysis Software Suite (COMPASS): a free and open-source software pipeline for high-throughput analysis of proteomics data, designed around the Open Mass Spectrometry Search Algorithm. We detail a synergistic set of tools for protein database generation, spectral reduction, peptide false discovery rate analysis, peptide quantitation via isobaric labeling, protein parsimony and protein false discovery rate analysis, and protein quantitation. We strive for maximum ease of use, utilizing graphical user interfaces and working with data files in the original instrument vendor format. Results are stored in plain text comma-separated values files, which are easy to view and manipulate with a text editor or spreadsheet program. We illustrate the operation and efficacy of COMPASS through the use of two LC–MS/MS datasets. The first is a dataset of a highly annotated mixture of standard proteins and manually validated contaminants that exhibits the identification workflow. The second is a dataset of yeast peptides, labeled with isobaric stable isotope tags and mixed in known ratios, to demonstrate the quantitative workflow. For these two datasets, COMPASS performs equivalently or better than the current de facto standard, the Trans-Proteomic Pipeline.
Keywords: Informatics, Protein identification, Protein quantitation, Proteomics, Software
1 Introduction
LC–MS/MS is the analytical tool of choice for assessing the protein content of a biological sample [1]. Over the past fifteen years, several database search algorithms have been developed for the pivotal task of matching experimental tandem mass spectra to peptide sequences through the use of a protein database, such as Sequest [2] and Mascot [3]. More recently, open-source software such as X!Tandem [4] and the Open Mass Spectrometry Search Algorithm (OMSSA) [5] has been released for this purpose. These free alternatives are competitive with their commercial counterparts [6] and have been steadily gaining popularity. However, a variety of common tasks require software in addition to database searching. These include protein database generation, spectral reduction, peptide false discovery rate (FDR) analysis, peptide quantitation, protein parsimony and protein FDR analysis, and protein quantitation. As many of these tasks are relatively recent additions to data processing workflows, software supporting them is far less mature. Nonetheless, they are essential for contemporary proteomics. FDR analysis, for example, is critical for maximizing sensitivity while simultaneously controlling specificity. Tools for performing these discrete tasks are sometimes freely available, but often from disparate sources and are usually not explicitly designed to work together [7]. Additionally, they are often intended for older database search algorithms (i.e., Sequest and Mascot) rather than the newer alternatives.
A software suite, comprising most of the tools necessary for typical proteomic data analysis, resolves this problem. Built on the pioneering algorithms PeptideProphet [8] and ProteinProphet [9], the Trans-Proteomics Pipeline (TPP) [10–12] is the de facto standard for such a software suite. The TPP, designed around open extensible markup language (XML) files [13, 14], admirably strives for maximum flexibility, with the ability to read input data files in a variety of instrument vendor formats. The TPP is primarily designed for peptide identification with the commercial tools Sequest or Mascot, although it has recently been adapted [11] to support OMSSA and various other search tools including SpectraST [15, 16], an open-source spectrum library algorithm.
Here we describe a free and open-source software package for use with OMSSA [5], which provides excellent results, speed, scalability, and flexibility. The software currently accepts Thermo Scientific .raw format files as input, but is readily adaptable to data files in other formats. The main output format is simple, plain text comma-separated values (CSV) files, which are easy to view and manipulate with a text editor or spreadsheet program. Unlike XML, CSV files are very intuitive for non-programmers, and require only standard office software to work with efficiently. The CSV files are originally an output of OMSSA, and are extended by simply appending columns with additional data. At later stages of the workflow, peptide- and protein-centric CSV files are created by the software to complement OMSSA’s original spectrum-centric CSV output files. All of the software in this suite is designed to work in Microsoft Windows (with the exception of OMSSA, which is cross-platform), and all programs have a graphical user interface (GUI) for ease of use. The suite contrasts with most other proteomics packages, such as Virtual Expert Mass Spectrometrist [17] and Proteios [18], in that it intended to be operated autonomously on a single desktop computer as opposed to a client–server model which can require considerable administration.
Note we aim to provide neither the absolute state of the art in proteomic data analysis, nor the tools for every possible analytical task. Rather, we intend to make available easy-to-use software for automatically applying the commonly accepted rules for interpretation of shotgun proteomics data, a chore which can be quite daunting without viable software. We anticipate that this free, open-source suite will constitute the backbone of software infrastructure for labs looking to perform high-throughput proteomics with OMSSA as the primary database search algorithm. The software is available at http://www.chem.wisc.edu/~coon/software.php#compass.
2 Materials and methods
2.1 Software
2.1.1 Development
All software (with the exception of OMSSA [5], which was developed by the National Center for Biotechnology Information) was developed in C# with Microsoft Visual Studio 2005/2008/2010 and the Microsoft .NET Framework version 2.0/3.5 (freely available at http://www.microsoft.com/NET/). Access to data in the proprietary .raw file format was enabled by the XRawfile Component Object Model (COM) library (XRawfile2.dll, installed automatically with Thermo Xcalibur).
2.1.2 Protein database generation
Database Maker creates protein databases for target–decoy searching [19]. Each protein sequence in an input .fasta text file is converted to a decoy version of the same length by reversing, shuffling, or generating random amino acids (the N-terminus can optionally be excluded to account for initiator methionines). The resulting concatenated target–decoy .fasta protein database is automatically converted to the basic local alignment search tool format for use with OMSSA.
2.1.3 Spectral reduction
DTA Generator reduces LC–MS/MS data to merged .dta text files for database searching with OMSSA. To facilitate different search parameters, spectra are automatically split into separate files for each combination of fragmentation method and MS/MS mass analyzer. For each MS/MS spectrum, if the precursor charge state was determined by the instrument firmware, only a single spectrum at that charge state is generated. If the charge is unknown (either due to ambiguous or low-resolution MS1 data), a spectrum is generated for each precursor charge state in a user-defined range. Removal of remaining precursor is optional, as well as electron-transfer dissociation (ETD) pre-processing to remove precursor, charge-reduced precursors, and neutral losses from charge-reduced precursors [20].
