Summary
Cancer cells neutralize p53 by deletion, mutation, proteasomal degradation, or sequestration to achieve a pathologic survival advantage. Targeting the E3 ubiquitin ligase HDM2 can lead to a therapeutic surge in p53 levels. However, the efficacy of HDM2 inhibition can be compromised by overexpression of HDMX, an HDM2 homologue that binds and sequesters p53. Here we report that a stapled p53 helix preferentially targets HDMX, blocks the formation of inhibitory p53-HDMX complexes, induces p53-dependent transcriptional upregulation, and thereby overcomes HDMX-mediated cancer resistance in vitro and in vivo. Importantly, our analysis of p53 interaction dynamics provides a blueprint for reactivating the p53 pathway in cancer by matching HDM2, HDMX, or dual inhibitors to the appropriate cellular context.
Introduction
p53 plays an essential regulatory role in the development and homeostasis of cells and tissues (Vousden and Lane, 2007). As “guardian of the genome,” p53 prevents the emergence of genetically variant clones by activating defense mechanisms, such as induction of senescent-like arrest and apoptotic programs, to prevent replication of defective cells (Lane, 1992; Vogelstein et al., 2000). The critical importance of this protective functionality is underscored by the diversity of molecular strategies employed by cancer cells to subvert p53 activity, such as deletions (Baker et al., 1989), mutations (Baker et al., 1989), protein destruction (Honda et al., 1997; Momand et al., 1992), or protein sequestration (Moll et al., 1995).
Restoration of p53 activity remains an important goal in the quest for more effective cancer therapeutics (Brown et al., 2009). Re-establishing wild-type p53 functionality in the context of genetic deletion or mutation has been especially daunting. However, when wild-type p53 is present, targeting its negative regulators holds promise to reinstate the p53 tumor suppressor pathway (Toledo and Wahl, 2006; Wade and Wahl, 2009). Genetic studies demonstrated that MDM2 and MDM4/MDMX exert protein interaction-mediated control over p53 activity through largely non-overlapping mechanisms (Itahana et al., 2007; Jones et al., 1995; Montes de Oca Luna et al., 1995; Parant et al., 2001). MDM2 is an E3 ubiquitin ligase that directly binds to and mediates ubiquitylation of p53, targeting it for proteasomal degradation (Honda et al., 1997; Tao and Levine, 1999). In contrast, MDM4/MDMX inhibits p53 transactivation through protein interaction-mediated sequestration (Ohtsubo et al., 2009; Shvarts et al., 1996). Certain human cancer cells exploit this natural regulatory axis by overexpressing HDM2 or HDMX to suppress p53 (Danovi et al., 2004; Fakharzadeh et al., 1991; Laurie et al., 2009; Oliner et al., 1992; Ramos et al., 2001), prompting a pharmacologic quest to target these negative regulators for cancer therapy.
The crystal structure of the p53-HDM2 binding interface revealed that a hydrophobic cleft on the N-terminal surface of HDM2 directly engages the p53 transactivation domain, which forms an amphipathic α-helix (Kussie et al., 1996). This seminal structural insight led to the development of a series of small molecules and peptides that target the p53-binding pocket of HDM2, disrupt the p53-HDM2 interaction, stabilize p53, and thereby reduce cancer cell viability (Bernal et al., 2007; Grasberger et al., 2005; Koblish et al., 2006; Kritzer et al., 2004; Shangary et al., 2008; Vassilev et al., 2004; Yin et al., 2005). For example, Nutlin-3 is a small molecule that inhibits HDM2, triggers cell cycle arrest and apoptosis, and exhibits anti-tumor efficacy in an osteosarcoma murine xenograft model (Vassilev et al., 2004). Whereas select cancer cell lines readily undergo single agent Nutlin-3-induced apoptosis (Drakos et al., 2007; Tabe et al., 2009), expression of HDMX, which does not bind Nutlin-3, can prevent activation of cell death programs and engender resistance (Hu et al., 2006; Patton et al., 2006; Wade et al., 2006). Given the importance of HDMX in regulating p53 dynamics and its emerging role in the pathogenesis, maintenance, and chemoresistance of human cancer (Danovi et al., 2004; Laurie et al., 2006; Ramos et al., 2001; Wade et al., 2006; Wang et al., 2007), the development of compounds to investigate and target HDMX in cells has become a pressing therapeutic goal (Harker et al., 2009; Hayashi et al., 2009; Hu et al., 2007; Kallen et al., 2009; Li et al., 2008; Michel et al., 2009; Pazgier et al., 2009; Reed et al., 2010).
We have applied a chemical strategy termed “hydrocarbon stapling” that installs an all-hydrocarbon crosslink within synthetic peptides to restore their α-helical structure, confer protease resistance, and promote cellular uptake (Bird et al., 2010; Schafmeister et al., 2000; Walensky et al., 2004). The resultant “stapled peptides” recapitulate the biological function of natural α-helical domains and have been deployed to interrogate and modulate intracellular protein interactions for mechanistic analyses and potential therapeutic benefit (Gavathiotis et al., 2008; Stewart et al., 2010; Walensky et al., 2004). Using this approach, we previously generated Stabilized Alpha-Helix of p53 (SAH-p53) peptides modeled after the transactivation domain of p53 and demonstrated that these compounds targeted HDM2 in situ and reactivated the p53 pathway in HDM2-overexpressing osteosarcoma cells (Bernal et al., 2007). As a recent structural analysis determined that HDMX engages the p53 transactivation α-helix in a manner similar to HDM2 (Popowicz et al., 2008), we examined the HDMX targeting capacity of our most effective HDM2 inhibitor, SAH-p53-8, and the functional consequences of HDMX inhibition.
Results
SAH-p53-8 is a Potent HDMX Binder
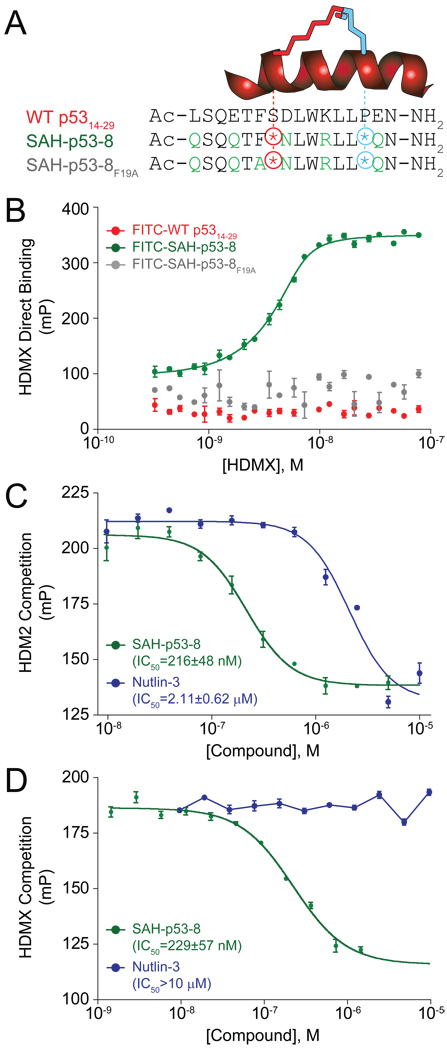
SAH-p53-8 was designed based on the peptide sequence of the p53 transactivation domain α-helix (Bernal et al., 2007) (Figure 1A). We replaced natural amino acids at positions S20 and P27 with synthetic olefinic residues, and generated the structurally reinforcing “hydrocarbon staple” by olefin metathesis (Bird et al., 2008). Additional residues not required for HDM2 interaction were also modified to improve peptide solubility and uptake (Bernal et al., 2007). Substitution of F19 (an essential amino acid for HDM2 interaction (Bottger et al., 1997)) with alanine yielded a negative control for biological experiments. HDMX binding was examined by fluorescence polarization (FP) using full length HDMX and FITC-labeled derivatives of wild-type p5314–29, SAH-p53-8, and SAH-p53-8F19A. FITC-SAH-p53-8 displayed strong affinity for HDMX (KD = 2.3 ± 0.2 nM), surpassing that previously reported for HDM2 (KD = 55 ± 11 nM (Bernal et al., 2007)), whereas the FITC-wild-type p5314–29 peptide and the point mutant FITC-SAH-p53-8F19A showed no interaction in this dose range (Figure 1B).
Figure 1. Primary sequence and HDM2/HDMX binding activity of SAH-p53-8.
(A) Composition of wild-type p5314–29, SAH-p53-8, and SAH-p53-8F19A peptides. (B) Direct binding of FITC-peptides to recombinant HDMX as measured by fluorescence polarization. Competition of SAH-p53-8 and Nutlin-3 with FITC-SAH-p53-8 for binding to HDM2 (C) and HDMX (D). mP: units of milli-polarization. Data are mean +/− s.e.m. for experiments performed in at least triplicate.
