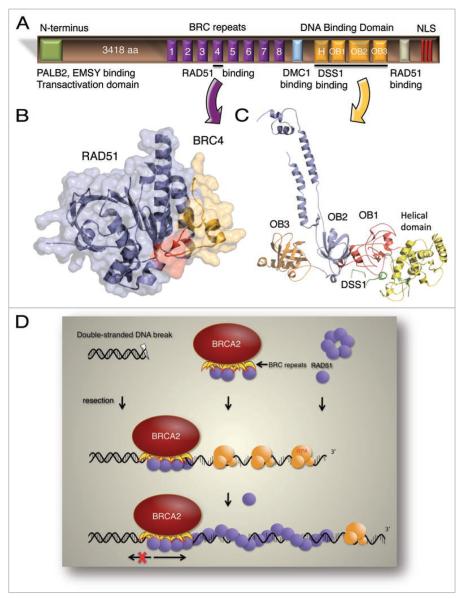
The breast cancer susceptibility proteins, BRCA1 and BRCA2, are encoded by tumor suppressor genes whose mutation results in half of the genetically predisposed cancers of the breast and ovary.1 BRCA2 and RAD51 are directly involved in DNA repair by homologous recombination (HR), a cellular process that accurately repairs double-stranded DNA breaks (DSB). RAD51 protein promotes the homology search and DNA strand exchange steps that are central to repair by HR.2
The finding that BRCA2 interacts with RAD51 provided the first evidence for involvement of BRCA2 in DNA repair.3 Human BRCA2 is 3,418 amino acids in size and contains several functional domains (Fig. 1A). The N-terminal region harbors interaction sites for PALB2,4 and EMSY,5 and a transactivation domain.6 Located in the central portion of the protein are eight BRC repeats, each about 35 amino acids in length. BRCA2 binds RAD51 primarily through these conserved motifs (Fig. 1B); this interaction is required for formation of RAD51 foci at DNA breaks in vivo.7 Downstream of the BRC repeats, there is a binding site for DMC1, the meiotic homolog of RAD51, implicating BRCA2 function in meiosis as well.8,9 In addition to the BRC repeats, there is an unrelated RAD51 binding sequence at the C-terminus of BRCA2; its phosphorylation by a cyclin-dependent kinase appears to regulate the interaction with RAD51, suggesting a cell cycle-dependent response to DNA damage.10 The C-terminal region of BRCA2 also contains a conserved DNA binding domain11 (Fig. 1C) and a binding site for DSS1 protein,12 a regulator of BRCA2. At the extreme C-terminus, there are three nuclear localization signals (NLS)11 (Fig. 1A).
Figure 1.
(A) Schematic representation of BRCA2 protein showing its functional domains. H, helical domain; OB, oligonucleotide-binding fold; NLS, nuclear localization signal. (B) Molecular surface and ribbon representation of RAD51 bound to BRC4 (PDB code 1n0w). RAD51 is shown in blue and BRC4 in orange. The highly conserved interacting region of BRC4 is highlighted in red. (C) Illustration of the DNA binding domain structure of mouse BRCA2 DBD-DSS1 (PDB code 1MIU). The helical domain is shown in yellow, and OB 1, 2 and 3 are shown in red, blue and orange, respectively. DSS1 protein is shown in green. (D) Model for the function of BRCA2 in the repair of DNA breaks. Through its DNA binding domain, BRCA2 targets RAD51 to the dsDNA-ssDNA junction of a resected DSB. Using its BRC repeats, BRCA2 directs RAD51 onto the ssDNa and blocks assembly onto dsDNa. This interaction likely provides a nucleus for nascent ATP-RAD51 filament formation. Subsequent growth of the nucleoprotein filament from this nucleus will displace RPA. Eventually, the RAD51-ssDNA filament will pair with an intact copy of the chromosome to continue the steps of DNA repair (not shown).
Although first identified in humans, BRCA2 homologs exist in many eukaryotes, including microbes, worms, plants and flies. The counterpart in Ustilago maydis is Brh2, and it can load RAD51 onto single-stranded DNA (ssDNA) at the junction with double-stranded DNA (dsDNA).13 One or more BRC repeats are present in every ortholog of BRCA2, and interestingly, the number can vary up to 15.14 In humans, the BRC-repeat domain spans ~1,200 amino acids, and at least 6 of the 8 BRC repeats directly interact with RAD51, with BRC4 having the highest affinity for RAD51.3 Mutations in individual BRC repeats that weaken RAD51 binding are associated with breast cancer progression (Breast Cancer Information Core Database, http://research.nhgri.nih.gov/bic/), highlighting the importance of the RAD51-BRCA2 interaction.