2.1.4 Peptide identification
OMSSA [5] (http://pubchem.ncbi.nlm.nih.gov/omssa/, version 2.1.7) was used for peptide identification by protein database search. The CSV output option (-oc) was used.
2.1.5 Peptide FDR analysis
FDR Optimizer calculates spectrum score and precursor mass error thresholds to maximize the number of target identifications at a given error rate. First, the best peptide–spectrum match (PSM) for each spectrum, as determined by expectation value (e-value), is extracted. The precursor mass error is determined by first finding the isolation center m/z peak (i.e., scan filter m/z) in the preceding MS1 spectrum. This isolation m/z is converted to neutral mass and compared to the monoisotopic mass of the identified peptide. The nearest multiple of 1.00335 Da (carbon-13 mass minus carbon-12 mass, the main contributor to peptide isotopic peaks) is subtracted, and this mass error is converted to ppm.
High-confidence identifications are leveraged to determine the systematic precursor mass error for post-acquisition recalibration. First, a preliminary 1% spectrum FDR threshold is established. The median precursor mass error of the PSMs below the FDR threshold is taken to be the systematic error, and this quantity is subtracted from every precursor mass error to yield an adjusted precursor mass error. This correction more effectively allows the use of a symmetric precursor mass error window, which greatly improves analysis speed.
To appropriately combine results from different searches, q-values [21, 22] are then computed for each PSM, without regard to precursor mass accuracy. The final FDR optimization considers q-values instead of e-values. Iteratively, each precursor mass error threshold is applied, and the q-value threshold is adjusted until the desired error rate is obtained. The thresholds yielding the maximum number of target identifications are used.
This program has many key options. First is the ability to select between low- or high-resolution precursor analysis. If an FT MS1 scan is available, the high-resolution option will perform a two-dimensional analysis utilizing both the q-value and precursor mass error of every PSM. The low-resolution version performs a simple one-dimensional analysis utilizing only q-value.
Another option is batch versus non-batch analysis. For experiments spanning multiple LC–MS/MS runs, it is often critical to establish a FDR for the entire dataset, requiring the batch version. Other times, such as when analyses are being compared, it is desirable to have a constant FDR threshold for each dataset, and non-batch is preferable. A final critical option is the ability to select between FDR analysis at the PSM or unique peptide level. If the maximum number of accepted spectra is desired, the former is preferable, but for most proteomic studies, the latter is more appropriate. Note that the software defines a unique peptide as a distinct combination of amino acid sequence and modifications, regardless of precursor charge state.
2.1.6 Peptide quantitation
TagQuant extracts and processes isobaric labeling quantitative information from MS/MS spectra. It is compatible with collision- and electron-based dissociation [23] of tandem mass tags (TMT) duplex [24] and 6-plex [25], and isobaric tags for relative and absolute quantitation (iTRAQ) 4-plex [26] and 8-plex [27]. Intensities of the reporter ions of interest are obtained from the raw data, and these values are subsequently denormalized by multiplying by the ion injection time to yield the number of ion counts detected, a quantity which can be fairly compared across different spectra and analyses. Purity correction is then applied, as has been previously published [28], using user-specified purity data provided by the manufacturer. Finally, normalization is performed such that the total intensity of each tag is equal, accounting for differences in sample mixing quantities.
2.1.7 Protein parsimony and protein FDR analysis
Protein Herder infers the most likely proteins identified based on the peptides validated by FDR Optimizer. All peptide sequences are first re-searched against the protein database to find all instances of those sequences in any protein, with enzyme specificity if provided. The Boyer–Moore string search algorithm [29] is used for optimal performance. Several filtering steps are then executed to apply the rules of parsimony to the identified protein list [30].
First, all sets of indistinguishable proteins, which are identified by the same collection of peptides, are combined into protein groups. Next, subset proteins, which are identified by fewer peptides than another protein and contain no unique peptides, are eliminated. At this stage, protein groups are sorted in ascending order by protein probability value (p-value), calculated as the product of the best p-value for each unique peptide [9]. Next, subsumable proteins, which are identified by a combination of the peptides that identify other proteins, are also eliminated. Finally, using decoy protein groups divided by target protein groups as the protein FDR, a protein p-value threshold is established to give a controlled error rate.
2.1.8 Protein quantitation
ProteinTagQuant combines peptide quantitation to yield protein quantitation. This is achieved by summing reporter ion intensities from TagQuant. Various criteria are available to filter out spectra that might provide dubious quantitation, e.g., high levels of precursor interference [31], peptides shared between multiple protein groups, or peptides containing modification sites.
2.2 Experimental
2.2.1 Identification dataset
The Institute for Systems Biology (ISB) standard protein mix sample [32] (mix “B”) was acquired following digestion with trypsin. Peptides were separated on a Waters nanoACQUITY UPLC (Milford, MA) with a self-packed 9 cm precolumn (75 μm i.d.) and a 25 cm analytical column (50 μm i.d.), both packed with Alltech Alltima 5 μm C18 particles (Deerfield, IL) [33]. The peptides were eluted with a gradient of 10 to 30% acetonitrile over two hours at a flow rate of 300 nL/min. The eluent was analyzed with LC–MS/MS on a Thermo Scientific LTQ Orbitrap Velos mass spectrometer (San Jose, CA/Bremen, Germany). The instrument method was 165 minutes and consisted of a 60,000 resolving power MS1 survey scan detected in the orbitrap followed by data-dependent top-10 MS2 detected in the ion trap, utilizing decision tree logic [34] to decide between resonant-excitation CAD and ETD [35] as the activation type. Precursor charge states that were unknown or +1 were excluded, and dynamic exclusion was enabled after one fragmentation event for 45 seconds.