We performed competition binding assays to test the capacity of acetyl-capped SAH-p53-8 to disrupt the high affinity complexes of FITC-SAH-p53-8 with HDM2 and HDMX. Both SAH-p53-8 and the selective HDM2-inhibitor Nutlin-3 effectively competed with FITC-SAH-p53-8 for HDM2 binding (Figure 1C). The relatively larger interaction surface of a stapled peptide compared to a small molecule may explain in part why SAH-p53-8 is more effective than Nutlin-3 in this assay. Importantly, only SAH-p53-8 was capable of dissociating the FITC-SAH-p53-8/HDMX interaction (Figure 1D). Taken together, these data demonstrate that SAH-p53-8 targets both HDM2 and HDMX in vitro, and exhibits a greater than 25-fold binding preference for HDMX over HDM2.
SAH-p53-8 Targets Both HDM2 and HDMX in Situ
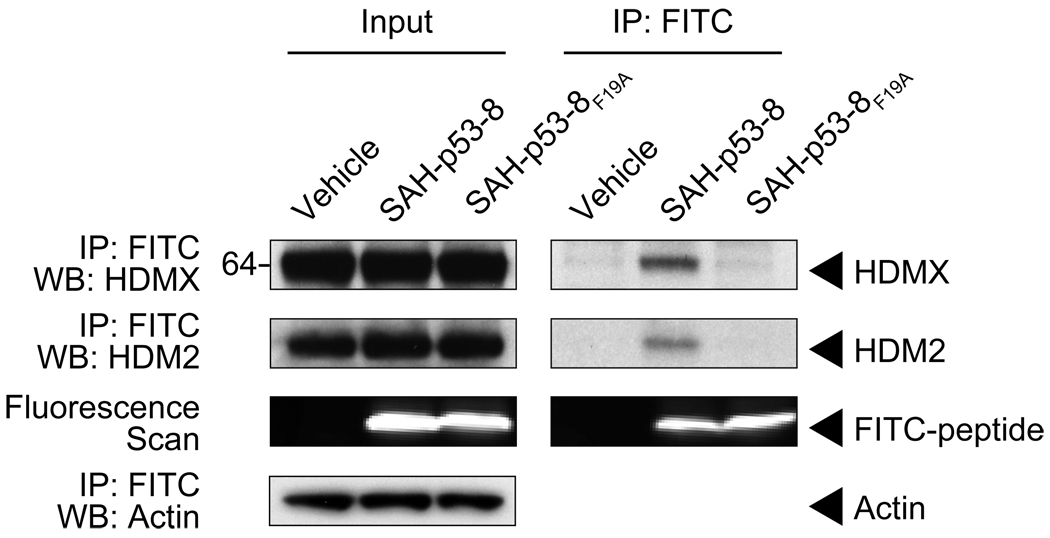
We investigated the capacity of SAH-p53-8 to target intracellular HDM2 and HDMX by conducting immunoprecipitation experiments in SJSA-X cells, an HDM2-overexpressing osteosarcoma cell line engineered to also overexpress HDMX (Wade et al., 2008). The cells were treated with vehicle, FITC-SAH-p53-8, or FITC-SAH-p53-8F19A for 12 hours. After cellular lysis, anti-FITC pull down was performed, followed by HDM2 and HDMX western analyses. Native HDM2 and HDMX specifically co-immunoprecipitated with FITC-SAH-p53-8 but not with the point mutant FITC-SAH-p53-8F19A peptide (Figure 2). A fluorescence scan of the western blot documented the presence of FITC-SAH-p53-8 and FITC-SAH-p53-8F19A in the cellular lysates, confirming that both SAH-p53 peptides efficiently penetrated intact SJSA-X cells. These data extend our in vitro findings by demonstrating that SAH-p53-8 can access both HDM2 and HDMX targets within cells.
Figure 2. SAH-p53-8 targets both HDM2 and HDMX in situ.
SJSA-X cells were treated for 12 hours with either vehicle, FITC-SAH-p53-8 (15 µM), or FITC-SAH-p53-8F19A (15 µM). Anti-FITC immunoprecipitates from cellular extracts were subjected to HDM2 and HDMX western analyses, with β-actin used as a loading control. The presence of FITC-labeled peptide in the extracts was detected by a fluorescence scan of the immunoblot.
SAH-p53-8 is Cytotoxic to Cancer Cells that Overexpress HDM2, HDMX, or Both Proteins
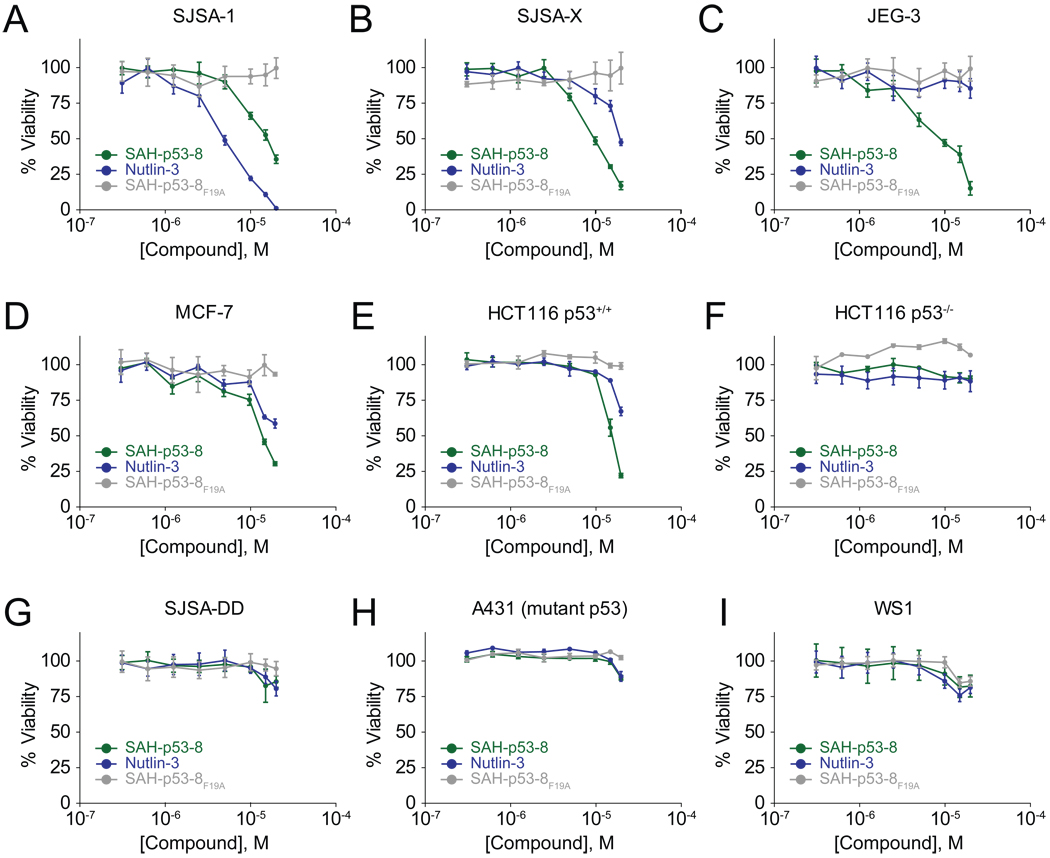
We evaluated the impact of SAH-p53-8 treatment on cell viability by treating a panel of solid tumor cells that express different levels of HDM2 and HDMX. The cell lines included: SJSA-1, an HDM2-overexpressing osteosarcoma; the engineered SJSA-X derivative that overexpresses both HDM2 and HDMX; the HDMX-overexpressing choriocarcinoma cell line JEG-3; and the HDM2- and HDMX-expressing breast and colon cancer cell lines MCF-7 and HCT116 (Figure 3) (Bunz et al., 1999; Chen et al., 2007; Drukteinis et al., 2005; Kwok et al., 1994; Wade et al., 2008; Xia et al., 2008). We examined the p53-dependence of the compounds using SJSA-DD cells that express the dominant negative p53-DD protein (Shaulian et al., 1992; Wade et al., 2008), HCT116 cells deficient in p53 (Bunz et al., 1999), and A431 melanoma cells bearing the R273H p53 mutation (Kwok et al., 1994). We also tested WS1 normal human fibroblasts to evaluate relative toxicity in tumor versus non-transformed cells.
Figure 3. Viability of cancer cells exposed to HDM2/HDMX inhibitors.
Cancer cell lines with differential expression levels of HDM2 and HDMX (A–E), deficient or dysfunctional p53 (F–H), and a non-tumor cell control (I) were treated with 0.3–20 µM Nutlin-3, SAH-p53-8, or SAH-p53-8F19A for 24 h. The cells were exposed to CellTiter-Glo™ reagent (Promega) and viability was assessed by ATP-induced chemiluminescence. Data are mean +/− s.e.m. for experiments performed in at least triplicate.