A seminal paper describing the crystal structure of RAD51-BRC4 revealed an important aspect of this interaction: BRC4 adopts a structure that mimics the interface of an adjacent RAD51 monomer.15 A stretch of highly conserved residues in the BRC repeat contact RAD51 via anti-parallel β-strand pairing (Fig. 1B). Consistent with the structure, overexpression of BRC4 in vivo blocks formation of RAD51 foci after exposure to ionizing radiation.15,16 Because RAD51 associates with BRC4 as a monomer in vitro,17 these collective results led to the conclusion that the BRC repeats block RAD51 nucleoprotein filament formation and that RAD51 polymerization is required for nuclear focus formation.14,15
However, these facts were seemingly contradictory: BRCA2 was a positive regulator of RAD51-promoted HR, but the BRC repeats reduced formation of RAD51 foci in vivo. Furthermore, a fusion of BRC4 tethered to RPA DBD was functional in vivo, implying that the BRC repeats needed to be targeted to the DNA in the cell.18 In vitro studies were equally confusing: the BRC repeats could apparently disrupt RAD51 nucleoprotein filaments,17,19 but the region containing BRC repeats 1-8 could stimulate RAD51-mediated DNA strand exchange.20
A recent study literally throws light on these puzzling results. Using both optical trapping to visualize individual RAD51 nucleoprotein filaments and ensemble studies, the BRC repeats were shown to modulate the DNA binding preferences of RAD51.21 In addition, whereas previous in vitro studies focused primarily on RAD51 nucleoprotein filaments formed on dsDNA,17,19 Carreira et al. examined the effects of the BRC repeats on RAD51 binding to both ssDNA and dsDNA.21 Strikingly, they found that either BRC4 alone or a region containing BRC repeats 1 through 8 stimulate the binding of RAD51 to ssDNA. Furthermore, BRC4 blocks ATP hydrolysis by RAD51, permitting accumulation of the active ATP-bound nucleoprotein filament. A potential explanation for this finding resides in the structural observation that the nucleotide-binding site of BRC4-bound RAD51 adopts a more closed conformation than that of RAD51 alone,15 suggesting that BRC4 could lock-down the ATP-bound state. This behavior may be a general property of the BRC repeats because the repeat from the BRCA2 homolog of Caenorhabditis elegans also inhibits ATP hydrolysis by RAD51.22 Moreover, human BRC4 stabilizes the inactive ADP form of the RAD51 nucleoprotein filament, yet permits exchange of ADP for ATP within the filament to result in a net accumulation of the active ATP-bound form of the RAD51-ssDNA complex.21 Thus, the first important finding from this new study is that the BRC repeats promote assembly of the active form of RAD51 nucleoprotein filaments on ssDNA.
In contrast to stabilizing and activating the RAD51-ssDNA filaments, previous reports showed that the BRC repeats disrupt RAD51 nucleoprotein filaments on duplex DNA when ATP hydrolysis is occuring.17,19 These findings led to the misconception that the BRC repeats disassemble such filaments. However, single-molecule visualization revealed that, instead of dissociating RAD51 nucleoprotein filaments, the BRC repeats block RAD51 nucleoprotein filament formation on dsDNA by slowing nucleation.21 In the ensemble studies, filaments were seen to dissociate, but only when filament turnover was permitted. Thus, the BRC repeats were not disassembling the filaments but rather were preventing their re-assembly by blocking subsequent nucleation. Therefore, the second important finding from this new study is that the BRC repeats prevent RAD51 assembly onto dsDNA. A recent study on the BRC repeats of BRCA2 support these conclusions and further extend the findings.23
Several variables determine whether a BRC repeat stabilizes or destabilizes a RAD51-DNA complex: the specific BRC repeat, its concentration, the nucleotide cofactor, whether ATP hydrolysis is occurring, and the DNA species (ssDNA or dsDNA). Appreciating this complex regulatory behavior and the dynamic aspects of the RAD51 filaments permits reconciliation of seemingly conflicting reports on BRC repeat activity and function.
The ability of the BRC repeats to differentially promote formation of RAD51 complexes with ssDNA, but block formation of complexes with dsDNA, can facilitate the appropriate loading of RAD51 onto ssDNA, rather than dsDNA, at DSBs. Indeed, the functional relevance of this differential modulation was validated when the BRC repeats were shown to stimulate RAD51-promoted DNA strand exchange in vitro.20,21 Collectively, these findings define the BRC repeats of BRCA2 as regulators of RAD51 DNA-binding selectivity.