This dataset was searched against the ISB database of 18 standard proteins + 92 contaminant proteins + 1709 Haemophilus influenzae Rd proteins as background (http://regis-web.systemsbiology.net/PublicDatasets/database/18mix_db_plus_contaminants_20081209.fasta) using OMSSA 2.1.7. Full trypsin enzymatic specificity was required, allowing up to three missed cleavages. Carbamidomethylation of cysteines (+57 Da) was specified as a fixed modification, while oxidation of methionines (+16 Da) was specified as a variable modification. An average mass tolerance of ±5 Da was used for precursors, while a monoisotopic mass tolerance of ±0.5 Da was used for products.
For TPP analysis, the data was searched with Sequest (version 27 from the University of Washington) or OMSSA (version 2.1.9) using either a ±5 Da average precursor mass tolerance or a ±10 ppm monoisotopic precursor mass tolerance and a monoisotopic fragment bin size of 0.38 Da (Sequest) or a monoisotopic product mass tolerance of ±0.5 Da (OMSSA). Results were filtered with PeptideProphet, iProphet and ProteinProphet from TPP 4.3 rev 1. The accurate mass, non-parametric model, and decoy estimation options were used in PeptideProphet.
2.2.2 Quantitation dataset
BY4741 wild-type yeast were grown in yeast extract peptone dextrose media to mid-log phase (OD600 = 0.6). Proteins were chemically extracted with YPer (Thermo Scientific Pierce; Rockford, IL), and digested with Promega sequencing-grade modified trypsin (Madison, WI) at a 1:50 enzyme:substrate ratio at 37 °C overnight and quenched by acidification with TFA. Peptides were desalted and labeled with Thermo Scientific Pierce TMTsixplex (Rockford, IL; lot number KD130680A), with intermittent mixing at room temperature, and quenched following an hour of incubation. Peptides labeled with tags of nominal m/z 126 through 131 were mixed in ratios of 1: 5: 2: 1.5: 1: 3, respectively.
Peptides were separated on a Waters nanoACQUITY UPLC (Milford, MA) with a self-packed 9 cm precolumn (75 μm i.d.) and a 30 cm analytical column (50 μm i.d.), both packed with Alltech Alltima 5 μm C18 particles (Deerfield, IL) [33]. The peptides were eluted with a gradient of 5 to 30% acetonitrile over two hours at a flow rate of 300 nL/min. The eluent was analyzed with LC–MS/MS on a Thermo Scientific LTQ Orbitrap Velos mass spectrometer (San Jose, CA/Bremen, Germany). The instrument method was 165 minutes and consisted of a 30,000 resolving power MS1 survey scan followed by data-dependent top-10 higher-energy collision dissociation (HCD) MS2 at 7,500 resolving power, all detected in the orbitrap. Precursor charges states that were unknown or +1 were excluded, and dynamic exclusion was enabled after one fragmentation event for 45 seconds.
This dataset was searched against the Saccharomyces Genome Database [36] (http://www.yeastgenome.org/; January 5, 2010 release; “all” file including verified, uncharacterized, and dubious open reading frames, and pseudogenes). Full trypsin enzymatic specificity was required, allowing up to three missed cleavages. Carbamidomethylation of cysteines (+57 Da) and TMT 6-plex on peptide N-termini and lysines (+229 Da) were specified as fixed modifications, while oxidation of methionines (+16 Da) and TMT 6-plex (+229 Da) on tyrosines were specified as variable modifications. An average mass tolerance of ±5 Da was used for precursors, while a monoisotopic mass tolerance of ±0.01 Da was used for products. For TPP analysis, the data was searched with Sequest (version 27 from the University of Washington) or OMSSA (2.1.9) using either a ±5 Da average precursor mass tolerance or a ±10 ppm monoisotopic precursor mass tolerance and a monoisotopic fragment bin size of 0.01 Da (Sequest) or a monoisotopic product mass tolerance of ±0.01 Da (OMSSA). Results were filtered with PeptideProphet, iProphet and ProteinProphet from TPP 4.3 rev 1. The accurate mass, non-parametric model, and decoy estimation options were used in PeptideProphet. The TPP component Libra was used for isobaric label quantitation.
3 Results and discussion
3.1 Data analysis workflow
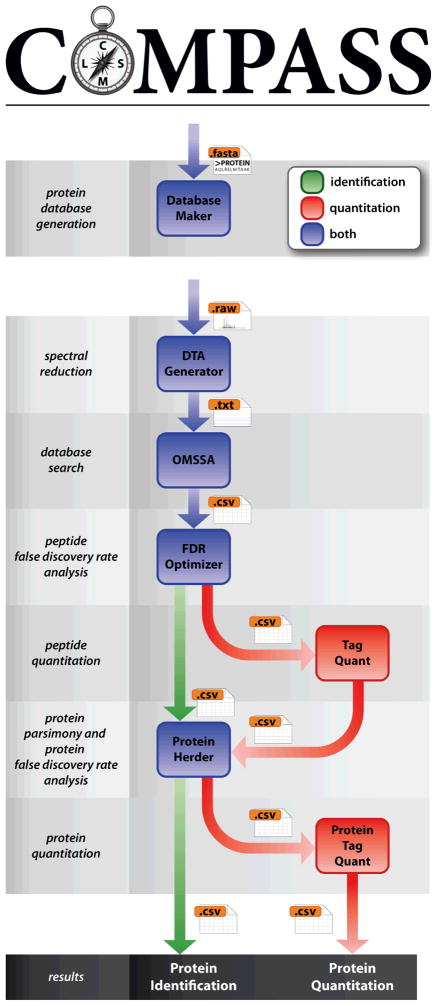
Figure 1 depicts the two basic workflows of COMPASS—identification and quantitation. Independently of the LC–MS/MS data, a protein database is generated with Database Maker. This step is only performed once per .fasta (e.g., when an updated protein database is released). Although several methods exist for performing target–decoy searches [19, 37], simple protein sequence reversal was the first [38] and is most straightforward. Other decoy methods are similarly effective but require more effort for database generation and/or post-search correction (i.e., with random databases, the increased number of decoy peptides relative to target peptides must be compensated for). A search against a concatenated database—the approach assumed by COMPASS—as opposed to separate target and decoy database searches, is also the simpler and arguably more effective approach [19].