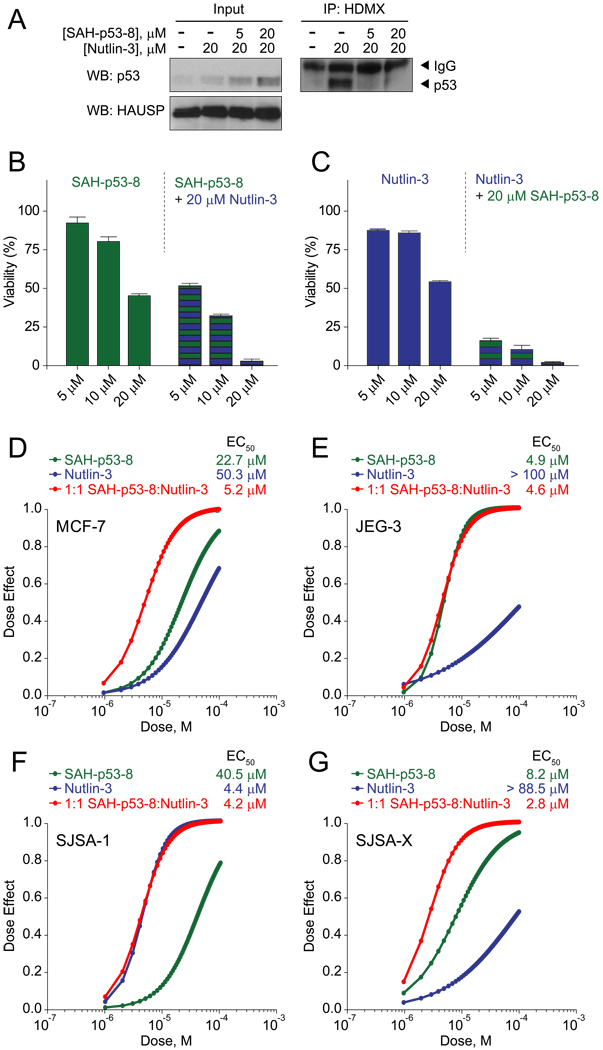
Cultured cells were treated with serial dilutions of Nutlin-3, SAH-p53-8, or the SAH-p53-8F19A point mutant control. Whereas SJSA-1 cells were very sensitive to treatment with Nutlin-3, SJSA-X and JEG-3 cells showed little to no response, consistent with the ability of Nutlin-3 to target HDM2 but not HDMX (Figure 3A–C). MCF-7 and HCT116 cells were modestly sensitive to Nutlin-3 treatment, consistent with the co-expression of HDM2 and HDMX in these cells (Fig. 3D, 3E). In contrast, SAH-p53-8 caused dose-dependent inhibition of cell viability in all five cell lines, suggesting that SAH-p53-8 is capable of reactivating the p53 pathway when cells express elevated levels of HDM2, HDMX, or both proteins (Figure 3A–E). Of note, despite the relatively enhanced capacity of SAH-p53-8 to displace the p53 transactivation helix from HDM2 compared to Nutlin-3 in vitro (Figure 1C), Nutlin-3 was more cytotoxic than SAH-p53-8 in SJSA-1 cells (Figure 3A). This apparent discrepancy likely derives from (1) the preferential HDMX-binding activity of SAH-p53-8 compared to HDM2, thus lowering the effective concentration of SAH-p53-8 available for HDM2-targeting, and (2) the differential efficiencies of cellular import mechanisms for stapled peptides (i.e. pinocytosis (Bernal et al., 2007; Walensky et al., 2004)) and small molecules (i.e. diffusion). Consistent with the in vitro binding data, which revealed a binding preference of SAH-p53-8 for HDMX over HDM2 (Figure 1A) (Bernal et al., 2007), SJSA-X cells were more susceptible to SAH-p53-8 than SJSA-1 cells (Figure 3A, 3B). Strikingly, the HDMX-overexpressing JEG-3 cells were most sensitive to SAH-p53-8 but most resistant to Nutlin-3 (Figure 3C), a key finding that formed the basis for our mechanistic analysis below.
Importantly, we first confirmed that SAH-p53-8 cytoxicity is specifically dependent on wild-type p53 protein activity and does not significantly affect the viability of normal fibroblasts. Genetic deletion of p53 from HCT116 cells (Bunz et al., 1999) or overexpression of a dominant negative form of p53 in SJSA-1 cells (Shaulian et al., 1992; Wade et al., 2008) rendered both cell types completely insensitive to Nutlin-3 and SAH-p53-8 (Figure 3F, 3G). The A431 melanoma cell line, which bears an inactivating p53 point mutation, was similarly unaffected by the treatments. We also found that SAH-p53-8, like Nutlin-3, had no impact on the viability of normal human fibroblasts (Figure 3I). As a further measure of specificity, the mutant peptide SAH-p53-8F19A is inactive in all cell lines tested (Figure 3A–I). These data further indicate that the anti-tumor cell activity of SAH-p53-8 is peptide-sequence dependent and derives from its intracellular targeting of HDM2 and HDMX.
SAH-p53-8 Blocks HDMX-mediated Sequestration of p53 and Reactivates the p53 Tumor Suppressor Pathway
We performed immunoprecipitation studies in JEG-3 cells to interrogate whether the apparent pharmacologic advantage of SAH-p53-8 in Nutlin-3-resistant cells derives from HDMX targeting. After 6 hour treatment with vehicle, SAH-p53-8, or Nutlin-3, cellular extracts were prepared and subjected to anti-HDMX pull down, followed by p53 western analysis. We found that JEG-3 cells had robust levels of p53 protein, which co-immunoprecipitated with HDMX (Figure 4A). Whereas an increase in p53 levels was observed upon treatment with either SAH-p53-8 or Nutlin-3 (Figure 4A), only SAH-p53-8 treatment impacted JEG-3 cell viability (Figure 3C). We examined whether SAH-p53-8 treatment prevents HDMX-mediated sequestration of p53, especially when p53 levels are further boosted by HDM2 blockade. Indeed, SAH-p53-8 effectively blocked the formation of p53-HDMX complexes (Figure 4A). In contrast, p53-HDMX complexes were preserved, if not increased, in Nutlin-3 treated cells. To stabilize p53 levels even further, we treated cells with the proteasome inhibitor MG-132 and correspondingly observed increased p53-HDMX complex, which was dose-responsively inhibited by SAH-p53-8 treatment (Figure 4B). To confirm this observation in a cellular context, we employed a proximity ligation in situ assay, or P-LISA, and directly monitored p53-HDMX complex formation and pharmacologic dissociation in cells (Söderberg et al., 2006) (Figure 4C–E). U2OS osteosarcoma cells containing a doxycycline-inducible HA-HDMX construct (Wang et al., 2007) were treated with doxycycline in the presence or absence of SAH-p53-8, Nutlin-3, or both. In the presence of doxycycline alone, the HA-HDMX produced binds to endogenous p53, leading to the formation of detectable but low intensity P-LISA foci (Figure 4D, 4E). The combination of doxycycline and Nutlin-3 treatment generated a robust P-LISA signal, which represents abundant formation of p53/HA-HDMX complexes (Figure 4D, 4E). Despite similar levels of p53 induction in response to single agent SAH-p53-8 or Nutlin-3 treatment (Figure 4C), the P-LISA signal generated by treatment with doxycycline was blocked upon co-treatment with SAH-p53-8 (Figure 4D, 4E). Furthermore, combining SAH-p53-8 with doxycycline/Nutlin-3 significantly reduced the P-LISA signal generated by the doxycycline/Nutlin-3 combination alone (Figure 4D, 4E). Taken together, the immunoprecipitation and P-LISA data document that SAH-p53-8, but not Nutlin-3, inhibits the formation of p53-HDMX complexes due to its capacity to target intracellular HDMX.
Figure 4. SAH-p53-8 blocks the formation of p53-HDMX complexes.
(A) JEG-3 choriocarcinoma cells were exposed to vehicle, 20 µM SAH-p53-8, or 20 µM Nutlin-3 for 6 h. Cellular extracts and anti-HDMX immunoprecipitates were subjected to SDS-PAGE and analyzed by western blotting for p53, HDMX, and HAUSP. (B) JEG-3 cells were treated with 0–20 µM SAH-p53-8 in the presence of 10 µM MG-132 for 6 h. Cellular extracts and anti-HDMX immunoprecipitates were analyzed by western blotting for p53, HDMX, and HAUSP. (C) Doxycycline-inducible U2OS cells were treated with or without doxycycline in the presence of Nutlin-3 (10 µM), SAH-p53-8 (10 µM), or both, and induction of HA-HDMX and p53 was detected and quantitated by immunofluorescence. Scale bar, 10 µm (D) Cultured U2OS cells were treated with or without doxycycline in the presence of Nutlin-3, SAH-p53-8, or both and then processed for P-LISA. Scale bar, 10 µm (E) Quantitation of p53-HDMX complexes as detected by P-LISA. Data are mean +/− s.d. *p=0.0003, **p=0.0001, unpaired t-test with Welch’s correction.