The proposed role for BRCA2 in HR places BRCA2 at the ssDNA-dsDNA junction of a DSB where it targets RAD51 to assemble into a nucleoprotein filament on the ssDNA to form a crucial intermediate in the repair of a DSB. So how does BRCA2 achieve this targeting? It is reasonable to assume that the DNA binding domain (Fig. 1C) provides the specificity for binding to the ssDNA/dsDNA junction.13 However, RAD51 nucleoprotein self-assembly is slow due to a rate-limiting nucleation step that requires 2–3 monomers to initiate filament formation.24 The BRC repeats can serve as a nucleation site by binding at least 2–3 RAD51 molecules, and by correctly orienting them for efficient nucleus formation (Fig. 1D). Although, in principle one BRC repeat bound to a dimeric form of RAD51 could suffice,13,25 the use of more BRC repeats could greatly increase the probability of nucleation and/or enhance the stability of the nascent filament. Additionally, the extra repeats may help enforce targeting RAD51 to the ssDNA at the junction, rather than to the dsDNA, by slowing or preventing assembly into the dsDNA region. Finally, by locally inhibiting ATP hydrolysis by the nascent RAD51 nucleus, the turnover of this unstable but key intermediate is blocked, further ensuring the likelihood of filament growth away from the BRCA2 binding locus. The realization that BRCA2 is a multi-domain multi-functional protein, which acts in concert with other regulatory proteins such as DSS1, illuminates another layer of complexity in the regulation of HR. Further clarification must await experiments with the full-length BRCA2 protein to more clearly understand how BRCA2 exerts its control on the formation of RAD51-DNA complexes and their function in recombinational DNA repair.
Acknowledgements
We thank the following members of the Kowalczykowski laboratory: Ichiro Amitani, Elda Cannavo, Christopher Dombrowski, Anthony Forget, Jovencio Hilario, Ryan Jensen, Taeho Kim, Katsumi Morimatsu, Amitabh Nimonkar, Behzad Rad and Lisa Vancelette for comments and discussions. This work was supported by NIH grants GM-62653 and GM-64745 to S.C.K. and a postdoctoral fellowship from the Ministerio de Educacion y Ciencia (Spain) to A.C.
References
- 1.Wooster R, et al. N Engl J Med. 2003;348:2339–47. doi: 10.1056/NEJMra012284. [DOI] [PubMed] [Google Scholar]
- 2.Bianco PR, et al. Front Biosci. 1998;3:570–603. doi: 10.2741/a304. [DOI] [PubMed] [Google Scholar]
- 3.Wong AKC, et al. J Biol Chem. 1997;272:31941–4. doi: 10.1074/jbc.272.51.31941. [DOI] [PubMed] [Google Scholar]
- 4.Xia B, et al. Mol Cell. 2006;22:719–29. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 5.Hughes-Davies L, et al. Cell. 2003;115:523–35. doi: 10.1016/s0092-8674(03)00930-9. [DOI] [PubMed] [Google Scholar]
- 6.Milner J, et al. Nature. 1997;386:772–3. doi: 10.1038/386772a0. [DOI] [PubMed] [Google Scholar]
- 7.Yuan SS, et al. Cancer Res. 1999;59:3547–51. [PubMed] [Google Scholar]
- 8.Siaud N, et al. EMBO J. 2004;23:1392–401. doi: 10.1038/sj.emboj.7600146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thorslund T, et al. EMBO J. 2007;26:2915–22. doi: 10.1038/sj.emboj.7601739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esashi F, et al. Nature. 2005;434:598–604. doi: 10.1038/nature03404. [DOI] [PubMed] [Google Scholar]
- 11.Yang H, et al. Science. 2002;297:1837–48. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
- 12.Marston NJ, et al. Mol Cell Biol. 1999;19:4633–42. doi: 10.1128/mcb.19.7.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang H, et al. Nature. 2005;433:653–7. doi: 10.1038/nature03234. [DOI] [PubMed] [Google Scholar]
- 14.Lo T, et al. DNA Repair (Amst) 2003;2:1015–28. doi: 10.1016/s1568-7864(03)00097-1. [DOI] [PubMed] [Google Scholar]
- 15.Pellegrini L, et al. Nature. 2002;420:287–93. doi: 10.1038/nature01230. [DOI] [PubMed] [Google Scholar]
- 16.Chen CF, et al. J Biol Chem. 1999;274:32931–5. doi: 10.1074/jbc.274.46.32931. [DOI] [PubMed] [Google Scholar]
- 17.Davies AA, et al. Mol Cell. 2001;7:273–82. doi: 10.1016/s1097-2765(01)00175-7. [DOI] [PubMed] [Google Scholar]
- 18.Saeki H, et al. Proc Natl Acad Sci USA. 2006;103:8768–73. doi: 10.1073/pnas.0600298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galkin VE, et al. Proc Natl Acad Sci USA. 2005;102:8537–42. doi: 10.1073/pnas.0407266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shivji MK, et al. Nucleic Acids Res. 2006;34:4000–11. doi: 10.1093/nar/gkl505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carreira A, et al. Cell. 2009;136:1032–43. doi: 10.1016/j.cell.2009.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petalcorin MI, et al. Proc Natl Acad Sci USA. 2007;104:8299–304. doi: 10.1073/pnas.0702805104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shivji MK, et al. Proc Natl Acad Sci USA. 2009;106:13254–9. doi: 10.1073/pnas.0906208106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hilario J, et al. Proc Natl Acad Sci USA. 2009;106:361–8. doi: 10.1073/pnas.0811965106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spies M, et al. Mol Cell. 2006;21:573–80. doi: 10.1016/j.molcel.2006.01.007. [DOI] [PubMed] [Google Scholar]