Figure 1.
Identification and quantitation workflow of COMPASS. Database Maker generates BLAST-formatted protein databases for OMSSA. DTA Generator converts raw instrument data to text files for searching with OMSSA. FDR Optimizer performs FDR analysis at the spectrum/peptide level, followed by protein parsimony and FDR analysis at the protein level with Protein Herder. For quantitation, the workflow is supplemented by TagQuant, which performs spectrum/peptide-level quantitation, and ProteinTagQuant, which performs protein-level quantitation.
DTA Generator processes instrument data from LC–MS/MS analyses. This software reduces the raw data to text formats usable by search algorithms. Although OMSSA is our focus, individual .dta files for Sequest or .mgf files for Mascot are additional output options. Database searching can be performed with OMSSA using either the command-line interface or the NCBI OMSSA Browser, with the only requirement that CSV output must be specified for use with the rest of the workflow. We have also developed our own GUI for OMSSA, named the OMSSA Navigator. This software translates between textual and graphical search parameters, and also validates user input, in real time.
FDR analysis is then performed at the spectrum/peptide level with FDR Optimizer. Two important considerations come into play at this step: when to apply the FDR threshold and whether it should be applied based on PSMs or unique peptides. For typical experiments, FDR analysis should be performed once for all datasets simultaneously (batch option) at the unique peptide level. Performing FDR analysis for each dataset independently will likely overestimate the number of identifications at the reported error rate, as target identifications are more likely to repeat in different analyses, while decoy identifications are more random. The same is true for PSMs, as target PSMs typically reduce more drastically to unique peptides than decoy PSMs.
At this point the identification and quantitation workflows diverge. For isobaric label-based experiments, quantitative data is extracted with TagQuant. The workflow continues as normal with the next stage, simply using new results files which have extra columns of quantitative data appended.
Next, Protein Herder infers the minimum set of proteins which can explain the list of confidently identified peptides. Previously established principles of parsimony [30] are applied to eliminate proteins whose peptides could be better explained by the presence of other proteins. Proteins which are indistinguishable, given the peptides that identify them, are combined into protein groups. The final parsimonious list of protein groups is then filtered to the user-specified FDR.
Finally, for quantitative datasets, peptide quantitation must be combined to yield protein quantitation. ProteinTagQuant accomplishes this task by summing quantitation from the peptides that make up a protein group, effectively weighting peptide quantitation by reporter tag signal abundance. Various filtering options are available to improve quantitation by removing data from peptides known to be problematic based on several criteria.
3.2 Identification dataset
With this dataset, we aim to demonstrate and validate the basic peptide and protein identification workflow of COMPASS. To accomplish this, we use a relatively simple, manually annotated sample—the ISB standard 18 protein mix [32], for which all 18 standard proteins have been identified and many contaminant proteins have been manually validated—to prove its efficacy. The sample was interrogated by nanoflow LC–MS/MS using a CAD/ETD decision-tree method [34].
One of the most critical components of contemporary shotgun proteomics is FDR analysis at the spectrum/peptide level, typically achieved using a target–decoy search strategy. Because an incorrect PSM is equally likely to match to a target or decoy sequence, the distribution of scores for decoy hits can be used as a surrogate for incorrect target hits by which one can estimate the number of false positives and thus, FDR. The advent of linear ion trap–Fourier transform hybrid mass spectrometers [39, 40] enhanced peptide identification by enabling the detection and selection of precursors for activation from high-mass accuracy MS1 spectra. This process yields ppm-level precursor mass errors, which provide a highly orthogonal dimension for FDR filtering [41].
The MacCoss lab has shown that wide precursor mass tolerance searches followed by filtering is preferable to narrow searches [42], and COMPASS uses this approach. For maximum sensitivity, DTA Generator outputs the isolation center m/z as the precursor and OMSSA searches should be performed with a wide precursor mass tolerance (i.e., up to ±5 Da) to ensure that the correct peptide will be considered even if an isotopic peak has been selected. This strategy avoids determination of the precursor monoisotopic m/z, which is an error-prone process that, when coupled with narrow precursor mass tolerance searches, can lead to the loss of identifications. The COMPASS workflow is explicitly designed to avoid these pitfalls.
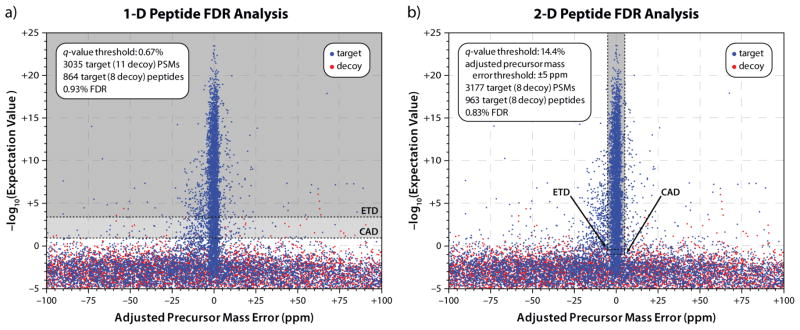
The post-search filtering process, performed by FDR Optimizer, is demonstrated in Figure 2. Precursor mass error—the x-axis—is a metric for how well the MS1 information matches the candidate peptide, while −log10(e-value)—the y-axis—measures how well the MS2 data matches the candidate peptide. With low-resolution MS1 data, only the y-axis is used due to poor measurement precision of the x-axis, leading to the acceptance of many matches that unknowingly have high precursor mass error, and thus are unlikely to be correct (Figure 2a). Performing FDR analysis using only q-values yields 864 unique target peptides at 1% FDR. However, when precursor mass error is used as a filter, the number of peptides taken from the dense region around 0 ppm precursor mass error is increased (Figure 2b). Peptides with worse-matching MS/MS spectra, but low precursor mass errors, can be accepted, leading to an 11.5% increase in the number of identifications, to 963 unique target peptides.