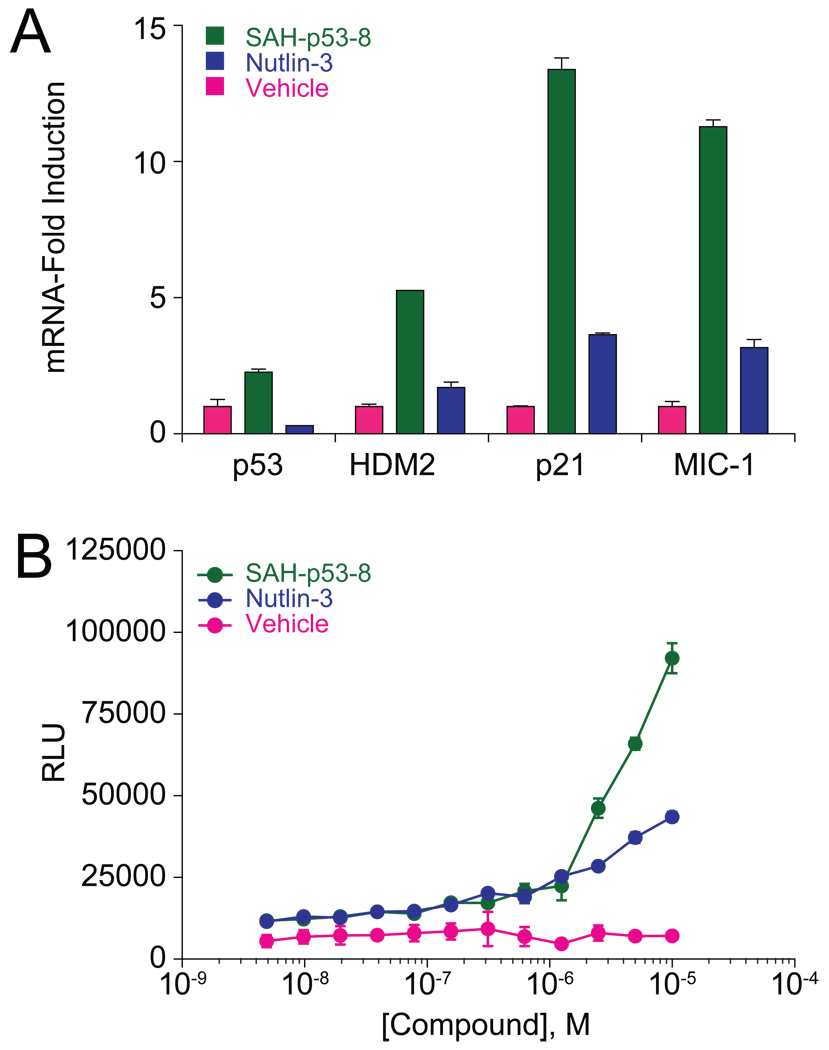
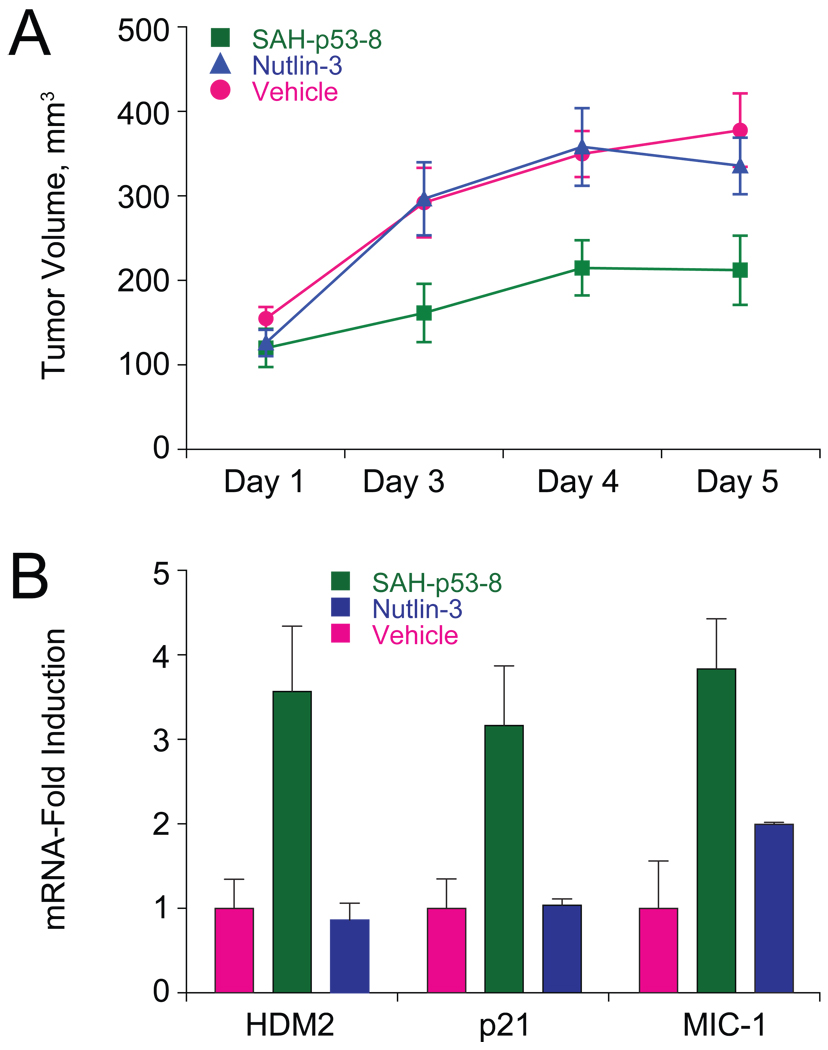
To link the pharmacologic disruption of the p53-HDMX protein complex by SAH-p53-8 with reactivation of the p53 tumor suppressor pathway, we monitored the transcriptional activation of p53 targets by qPCR analysis and the induction of apoptosis using a caspase-3/7 activation assay. JEG-3 cells were treated with vehicle, Nutlin-3 (20 µM), or SAH-p53-8 (20 µM) for 6 h at 37°C, followed by RNA isolation, reverse transcription, and qPCR analysis of the derived cDNA substrate using HDM2, p21, and MIC-1 primers (Figure 5A). Whereas Nutlin-3 induced modest (1.5–4 fold) upregulation of p53 transcriptional targets compared to vehicle, SAH-p53-8 triggered significantly higher (5–14 fold) mRNA levels for the same target genes. Correspondingly, SAH-p53-8 treatment caused dose-responsive activation of caspase-3/7, whereas Nutlin-3 had little effect (Figure 5B). Thus, targeted disruption of the p53-HDMX complex by SAH-p53-8 in Nutlin-3-resistant JEG-3 cells coincides with upregulation of p53 transcriptional targets, caspase-3/7 activation, and reduced tumor cell viability.
Figure 5. Reactivation of p53-dependent Transcription and Apoptosis by SAH-p53-8.
(A) JEG-3 cells were treated with vehicle, SAH-p53-8 (20 µM), or Nutlin-3 (20 µM) and transcriptional upregulation of HDM2, p21, and MIC-1 was evaluated by qPCR analysis. (B) JEG-3 cells were treated with vehicle, or 0.3–10 µM SAH-p53-8 or Nutlin-3 for 12 hours, followed by exposure to Caspase-3/7-Glo™ reagent (Promega). Caspase-3/7 activation was assessed by monitoring the cleavage of proluminescent caspase-3/7 substrate. Data are mean +/− s.e.m. for experiments performed in at least triplicate.
Suppression of JEG-3 Tumor Growth by Reactivation of the p53 Pathway in Vivo
To determine if SAH-p53-8 could modulate the p53 pathway in vivo and thereby inhibit tumor growth, we compared the activity of vehicle, Nutlin-3, and SAH-p53-8 in a JEG-3 murine xenograft model. JEG-3 xenografts were established by injecting 107 cells subcutaneously into the flanks of NOD-SCID-IL2Rγnull (NSG) mice. When tumors reached an average volume of 100 mm3 as determined by caliper measurements, cohorts (n=7) were treated intravenously with vehicle (5% DMSO in D5W), SAH-p53-8 (10 mg kg−1), or Nutlin-3 (10 mg kg−1) daily for 4 days. Whereas the tumor growth rate was not affected by Nutlin-3 treatment, SAH-p53-8 significantly suppressed tumor growth, achieving and maintaining a 37–46% reduction in tumor burden compared to vehicle and Nutlin-3 throughout the 5-day evaluation period (Figure 6A). On day 5, the tumors were excised, flash frozen, and RNA extracted for qPCR analysis using the HDM2, p21, and MIC-1 primer sets. Like the corresponding in vitro study performed with cultured JEG-3 cells, SAH-p53-8 induced statistically significant transcriptional activation of HDM2, p21, and MIC-1 in the tumors of treated mice (Figure 6B). Histologic examination of SAH-p53-8-treated mice showed no obvious toxicity of the compound to normal tissues, consistent with the inactivity of SAH-p53-8 in cell viability assays using cultured WS1 fibroblasts (Figure 3I). These in vivo data underscore the pharmacologic potential of HDMX targeting to functionally suppress tumor growth by reactivating the p53 pathway in the context of HDMX-mediated p53 suppression and Nutlin-3 resistance.
Figure 6. SAH-p53-8 Overcomes HDMX-mediated p53 Suppression and Blocks Tumor Growth in Vivo.
(A) Cohorts (n=7) of JEG-3 xenograft mice were treated with vehicle (5% DMSO in D5W) or 10 mg kg−1 of SAH-p53-8 or Nutlin-3 by intravenous injection daily for 4 days and tumor volume was monitored by caliper measurement on days 1, 3, 4 and 5. Data are mean +/− s.d. (D3: SAH/Veh, p=0.032; SAH/Nut, p=0.032, Nut/Veh, p=0.94; D4: SAH/Veh, p=0.008; SAH/Nut, p=0.026, Nut/Veh, p=0.88; D5: SAH/Veh, p=0.017; SAH/Nut, p=0.037, Nut/Veh, p=0.46). (B) RNA was prepared from the excised JEG-3 tumors and qPCR analysis was performed to measure levels of the p53 transcriptional targets HDM2, p21, and MIC-1. Data are mean +/− s.e.m.
Blueprint for Matching HDM2, HDMX, or Dual Inhibitors to Susceptible Cancer Cells
Because HDMX targeting by SAH-p53-8 was most effective in reducing tumor cell viability when basal p53 levels were naturally elevated, we performed synergy studies in Nutlin-3-resistant cells to test whether SAH-p53-8-mediated HDMX inhibition could resensitize cells to Nutlin-3-mediated p53 upregulation. In cells with high HDMX, the pool of p53 induced by Nutlin-3 treatment can be sequestered in p53-HDMX complexes, subverting Nutlin-3 activity. We hypothesized that HDMX blockade by SAH-p53-8 could restore Nutlin-3 activity by preventing HDMX-mediated p53 sequestration. The breast adenocarcinoma cell line MCF-7 overexpresses HDMX (Danovi et al., 2004) and is only modestly sensitive to Nutlin-3 treatment (Wade et al., 2008). Whereas Nutlin-3 can elevate p53 levels in MCF-7 cells, this increase in p53 coincides with the appearance of the inhibitory p53-HDMX complex, limiting Nutlin-3 cytotoxicity (Figure 7A). However, by employing Nutlin-3, we pharmacologically transform MCF-7 cells into a JEG-3-like state that manifests increased abundance of p53 in complex with HDMX. When combined, SAH-p53-8 and Nutlin-3 sensitize one another in a dose-responsive fashion (Figure 7B, 7C), with synergy confirmed by CalcuSyn dose-effect analysis (Chou, 2006) (Figure 7D). The enhanced cytotoxicity observed upon co-treatment with SAH-p53-8 correlates with its blockade of p53-HDMX complex formation (Figure 7A), consistent with the added capacity of SAH-p53-8 to target HDMX, a key source of resistance in MCF-7 cells. Importantly, Nutlin-3 does not synergize with SAH-p53-8 in JEG-3 cells (Figure 7E), since endogenous p53 levels are already elevated. Conversely, when HDMX and p53 expression levels are low and the cellular phenotype is instead driven by HDM2 expression, as in SJSA-1 cells, co-treatment with SAH-p53-8 provides no added benefit to Nutlin-3 (Figure 7F). However, induction of HDMX expression in this isogenic cell line completely alters the synergy results, rendering Nutlin-3 ineffective as a single agent, restoring susceptibility to SAH-p53-8, and re-establishing the synergistic benefit of combining Nutlin-3 with SAH-p53-8 treatment (Figure 7G). These synergy analyses provide a mechanistic framework for determining how to optimally apply HDM2- and HDMX-targeting agents to reactivate the p53 pathway in cancer. Indeed, HDMX targeting is maximally effective when p53 levels are naturally or pharmacologically increased, as reflected by the presence of detectable p53-HDMX complexes, a potentially valuable biomarker for predicting therapeutic efficacy and monitoring the pharmacodynamic effects of treatment.
Figure 7. Pharmacologic Induction of p53-HDMX Complexes Sensitizes Nutlin-3-Resistant Cancer Cells to HDMX Inhibition.
(A) MCF-7 breast adenocarcinoma cells were exposed to vehicle, Nutlin-3 (20 µM), or Nutlin-3 in combination with 5 or 20 µM SAH-p53-8 in the presence of 10 µM MG-132 for 6 h. Cellular extracts and anti-HDMX immunoprecipitates were subjected to SDS-PAGE and analyzed by western blotting for p53, HDMX, and HAUSP. For synergy studies, MCF-7 cells were treated with 5–20 µM SAH-p53-8 with or without 20 µM Nutlin-3 (B) or 5–20 µM Nutlin-3 with or without 20 µM SAH-p53-8 (C), and cell viability measured at 24 hours by CellTiter-Glo™ assay. Data are mean +/− s.e.m. for experiments performed in at least triplicate. (D–G) Dose-effect synergy analyses of MCF-7 (D), JEG-3 (E), SJSA-1 (F), and SJSA-X (G) cells treated with 0.5–20 µM SAH-p53-8, Nutlin-3, or an equimolar combination.
Discussion
p53 lies at the crossroads of an intricate signal transduction network that regulates cell cycle progression, apoptosis, senescence, and a host of other homeostatic functions (Brown et al., 2009). As such, p53 levels are governed by a cascade of transcriptional, post-translational, and protein interaction-based control systems. For example, in response to DNA damage, the ATM kinase phosphorylates p53, reducing its binding to HDM2 and HDMX and thereby enhancing p53 stability and activity (Shieh et al., 1997; Siliciano et al., 1997). HDM2 and HDMX are likewise phosphorylated, which leads to increased ubiquitylation of both negative regulators, targeting them for accelerated proteasomal degradation. This heightened turnover of HDM2 and HDMX also contributes to p53 stabilization and activation (Meulmeester et al., 2005; Pan and Chen, 2003; Stommel and Wahl, 2004). Dissociation of the deubiquitylating enzyme HAUSP participates in the switch of HDM2-mediated ubiquitylation activity from p53 to itself and HDMX (Meulmeester et al., 2005). These responses are ultimately tempered by the transcriptional auto-inhibitory feedback loop of p53, which restores HDM2 levels and downregulates p53 activity. Whereas the relative protein levels and regulatory mechanisms of p53, HDM2, and HDMX vary among cells, overexpression of HDM2 and HDMX has emerged as a formidable barrier to reactivation of the p53 tumor suppressor pathway in cancer (Marine et al., 2007; Toledo and Wahl, 2006).
Restoring p53 activity by targeting its negative regulators is a promising strategy for treating cancers that retain wild-type p53. We previously developed SAH-p53-8 as an HDM2-targeting agent, but now find that it has a 25-fold greater binding preference for HDMX. SAH-p53-8 targets HDMX in cells, blocks formation of the p53-HDMX interaction, and thereby restores the p53 pathway, as evidenced by transcriptional upregulation of p53 targets and reduction of tumor cell viability. The on-target specificity of SAH-p53-8 activity is highlighted by the explicit p53-dependence of its effects and the complete abrogation of functional activity by single point mutagenesis of a critical residue at the α-helix binding interface. Importantly, intravenous administration of SAH-p53-8 to mice bearing an HDMX-expressing and Nutlin-3-resistant cancer also triggered upregulation of p53 transcriptional targets and suppressed tumor growth.
In addition to validating an intracellular HDMX inhibitor using in vitro and in vivo analyses, what has also emerged from this study is a mechanism-based framework for (1) determining which cancer cells will be susceptible to single agent HDM2 or HDMX inhibition and (2) overcoming p53 suppression in a resistant cell through synergistic HDM2 and HDMX targeting. Whereas sensitivity to single agent HDM2 inhibition is predominantly determined by the presence or absence of HDMX (Figure 8A, 8B), susceptibility to HDMX targeting is dependent upon the cellular level of p53 (Figure 8C, 8D). Our analysis of SAH-p53-8 activity in HDMX-expressing, Nutlin-3-resistant cells revealed that a maximal response was achieved when baseline p53 levels were sufficiently elevated to detect native p53-HDMX complexes, such as in JEG-3 cells. The presence of these complexes indicates that a reservoir of p53 can be promptly released by pharmacologic disruption of the p53-HDMX interaction. Thus, cancers that sequester p53 via HDMX overexpression may be particularly sensitive to SAH-p53-8 or other HDMX-specific antagonists in development. When cellular levels of HDMX are high, HDM2 antagonists are less effective and when p53 levels are low, targeting HDMX is essentially inconsequential. In this resistant context, pharmacologic induction of p53 by HDM2 antagonists combined with HDMX blockade to inhibit p53 sequestration optimally reactivates the p53 pathway (Figure 8E). Thus, we find that targeting HDMX can overcome HDMX-mediated p53 suppression and resistance to selective HDM2 inhibition, while dual targeting of HDM2 and HDMX can maximize therapeutic reactivation of the p53 tumor suppressor pathway in cancers that retain wild-type p53 but maintain pathologically low levels of p53 expression. Importantly, monitoring cellular levels of p53-HDMX complex can both predict cancer cell susceptibility to single agent HDMX inhibition and determine the efficacy of HDM2-mediated p53 upregulation, which forms the basis for enhancing the therapeutic impact of dual HDM2/HDMX targeting.
Figure 8. Molecular Determinants of Cancer Cell Susceptibility to HDM2, HDMX, and Dual Inhibition.