Figure 2.
Comparison of one- (a) versus two-dimensional (b) FDR analysis at the peptide level. Without high-mass accuracy precursor detection, e-value is the sole discriminant between correct and incorrect PSMs. As a result, many PSMs with high precursor mass error, and therefore, less likelihood of being correct, are accepted. The addition of precursor mass accuracy as a secondary discriminant allows the acceptance of spectra with worse e-values, giving a higher number of PSMs and unique peptides at the same FDR. In both cases, the q-value threshold corresponds to slightly better (i.e., lower) e-values for ETD (upper dashed line) than CAD (lower dashed line).
Carrying out FDR analysis using q-values, not e-values, is a critical distinction for combining diverse search results. In this case, although it is a single LC–MS/MS analysis, two different fragmentation methods—CAD and ETD—are utilized. Because OMSSA e-values can have slightly different meanings for different data types or search parameters, the software calculates the q-value for each PSM. Q-values are a more empirical measure of confidence in a given identification, and therefore are better suited for combining results. In both Figure 2a and 2b, a single q-value threshold represents an e-value threshold for CAD results that is slightly lower than that for ETD results.
The well-annotated nature of the sample analyzed in this dataset enables validation of the FDR analysis employed by COMPASS. The database provides two different levels of decoy databases; therefore, after the normal FDR rules are applied, additional “background” proteins from an unrelated organism (H. influenzae, in this case) remain to verify the estimated error rate. In this case, out of the 3,177 accepted PSMs, only 17 (0.54%) were from H. influenzae proteins. At the unique peptide level, 17 out of 963 were from H. influenzae, for an actual error rate of 1.8%, close to the expected 1%.
Protein-level analysis is shown in Table 1. In total, 34 proteins were identified at a 1% FDR. All 18 of the standard proteins were identified with very high confidence, ranging from 17 to 399 PSMs and from 4 to 92 unique peptides. Furthermore, though they tend to overestimate confidence, protein p-values were at worst 10−66, and many were effectively zero due to numeric underflow (i.e., many small numbers multiplied until the computer assumes zero). The minimum protein sequence coverage was about 20%, ranging all the way up to 95%. Additionally, 15 contaminant proteins that have previously been manually validated were identified.
Table 1.
Parsimonious proteins detected at a 1% FDR from the identification dataset. White rows correspond to the standard proteins (18), light grey rows correspond to known contaminants (15), and dark grey rows correspond to background proteins from Haemophilus influenzae Rd. (1)
| Protein | Organism | PSMs | Peptides | Sequence Coverage | P-value |
|---|---|---|---|---|---|
| serotransferrin precursor | cow | 399 | 92 | 85% | 0 |
| glycogen phosphorylase, muscle form | rabbit | 249 | 76 | 65% | 0 |
| serum albumin precursor | cow | 303 | 73 | 84% | 0 |
| alkaline phosphatase precursor | Escherichia coli | 358 | 61 | 95% | 0 |
| beta-galactosidase | Escherichia coli | 222 | 60 | 56% | 0 |
| catalase | cow | 212 | 38 | 65% | 0 |
| glyceraldehyde 3-phosphate dehydrogenase | rabbit | 216 | 33 | 62% | 0 |
| alpha-amylase | Bacillus licheniformis | 168 | 31 | 50% | 0 |
| cytochrome C | cow | 122 | 30 | 83% | 0 |
| carbonic anhydrase II | cow | 147 | 23 | 63% | 0 |
| mannose-6-phosphate isomerase | Escherichia coli | 94 | 19 | 55% | 0 |
| trypsin | pig | 31 | 5 | 25% | 0 |
| actin, aortic smooth muscle | cow | 121 | 25 | 58% | 8×10−301 |
| beta-lactoglobulin precursor | cow | 76 | 21 | 63% | 2×10−289 |
| myoglobin | horse | 36 | 15 | 67% | 2×10−210 |
| troponin I, fast skeletal muscle | rabbit | 41 | 15 | 66% | 1×10−188 |
| myosin light chain 1, skeletal muscle isoform | rabbit | 31 | 15 | 63% | 2×10−156 |
| troponin C, skeletal muscle | rabbit | 29 | 11 | 61% | 1×10−148 |
| ovalbumin | chicken | 64 | 12 | 28% | 9×10−130 |
| alpha-S2-casein precursor | cow | 24 | 11 | 37% | 6×10−123 |
| glucoamylase precursor | Aspergillus awamori | 24 | 7 | 17% | 3×10−102 |
| alpha-lactalbumin precursor | cow | 17 | 6 | 46% | 5×10−83 |
| ubiquitin | cow | 13 | 9 | 95% | 5×10−80 |
| alpha-S1-casein precursor | cow | 9 | 8 | 30% | 1×10−79 |
| beta casein precursor | cow | 18 | 4 | 20% | 1×10−66 |
| transthyretin precursor | cow | 11 | 7 | 61% | 1×10−62 |
| myosin regulatory light chain 2, skeletal muscle isoform type 2 | rabbit | 14 | 5 | 25% | 2×10−61 |
| UPF0076 protein yjgF | Escherichia coli | 13 | 4 | 54% | 1×10−42 |
| hemoglobin subunit alpha-1/2 | rabbit | 7 | 4 | 40% | 1×10−38 |
| fructose-bisphosphate aldolase A | rabbit | 3 | 3 | 16% | 3×10−29 |
| hemoglobin subunit beta-1/2 | rabbit | 4 | 3 | 21% | 3×10−26 |
| kappa-casein precursor | cow | 4 | 2 | 15% | 6×10−25 |
| aldehyde dehydrogenase, mitochondrial | hamster | 2 | 2 | 4% | 1×10−12 |
| queosine biosynthesis protein | Haemophilus influenzae Rd | 1 | 1 | 4% | 2×10−6 |
As one out of the 34 proteins identified at a 1% FDR was actually a background H. influenzae protein, the true error rate for this dataset was 2.9%. However, this value reflects the low number of proteins present in the sample, and in fact one background protein is expected to be accepted at a 1% protein FDR statistically (negative binomial distribution; r = 1, p = 0.5). For realistic proteomic datasets with hundreds or even thousands of identified proteins, this issue is much less significant.