In the context of HDM2-driven p53 suppression, HDM2 inhibition triggers a surge in p53 levels and induces cancer cell death (A). However, if HDMX is present, p53 can become sequestered in p53-HDMX complexes, limiting the cellular response to HDM2 inhibition (B). When p53-HDMX complex levels are elevated, HDMX inhibition reactivates the p53 pathway and induces cell death (C). However, if cellular levels of p53 are low, targeting HDMX has little to no effect on cell viability (D). When p53 levels are suppressed by HDM2, and HDMX is also expressed, maximal reactivation of the p53 pathway is achieved by combined blockade of HDM2 and HDMX, which elevates p53 levels and blocks formation of inhibitory p53-HDMX complexes, respectively (E). Thus, cancer cell susceptibility to pharmacologic inhibition of HDM2, HDMX, or both targets is determined by the respective levels and interactions of p53, HDM2, and HDMX.
Experimental Procedures
Peptide Synthesis
Peptide synthesis, olefin metathesis, FITC derivatization, reverse phase HPLC purification, and amino acid analysis were performed as previously reported (Bernal et al., 2007; Bird et al., 2008).
Fluorescence Polarization Binding Assays
FP assays were performed as previously described (Bernal et al., 2007; Pitter et al., 2008). Briefly, to determine dissociation constants for peptide-protein interactions, fluoresceinated peptides (25 nM) were incubated with full-length HDMX or HDM217–125 (25 pM – 100 nM), and fluorescence polarization measured at equilibrium on a SpectraMax M5 microplate reader (Molecular Devices). For competition assays, FITC-peptide (25 nM) was combined with a serial dilution of Nutlin-3 (EMD Chemicals) or unlabeled SAH-p53-8 followed by addition of HDM2 or HDMX protein (100 nM). IC50 values for FITC-peptide displacement were calculated by nonlinear regression analysis using Prism software (GraphPad). Recombinant HDM2 was generated as previously reported (Bernal et al., 2007). To generate recombinant HDMX, Escherichia coli BL21 (DE3) pLysS containing the plasmid encoding full-length HDMX with N-terminal thioredoxin, S, and hexahistidine tags, and an engineered TEV protease cleavage site (pET32-LIC vector, Novagen) were cultured in ampicillin- and chloramphenicol-containing Luria-Bertani broth and induced with 0.1 mM isopropyl β-d-thiogalactoside. The cells were harvested after 6 h by centrifugation for 20 min at 3200 rpm, resuspended in buffer A (20 mM Tris pH 8.0, 0.5 M NaCl) and lysed by sonication. Cellular debris was pelleted by centrifugation for 30 min at 15,000 rpm, and the supernatant was incubated with Ni-NTA agarose (Qiagen) for 2 h. The resin was washed with buffer A and eluted with a gradient of imidazole ranging from 5 mM to 500 mM. The fractions containing the eluted protein were concentrated and treated overnight with recombinant TEV S219V protease (Kapust et al., 2001) at 4°C. The reaction was concentrated to 2 mL and purified by size exclusion chromatography using a G200 column. Purity of the protein was assessed by SDS-PAGE and its identity confirmed by MALDI-TOF mass spectrometry.
Cell Viability Assays
Cultured cells were maintained in the indicated media supplemented with fetal calf serum and penicillin/streptomycin: SJSA-1, SJSA-X, SJSA-DD, A431 (DMEM); JEG-3, MCF-7, WS1 (EMEM); HCT116 (McCoy’s 5A). For viability analysis, cells (1 × 104) were seeded in 96-well plates overnight, washed in PBS, and then incubated with peptides, Nutlin-3 (EMD Chemicals), or both in OPTI-MEM™ at the indicated doses for 24 h. Cell viability was assayed by addition of CellTiter-Glo™ chemiluminescence reagent according to the manufacturer’s protocol (Promega) and luminescence measured using a Spectramax M5 microplate reader (Molecular Devices). Data are normalized to vehicle-treated controls. Synergy between SAH-p53-8 and Nutlin-3 was calculated using the Calcusyn software package (Chou, 2006) (Biosoft; Cambridge, UK). Viability assays were performed in at least triplicate.
Quantitative RT-PCR
JEG-3 cells (7.5 × 106) were seeded in 6-well plates and treated with vehicle or 20 µM SAH-p53-8 or Nutlin-3 (EMD Chemicals), and incubated for 6 h at 37°C. RNA samples were prepared using the RNEasy Mini kit (Qiagen). Total RNA was reverse transcribed to cDNA using Superscript III reverse transcriptase (Invitrogen). The derived cDNA was employed as the substrate to measure relative transcript levels by qRT-PCR on a PRISM 7700 system using SYBR Green PCR Master Mix (Applied Biosystems). Primers specific for HDM2, p21, MIC-1, and the UBB control were employed. Triplicate reactions were prepared in 96-well optical PCR plates. Threshold-cycle (Ct) values were automatically calculated for each replicate and used to determine the relative expression of the gene of interest relative to UBB for both treated and untreated samples by the 2−ΔΔCt method (Livak and Schmittgen, 2001).
Caspase-3/7 Activation Assay
JEG-3 cells (1 × 104) were seeded in 96-well plates overnight, washed in PBS, and then incubated with the indicated doses of SAH-p53-8 or Nutlin-3 (EMD Chemicals) in OPTI-MEM™ for 12 h. Caspase-3/7 activity was assayed by addition of Caspase-3/7-Glo™ chemiluminescence reagent according to the manufacturer’s protocol (Promega) and luminescence measured using a Spectramax M5 microplate reader (Molecular Devices).
Co-Immunoprecipitation Analysis
Anti-FITC pull down experiments were performed as previously described (Pitter et al., 2008; Walensky et al., 2006). Briefly, cells (1.5 × 106) were treated with FITC-SAH-p53 peptides (15 µM) and lysed in buffer B (0.1% Triton X-100, 50 mM Tris pH 7.4, 150 mM NaCl, 1 mM PMSF, EDTA-free protease inhibitor tablet [Roche], and 20 units/mL benzonase nuclease [EMD Chemicals]). Proteins were co-immunoprecipitated with goat-anti-FITC antibody (AbCam) and western analysis of electrophoresed proteins performed using mouse anti-HDM2 (IF2; EMD Chemicals), rabbit anti-HDMX (BL1258; Bethyl Laboratories), and β-actin (Sigma) antibodies. Fluorescence imaging of the blot was performed using a Typhoon 8600 Imaging System (Molecular Dynamics). For HDMX immunoprecipitation experiments, cells (1.5 × 106) were treated with either SAH-p53-8 or Nutlin-3 (EMD Chemicals) at the indicated doses in 0.5 mL OPTI-MEM™ (Invitrogen) with or without 10 µM MG-132 for 6 h. After washing with PBS (2 × 1 mL), the cells were lysed in buffer B. HDMX was immunoprecipitated with anti-HDMX antibody (BL1258; Bethyl Laboratories) and western analysis of electrophoresed protein was conducted using p53 (DO-1; EMD Chemicals or FL-393; Santa Cruz Biotechnology), HDMX (MDMX-82; Abcam), and HAUSP (anti-USP7, Bethyl Labs) antibodies, and Western Lightning chemiluminescence reagent (PerkinElmer).
P-LISA and Immunofluorescence
U2OS cells expressing a doxycycline-inducible HA-HDMX construct (Wang et al., 2007) were seeded onto coverslips and treated with doxycyline for 24 h. SAH-p53-8 (10 µM), enantiomeric Nutlin-3 (10 µM) (Roche), or both compounds were added for the final 8 h of treatment. The cells were fixed in 3.7% paraformaldehyde, washed in PBS, and permeabilized in 0.2% Triton X-100 for 5 min. Coverslips were then blocked in 10% normal goat serum in PBS (NGS) for 2 h. For P-LISA, primary antibodies HA.11 (BabCo, 1:500) and FL393 (Santa Cruz, 1:1000) were diluted in PBS/EDTA/0.2% Triton X-100/2% NGS and incubated at 4°C overnight. Following washes with TBS/0.05% Tween-20, a proximity ligation in situ assay (P-LISA) was performed according to the manufacturer’s protocol (Detection Kit 613, OLink Bioscience) with the following exception: goat anti-rabbit (minus) and anti-mouse (plus) P-LISA probes were diluted in NGS at 1:10 instead of 1:5. Coverslips were mounted on microscope slides and images acquired using OpenLab software (Improvision) and a Zeiss Axioplan 2 microscope. Nuclear foci (at least 100 cells per treatment) were quantified using Blobfinder software (Centre for Image Analysis, Uppsala University, Sweden). All exposure times and intensity thresholds were set based on doxycycline/Nutlin-3 co-treatment and kept constant for each treatment. The statistical significance of the observed differences in foci number among the treatment conditions was determined using the unpaired t-test with Welch’s correction. For standard immunofluorescence imaging of p53 and HDMX, the antibodies indicated above were again employed but following the PBS washes, the slides were incubated (1 h, room temperature) with goat anti-rabbit AF568 (1:1000) and goat anti-mouse AF488 (1:500) (Invitrogen/Molecular Probes) containing 1 µg/mL Hoechst. Density slices from each Hoechst image were generated in OpenLab, and used as masks to quantify the nuclear intensity of both p53 and HDMX. Total intensity was defined as average pixel intensity × nuclear area, and was corrected for nuclear size differences. Graphical representation and statistical analyses were performed using Microsoft Excel and Prism software (GraphPad).
JEG-3 Xenograft Study
JEG-3 xenografts were established by injecting 107 cells subcutaneously into the flanks of NOD-SCID-IL2Rγnull (NSG) mice (Jackson Labs). When tumors reached an average volume of 100 mm3, cohorts (n=7) were treated with vehicle (5% DMSO in D5W), SAH-p53-8 (10 mg/kg), or Nutlin-3 (10 mg/kg) (EMD Chemicals) once daily for 4 days by intravenous injection. Tumor size was measured with calipers on days 1, 3, 4 and 5. On day 5, the mice were euthanized, tumors excised, and a portion of each tumor flash frozen and subjected to RNA isolation for qRT-PCR analysis, as described above. The remainder of each animal was fixed in Bouin’s fixative and submitted to the Dana-Farber/Harvard Cancer Center (DF/HCC) Rodent Histopathology Core for complete necropsy. Animal experiments were approved by and performed in accordance with the guidelines and regulations set forth by the Institutional Animal Care and Use Committee of the Dana-Farber Cancer Institute.
Significance
As guardian of the genome, p53 protects the cell from genetic assault by triggering cell cycle arrest and apoptosis. Cancer cells disarm p53 by a variety of mechanisms, including overexpression of the p53 antagonists HDM2 and HDMX, making deregulation of the p53 pathway one of the most common defects in human cancer. We employed a stapled peptide helix based on the p53 transactivation domain to interrogate and overcome HDMX-mediated p53 antagonism in cancer. Whereas the formation of p53-HDMX complexes reflects a resistance mechanism to HDM2 targeting, it likewise serves as a biomarker for cancer cell susceptibility to HDMX inhibition. Pharmacologic disruption of the p53-HDMX interaction reactivated the p53 pathway and suppressed the growth of an HDM2 inhibitor-resistant tumor in vivo.
Acknowledgements
We thank Eric Smith for editorial and graphics assistance, Gregory Verdine for catalyzing the collaboration between L.D.W and G.M.W, Bert Vogelstein for supplying HCT116 p53+/+ and HCT116 p53−/− cell lines, Lyubomir Vassilev for providing G.M.W. with enantiomeric Nutlin-3 for P-LISA experiments, and Ulf Landegren, Ola Söderberg, and Eric Nyström for invaluable advice regarding P-LISA. This work was supported by NCI Career Transition Award 1K22CA128886 to F.B., NIH grant 5P01CA92625, the Todd J. Schwartz Pediatric Oncology Fund, and a Burroughs Wellcome Fund Career Award to L.D.W., and NIH Grants CA61449 and CA100845 to G.M.W.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baker SJ, Fearon ER, Nigro JM, Hamilton SR, Preisinger AC, Jessup JM, vanTuinen P, Ledbetter DH, Barker DF, Nakamura Y, et al. Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science. 1989;244:217–221. doi: 10.1126/science.2649981. [DOI] [PubMed] [Google Scholar]
- Bernal F, Tyler AF, Korsmeyer SJ, Walensky LD, Verdine GL. Reactivation of the p53 tumor suppressor pathway by a stapled p53 peptide. J Am Chem Soc. 2007;129:2456–2457. doi: 10.1021/ja0693587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird GH, Bernal F, Pitter K, Walensky LD. Synthesis and biophysical characterization of stabilized alpha-helices of BCL-2 domains. Methods Enzymol. 2008;446:369–386. doi: 10.1016/S0076-6879(08)01622-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird GH, Madani N, Perry AF, Princiotto AM, Supko JG, He X, Gavathiotis E, Sodroski JG, Walensky LD. Hydrocarbon double-stapling remedies the proteolytic instability of a lengthy peptide therapeutic. Proc Natl Acad Sci U S A. 2010;107:14093–14098. doi: 10.1073/pnas.1002713107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttger A, Böttger V, Garcia-Echeverria C, Chène P, Hochkeppel HK, Sampson W, Ang K, Howard SF, Picksley SM, Lane DP. Molecular characterization of the hdm2-p53 interaction. J Mol Biol. 1997;269:744–756. doi: 10.1006/jmbi.1997.1078. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Lain S, Verma CS, Fersht AR, Lane DP. Awakening guardian angels: drugging the p53 pathway. Nat Rev Cancer. 2009;9:862–873. doi: 10.1038/nrc2763. [DOI] [PubMed] [Google Scholar]
- Bunz F, Hwang PM, Torrance C, Waldman T, Zhang Y, Dillehay L, Williams J, Lengauer C, Kinzler KW, Vogelstein B. Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J Clin Invest. 1999;104:263–269. doi: 10.1172/JCI6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Qian H, Wang H, Zhang X, Fu M, Liang X, Ma Y, Zhan Q, Lin C, Xiang Y. Ad-PUMA sensitizes drug-resistant choriocarcinoma cells to chemotherapeutic agents. Gynecol Oncol. 2007;107:505–512. doi: 10.1016/j.ygyno.2007.08.007. [DOI] [PubMed] [Google Scholar]
- Chou T-C. Theoretical Basis, Experimental Design, and Computerized Simulation of Synergism and Antagonism in Drug Combination Studies. Pharmacol Rev. 2006;58:621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- Danovi D, Meulmeester E, Pasini D, Migliorini D, Capra M, Frenk R, de Graaf P, Francoz S, Gasparini P, Gobbi A, et al. Amplification of Mdmx (or Mdm4) directly contributes to tumor formation by inhibiting p53 tumor suppressor activity. Mol Cell Biol. 2004;24:5835–5843. doi: 10.1128/MCB.24.13.5835-5843.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakos E, Thomaides A, Medeiros LJ, Li J, Leventaki V, Konopleva M, Andreeff M, Rassidakis GZ. Inhibition of p53-murine double minute 2 interaction by nutlin-3A stabilizes p53 and induces cell cycle arrest and apoptosis in Hodgkin lymphoma. Clin Cancer Res. 2007;13:3380–3387. doi: 10.1158/1078-0432.CCR-06-2581. [DOI] [PubMed] [Google Scholar]
- Drukteinis JS, Medrano T, Ablordeppey EA, Kitzman JM, Shiverick KT. Benzo[a]pyrene, but not 2,3,7,8-TCDD, induces G2/M cell cycle arrest, p21CIP1 and p53 phosphorylation in human choriocarcinoma JEG-3 cells: a distinct signaling pathway. Placenta. 2005;26 Suppl A:S87–S95. doi: 10.1016/j.placenta.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Fakharzadeh SS, Trusko SP, George DL. Tumorigenic potential associated with enhanced expression of a gene that is amplified in a mouse tumor cell line. EMBO J. 1991;10:1565–1569. doi: 10.1002/j.1460-2075.1991.tb07676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavathiotis E, Suzuki M, Davis ML, Pitter K, Bird GH, Katz SG, Tu HC, Kim H, Cheng EH, Tjandra N, Walensky LD. BAX activation is initiated at a novel interaction site. Nature. 2008;455:1076–1081. doi: 10.1038/nature07396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasberger BL, Lu T, Schubert C, Parks DJ, Carver TE, Koblish HK, Cummings MD, LaFrance LV, Milkiewicz KL, Calvo RR, et al. Discovery and cocrystal structure of benzodiazepinedione HDM2 antagonists that activate p53 in cells. J Med Chem. 2005;48:909–912. doi: 10.1021/jm049137g. [DOI] [PubMed] [Google Scholar]
- Harker EA, Daniels DS, Guarracino DA, Schepartz A. Beta-peptides with improved affinity for hDM2 and hDMX. Bioorg Med Chem. 2009;17:2038–2046. doi: 10.1016/j.bmc.2009.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi R, Wang D, Hara T, Iera JA, Durell SR, Appella DH. N-acylpolyamine inhibitors of HDM2 and HDMX binding to p53. Bioorg Med Chem. 2009;17:7884–7893. doi: 10.1016/j.bmc.2009.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- Hu B, Gilkes DM, Chen J. Efficient p53 activation and apoptosis by simultaneous disruption of binding to MDM2 and MDMX. Cancer Res. 2007;67:8810–8817. doi: 10.1158/0008-5472.CAN-07-1140. [DOI] [PubMed] [Google Scholar]
- Hu B, Gilkes DM, Farooqi B, Sebti SM, Chen J. MDMX overexpression prevents p53 activation by the MDM2 inhibitor Nutlin. J Biol Chem. 2006;281:33030–33035. doi: 10.1074/jbc.C600147200. [DOI] [PubMed] [Google Scholar]
- Itahana K, Mao H, Jin A, Itahana Y, Clegg HV, Lindström MS, Bhat, Krishna P, Godfrey VL, Evan GI, Zhang Y. Targeted Inactivation of Mdm2 RING Finger E3 Ubiquitin Ligase Activity in the Mouse Reveals Mechanistic Insights into p53 Regulation. Cancer Cell. 2007;12:355–366. doi: 10.1016/j.ccr.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Jones SN, Roe AE, Donehower LA, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378:206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- Kallen J, Goepfert A, Blechschmidt A, Izaac A, Geiser M, Tavares G, Ramage P, Furet P, Masuya K, Lisztwan J. Crystal structures of human MdmX (HdmX) in complex with p53 peptide analogues reveal surprising conformational changes. J Biol Chem. 2009;284:8812–8821. doi: 10.1074/jbc.M809096200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapust RB, Tozser J, Fox JD, Anderson DE, Cherry S, Copeland TD, Waugh DS. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 2001;14:993–1000. doi: 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