For comparison, this dataset was searched with the TPP using a ±5.0 Da average precursor mass tolerance, typical for COMPASS, and a ±10 ppm monoisotopic precursor mass tolerance, typical of most proteomic searches. The TPP searches were done using both Sequest and OMSSA. PSMs, peptide, and protein identifications at a 1% FDR are given in Table 2. COMPASS performs favorably in all metrics, in particular unique peptides for which it yielded the most across all analyses.
Table 2.
Comparison of PSMs, peptide, and protein identifications at a 1% FDR produced by COMPASS and the TPP for the identification and quantitation datasets. The best results for each quantity are bolded for both datasets
| Dataset | Software Suite | Search Algorithm | Precursor Mass Tolerance | PSMs | Peptides | Proteins (Non-background) |
|---|---|---|---|---|---|---|
| Identification | COMPASS | OMSSA | ±5 Da (average) | 3177 | 963 | 34 (33) |
| Identification | TPP | Sequest | ±5 Da (average) | 2786 | 733 | 35 (33) |
| Identification | TPP | Sequest | ±10 ppm (monoisotopic) | 2943 | 772 | 35 (34) |
| Identification | TPP | OMSSA | ±5 Da (average) | 3437 | 879 | 34 (34) |
| Identification | TPP | OMSSA | ±10 ppm (monoisotopic) | 1284 | 560 | 33 (31) |
| Quantitation | COMPASS | OMSSA | ±5 Da (average) | 9931 | 5832 | 917 |
| Quantitation | TPP | Sequest | ±5 Da (average) | 9693 | 5394 | 920 |
| Quantitation | TPP | Sequest | ±10 ppm (monoisotopic) | 9256 | 5163 | 869 |
| Quantitation | TPP | OMSSA | ±5 Da (average) | 9321 | 5223 | 929 |
| Quantitation | TPP | OMSSA | ±10 ppm (monoisotopic) | 6199 | 3905 | 757 |
3.3 Quantitation dataset
With this dataset we aim to demonstrate and validate the quantitative workflow of COMPASS. We achieve this by using yeast proteins, digested with trypsin, labeled with isobaric stable isotope tags, and mixed in known ratios. We used TMT 6-plex tags to label peptides mixed in ratios of 1: 1: 1.5: 2: 3: 5. The peptides were analyzed by LC–MS/MS utilizing a data-dependent top-10 HCD method, with all spectra acquired in the orbitrap. Analysis by COMPASS yielded 9,931 target PSMs and 5,832 unique target peptides at a 1% peptide FDR, translating to 917 target proteins at a 1% protein FDR.
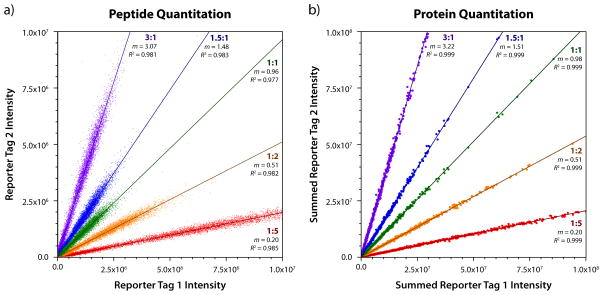
Accuracy and precision of quantitation can be evaluated by plotting the intensity of reporter tags on opposite axes, as shown for all 9,931 accepted PSMs in Figure 3. The slope (m) of each series represents the accuracy, while the coefficient of determination (R2) represents the precision. For PSMs, depicted in Figure 3a, the slopes for mixing ratios of 1, 1.5, 2, 3, and 5 had errors of −3.7%, −1.4%, +2.4%, +2.3%, and −0.3%, respectively. We note that this level of accuracy was achieved even without any tag intensity normalization, meaning that any imprecision in mixing amounts are reflected in these errors. The coefficients of determination were 0.977, 0.983, 0.982, 0.981, and 0.985, indicating excellent precision.
Figure 3.
Peptide (a) and protein (b) quantitation for the quantitation dataset. For each of the known ratios of 1: 1: 1.5: 2: 3: 5, the error between the observed and expected ratio was always less than 10%, even without any normalization to account for imprecision in sample mixing. Although the correlation was quite good at the peptide level, with a typical R2 of 0.98, summing gave superior quantitation at the protein level, with every ratio producing an R2 of 0.999.
Precision and accuracy of the quantitative analysis improves even further at the protein level, as shown for all 917 identified proteins in Figure 3b. By summing peptide quantitation data, results are weighted according to their abundance, which tends to correlate well with its reliability. The slopes for mixing ratios of 1, 1.5, 2, 3, and 5 had errors of −2.5%, +0.6%, +1.9%, +7.3%, and −2.2%, respectively. The precision was significantly higher at the protein level, with coefficients of determination of 0.999 for all series.
This dataset was also searched by the TPP using the same two precursor mass search types and search algorithms for comparison. Again, COMPASS performed favorably, in this case identifying the most PSMs and unique peptides across all analyses, and only slightly fewer proteins. TPP’s Libra and COMPASS’s TagQuant/ProteinTagQuant produced quantitation of similar quality.
3.4 Large-scale datasets
COMPASS has been used in multiple large-scale proteomic studies. In one study, 3,908 yeast proteins were identified at a 1% FDR, utilizing digestion with five different enzymes, fractionation by strong cation exchange (SCX) chromatography, and triplicate LC–MS/MS analysis [43]. In another study, human embryonic stem cells have been quantitatively compared to induced pluripotent stem cells and their somatic precursors, yielding 7,962 proteins at a 1% FDR, 6,179 of which were quantified by iTRAQ 4-plex [44]. Finally, in an investigation of environmental stress response, 2,973 yeast proteins were identified at a 1% FDR, of which 1,373 were quantified in biological triplicate with TMT 6-plex over a 240 minute time course following treatment with 0.7 M NaCl [45].
3.5 Software availability
All of the software described here—both as a Microsoft Windows installer and full source—code is available at http://www.chem.wisc.edu/~coon/software.php#compass. It is licensed under the GNU General Public License version 3.
4 Concluding remarks
The development of software for the analysis of mass spectral data from biological samples can present significant challenges. Complete analysis requires attention to many important components, many of which are not widely available. Our aim is to distribute an open-source companion platform for OMSSA, COMPASS, to facilitate typical proteomic analysis functions so that users can freely utilize a modern, competitive pipeline that is easy to use.
Supplementary Material
Acknowledgments
We acknowledge all current and former members of the Coon group for their use of and feedback on this software suite, which has greatly enhanced its development. We thank A.J. Bureta for figure illustrations and Alicia Williams for critical proofreading. We are grateful to Carly Holstein, Jimmy Eng, and Daniel Martin at the ISB for providing the standard protein mix sample and performing the TPP analyses, supported by the University of Washington’s Proteomics Resource (UWPR95794). This work was supported by the National Institutes of Health (NIH) R01 GM080148 and P01 GM081629 to J.J.C. D.H.P. acknowledges support from an NIH predoctoral traineeship—the Genomic Sciences Training Program, NIH 5T32HG002760.
Abbreviations
- COM
Component Object Model
- COMPASS
Coon OMSSA Proteomic Analysis Software Suite
- CSV
comma-separated values
- ETD
electron-transfer dissociation
- FDR
false discovery rate
- GUI
graphical user interface
- HCD
higher-energy collision dissociation
- ISB
Institute for Systems Biology
- iTRAQ
isobaric tags for relative and absolute quantitation
- OMSSA
Open Mass Spectrometry Search Algorithm
- TMT
tandem mass tags
- TPP
Trans-Proteomic Pipeline
- XML
extensible markup language
Footnotes
The authors have declared no conflict of interest.
Step-by-step instructions for processing the identification and quantitation datasets with COMPASS, as well as links to download the raw data and results, are provided in the Supporting Information.
References
- 1.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 2.Eng JK, McCormack AL, Yates JR. An Approach to Correlate Tandem Mass-Spectral Data of Peptides with Amino-Acid-Sequences in a Protein Database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 3.Perkins DN, Pappin DJC, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 4.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 5.Geer LY, Markey SP, Kowalak JA, Wagner L, et al. Open mass spectrometry search algorithm. J Proteome Res. 2004;3:958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 6.Balgley BM, Laudeman T, Yang L, Song T, Lee CS. Comparative evaluation of tandem MS search algorithms using a target-decoy search strategy. Mol Cell Proteomics. 2007;6:1599–1608. doi: 10.1074/mcp.M600469-MCP200. [DOI] [PubMed] [Google Scholar]
- 7.Mueller LN, Brusniak MY, Mani DR, Aebersold R. An assessment of software solutions for the analysis of mass spectrometry based quantitative proteomics data. J Proteome Res. 2008;7:51–61. doi: 10.1021/pr700758r. [DOI] [PubMed] [Google Scholar]
- 8.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 9.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 10.Keller A, Eng J, Zhang N, Li XJ, Aebersold R. A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol Syst Biol. 2005 doi: 10.1038/msb4100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deutsch EW, Mendoza L, Shteynberg D, Farrah T, et al. A guided tour of the Trans-Proteomic Pipeline. Proteomics. 2010;10:1150–1159. doi: 10.1002/pmic.200900375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deutsch EW, Shteynberg D, Lam H, Sun Z, et al. Trans-Proteomic Pipeline supports and improves analysis of electron transfer dissociation data sets. Proteomics. 2010;10:1190–1195. doi: 10.1002/pmic.200900567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pedrioli PGA, Eng JK, Hubley R, Vogelzang M, et al. A common open representation of mass spectrometry data and its application to proteomics research. Nat Biotechnol. 2004;22:1459–1466. doi: 10.1038/nbt1031. [DOI] [PubMed] [Google Scholar]
- 14.Lin SM, Zhu LH, Winter AQ, Sasinowski M, Kibbe WA. What is mzXML good for? Expert Rev Proteomic. 2005;2:839–845. doi: 10.1586/14789450.2.6.839. [DOI] [PubMed] [Google Scholar]
- 15.Lam H, Deutsch EW, Eddes JS, Eng JK, et al. Development and validation of a spectral library searching method for peptide identification from MS/MS. Proteomics. 2007;7:655–667. doi: 10.1002/pmic.200600625. [DOI] [PubMed] [Google Scholar]
- 16.Lam H, Deutsch EW, Eddes JS, Eng JK, et al. Building consensus spectral libraries for peptide identification in proteomics. Nat Methods. 2008;5:873–875. doi: 10.1038/nmeth.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodriguez-Suarez E, Gubb E, Alzueta IF, Falcon-Perez JM, et al. Virtual Expert Mass Spectrometrist: iTRAQ tool for database-dependent search, quantitation and result storage. Proteomics. 2010;10:1545–1556. doi: 10.1002/pmic.200900255. [DOI] [PubMed] [Google Scholar]
- 18.Hakkinen J, Vincic G, Mansson O, Warell K, Levander F. The Proteios Software Environment: An Extensible Multiuser Platform for Management and Analysis of Proteomics Data. J Proteome Res. 2009;8:3037–3043. doi: 10.1021/pr900189c. [DOI] [PubMed] [Google Scholar]
- 19.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 20.Good DM, Wenger CD, McAlister GC, Bai DL, et al. Post-Acquisition ETD Spectral Processing for Increased Peptide Identifications. J Am Soc Mass Spectrom. 2009;20:1435–1440. doi: 10.1016/j.jasms.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kall L, Storey JD, MacCoss MJ, Noble WS. Assigning significance to peptides identified by tandem mass spectrometry using decoy databases. J Proteome Res. 2008;7:29–34. doi: 10.1021/pr700600n. [DOI] [PubMed] [Google Scholar]
- 22.Kall L, Storey JD, MacCoss MJ, Noble WS. Posterior error probabilities and false discovery rates: Two sides of the same coin. J Proteome Res. 2008;7:40–44. doi: 10.1021/pr700739d. [DOI] [PubMed] [Google Scholar]
- 23.Phanstiel D, Zhang Y, Marto JA, Coon JJ. Peptide and protein quantification using iTRAQ with electron transfer dissociation. J Am Soc Mass Spectrom. 2008;19:1255–1262. doi: 10.1016/j.jasms.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson A, Schafer J, Kuhn K, Kienle S, et al. Tandem mass tags: A novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem. 2003;75:1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 25.Dayon L, Hainard A, Licker V, Turck N, et al. Relative quantification of proteins in human cerebrospinal fluids by MS/MS using 6-plex isobaric tags. Anal Chem. 2008;80:2921–2931. doi: 10.1021/ac702422x. [DOI] [PubMed] [Google Scholar]
- 26.Ross PL, Huang YLN, Marchese JN, Williamson B, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 27.Choe L, D’Ascenzo M, Relkin NR, Pappin D, et al. 8-Plex quantitation of changes in cerebrospinal fluid protein expression in subjects undergoing intravenous immunoglobulin treatment for Alzheimer’s disease. Proteomics. 2007;7:3651–3660. doi: 10.1002/pmic.200700316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shadforth IP, Dunkley TPJ, Lilley KS, Bessant C. i-Tracker: For quantitative proteomics using iTRAQ (TM) BMC Genomics. 2005:6. doi: 10.1186/1471-2164-6-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boyer RS, Moore JS. Fast String Searching Algorithm. Communications of the Acm. 1977;20:762–772. [Google Scholar]
- 30.Nesvizhskii AI, Aebersold R. Interpretation of shotgun proteomic data - The protein inference problem. Mol Cell Proteomics. 2005;4:1419–1440. doi: 10.1074/mcp.R500012-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Ow SY, Salim M, Noirel J, Evans C, et al. iTRAQ Underestimation in Simple and Complex Mixtures: “The Good, the Bad and the Ugly”. J Proteome Res. 2009;8:5347–5355. doi: 10.1021/pr900634c. [DOI] [PubMed] [Google Scholar]
- 32.Klimek J, Eddes JS, Hohmann L, Jackson J, et al. The standard protein mix database: A diverse data set to assist in the production of improved peptide and protein identification software tools. J Proteome Res. 2008;7:96–103. doi: 10.1021/pr070244j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin SE, Shabanowitz J, Hunt DF, Marto JA. Subfemtomole MS and MS/MS peptide sequence analysis using nano-HPLC micro-ESI Fourier transform ion cyclotron resonance mass spectrometry. Anal Chem. 2000;72:4266–4274. doi: 10.1021/ac000497v. [DOI] [PubMed] [Google Scholar]
- 34.Swaney DL, McAlister GC, Coon JJ. Decision tree-driven tandem mass spectrometry for shotgun proteomics. Nat Methods. 2008;5:959–964. doi: 10.1038/nmeth.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci U S A. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cherry JM, Adler C, Ball C, Chervitz SA, et al. SGD: Saccharomyces Genome Database. Nucleic Acids Res. 1998;26:73–79. doi: 10.1093/nar/26.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bianco L, Mead JA, Bessant C. Comparison of Novel Decoy Database Designs for Optimizing Protein Identification Searches Using ABRF sPRG2006 Standard MS/MS Data Sets. J Proteome Res. 2009;8:1782–1791. doi: 10.1021/pr800792z. [DOI] [PubMed] [Google Scholar]
- 38.Moore RE, Young MK, Lee TD. Qscore: An algorithm for evaluating SEQUEST database search results. J Am Soc Mass Spectrom. 2002;13:378–386. doi: 10.1016/S1044-0305(02)00352-5. [DOI] [PubMed] [Google Scholar]
- 39.Syka JEP, Marto JA, Bai DL, Horning S, et al. Novel linear quadrupole ion trap/FT mass spectrometer: Performance characterization and use in the comparative analysis of histone H3 post-translational modifications. J Proteome Res. 2004;3:621–626. doi: 10.1021/pr0499794. [DOI] [PubMed] [Google Scholar]
- 40.Makarov A, Denisov E, Kholomeev A, Baischun W, et al. Performance evaluation of a hybrid linear ion trap/orbitrap mass spectrometer. Anal Chem. 2006;78:2113–2120. doi: 10.1021/ac0518811. [DOI] [PubMed] [Google Scholar]
- 41.Dieguez-Acuna FJ, Gerber SA, Kodama S, Elias JE, et al. Characterization of mouse spleen cells by subtractive proteomics. Mol Cell Proteomics. 2005;4:1459–1470. doi: 10.1074/mcp.M500137-MCP200. [DOI] [PubMed] [Google Scholar]
- 42.Hsieh EJ, Hoopmann MR, MacLean B, MacCoss MJ. Comparison of Database Search Strategies for High Precursor Mass Accuracy MS/MS Data. J Proteome Res. 2010;9:1138–1143. doi: 10.1021/pr900816a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swaney DL, Wenger CD, Coon JJ. Value of Using Multiple Proteases for Large-Scale Mass Spectrometry-Based Proteomics. J Proteome Res. 2010;9:1323–1329. doi: 10.1021/pr900863u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Phanstiel DH, Brumbaugh J, Wenger CD, McAlister GC, et al. Comprehensive quantitative proteomic comparison of human ES, iPS, and somatic cells. [Google Scholar]
- 45.Lee MV, Topper SE, Hubler SL, Wenger CD, et al. A dynamic model of proteome changes reveals new roles for transcript alteration in yeast. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.