- Koblish HK, Zhao S, Franks CF, Donatelli RR, Tominovich RM, LaFrance LV, Leonard KA, Gushue JM, Parks DJ, Calvo RR, et al. Benzodiazepinedione inhibitors of the Hdm2:p53 complex suppress human tumor cell proliferation in vitro and sensitize tumors to doxorubicin in vivo. Mol Cancer Ther. 2006;5:160–169. doi: 10.1158/1535-7163.MCT-05-0199. [DOI] [PubMed] [Google Scholar]
- Kritzer JA, Lear JD, Hodsdon ME, Schepartz A. Helical β-Peptide Inhibitors of the p53-hDM2 Interaction. J Am Chem Soc. 2004;126:9468–9469. doi: 10.1021/ja031625a. [DOI] [PubMed] [Google Scholar]
- Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, Pavletich NP. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- Kwok TT, Mok CH, Menton-Brennan L. Up-regulation of a mutant form of p53 by doxorubicin in human squamous carcinoma cells. Cancer Res. 1994;54:2834–2836. [PubMed] [Google Scholar]
- Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- Laurie N, Mohan A, McEvoy J, Reed D, Zhang J, Schweers B, Ajioka I, Valentine V, Johnson D, Ellison D, Dyer MA. Changes in retinoblastoma cell adhesion associated with optic nerve invasion. Mol Cell Biol. 2009;29:6268–6282. doi: 10.1128/MCB.00374-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie NA, Donovan SL, Shih CS, Zhang J, Mills N, Fuller C, Teunisse A, Lam S, Ramos Y, Mohan A, et al. Inactivation of the p53 pathway in retinoblastoma. Nature. 2006;444:61–66. doi: 10.1038/nature05194. [DOI] [PubMed] [Google Scholar]
- Li C, Liu M, Monbo J, Zou G, Yuan W, Zella D, Lu WY, Lu W. Turning a scorpion toxin into an antitumor miniprotein. J Am Chem Soc. 2008;130:13546–13548. doi: 10.1021/ja8042036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Marine JC, Dyer MA, Jochemsen AG. MDMX: from bench to bedside. J Cell Sci. 2007;120:371–378. doi: 10.1242/jcs.03362. [DOI] [PubMed] [Google Scholar]
- Meulmeester E, Maurice MM, Boutell C, Teunisse AF, Ovaa H, Abraham TE, Dirks RW, Jochemsen AG. Loss of HAUSP-mediated deubiquitination contributes to DNA damage-induced destabilization of Hdmx and Hdm2. Mol Cell. 2005;18:565–576. doi: 10.1016/j.molcel.2005.04.024. [DOI] [PubMed] [Google Scholar]
- Michel J, Harker EA, Tirado-Rives J, Jorgensen WL, Schepartz A. In Silico Improvement of beta3-peptide inhibitors of p53 × hDM2 and p53 × hDMX. J Am Chem Soc. 2009;131:6356–6357. doi: 10.1021/ja901478e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll UM, LaQuaglia M, Benard J, Riou G. Wild-type p53 protein undergoes cytoplasmic sequestration in undifferentiated neuroblastomas but not in differentiated tumors. Proc Natl Acad Sci U S A. 1995;92:4407–4411. doi: 10.1073/pnas.92.10.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–205. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- Ohtsubo C, Shiokawa D, Kodama M, Gaiddon C, Nakagama H, Jochemsen AG, Taya Y, Okamoto K. Cytoplasmic tethering is involved in synergistic inhibition of p53 by Mdmx and Mdm2. Cancer Sci. 2009;100:1291–1299. doi: 10.1111/j.1349-7006.2009.01180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature. 1992;358:80–83. doi: 10.1038/358080a0. [DOI] [PubMed] [Google Scholar]
- Pan Y, Chen J. MDM2 promotes ubiquitination and degradation of MDMX. Mol Cell Biol. 2003;23:5113–5121. doi: 10.1128/MCB.23.15.5113-5121.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parant J, Chavez-Reyes A, Little NA, Yan W, Reinke V, Jochemsen AG, Lozano G. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat Genet. 2001;29:92–95. doi: 10.1038/ng714. [DOI] [PubMed] [Google Scholar]
- Patton JT, Mayo LD, Singhi AD, Gudkov AV, Stark GR, Jackson MW. Levels of HdmX expression dictate the sensitivity of normal and transformed cells to Nutlin-3. Cancer Res. 2006;66:3169–3176. doi: 10.1158/0008-5472.CAN-05-3832. [DOI] [PubMed] [Google Scholar]
- Pazgier M, Liu M, Zou G, Yuan W, Li C, Li J, Monbo J, Zella D, Tarasov SG, Lu W. Structural basis for high-affinity peptide inhibition of p53 interactions with MDM2 and MDMX. Proc Natl Acad Sci U S A. 2009;106:4665–4670. doi: 10.1073/pnas.0900947106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitter K, Bernal F, Labelle J, Walensky LD. Dissection of the BCL-2 family signaling network with stabilized alpha-helices of BCL-2 domains. Methods Enzymol. 2008;446:387–408. doi: 10.1016/S0076-6879(08)01623-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popowicz GM, Czarna A, Holak TA. Structure of the human Mdmx protein bound to the p53 tumor suppressor transactivation domain. Cell Cycle. 2008;7:2441–2443. doi: 10.4161/cc.6365. [DOI] [PubMed] [Google Scholar]
- Ramos YF, Stad R, Attema J, Peltenburg LT, van der Eb AJ, Jochemsen AG. Aberrant expression of HDMX proteins in tumor cells correlates with wild-type p53. Cancer Res. 2001;61:1839–1842. [PubMed] [Google Scholar]
- Reed D, Shen Y, Shelat AA, Arnold LA, Ferreira AM, Zhu F, Mills N, Smithson DC, Regni CA, Bashford D, et al. Identification and characterization of the first small molecule inhibitor of MDMX. J Biol Chem. 2010;285:10786–10796. doi: 10.1074/jbc.M109.056747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafmeister C, Po J, Verdine G. An all-hydrocarbon cross-linking system for enhancing the helicity and metabolic stability of peptides. J Am Chem Soc. 2000;122:5891–5892. [Google Scholar]
- Shangary S, Qin D, McEachern D, Liu M, Miller RS, Qiu S, Nikolovska-Coleska Z, Ding K, Wang G, Chen J, et al. Temporal activation of p53 by a specific MDM2 inhibitor is selectively toxic to tumors and leads to complete tumor growth inhibition. Proc Natl Acad Sci USA. 2008;105:3933–3938. doi: 10.1073/pnas.0708917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaulian E, Zauberman A, Ginsberg D, Oren M. Identification of a minimal transforming domain of p53: negative dominance through abrogation of sequence-specific DNA binding. Mol Cell Biol. 1992;12:5581–5592. doi: 10.1128/mcb.12.12.5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh SY, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- Shvarts A, Steegenga WT, Riteco N, van Laar T, Dekker P, Bazuine M, van Ham RC, van der Houven van Oordt W, Hateboer G, van der Eb AJ, Jochemsen AG. MDMX: a novel p53-binding protein with some functional properties of MDM2. EMBO J. 1996;15:5349–5357. [PMC free article] [PubMed] [Google Scholar]
- Siliciano JD, Canman CE, Taya Y, Sakaguchi K, Appella E, Kastan MB. DNA damage induces phosphorylation of the amino terminus of p53. Genes Dev. 1997;11:3471–3481. doi: 10.1101/gad.11.24.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderberg O, Gullberg M, Jarvius M, Ridderstråle K, Leuchowius K-J, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson L-G, Landegren U. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- Stewart ML, Fire E, Keating AE, Walensky LD. The MCL-1 BH3 helix is an exclusive MCL-1 inhibitor and apoptosis sensitizer. Nat Chem Biol. 2010;6:595–601. doi: 10.1038/nchembio.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stommel JM, Wahl GM. Accelerated MDM2 auto-degradation induced by DNA-damage kinases is required for p53 activation. EMBO J. 2004;23:1547–1556. doi: 10.1038/sj.emboj.7600145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabe Y, Sebasigari D, Jin L, Rudelius M, Davies-Hill T, Miyake K, Miida T, Pittaluga S, Raffeld M. MDM2 antagonist nutlin-3 displays antiproliferative and proapoptotic activity in mantle cell lymphoma. Clin Cancer Res. 2009;15:933–942. doi: 10.1158/1078-0432.CCR-08-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao W, Levine AJ. Nucleocytoplasmic shuttling of oncoprotein Hdm2 is required for Hdm2-mediated degradation of p53. Proc Natl Acad Sci U S A. 1999;96:3077–3080. doi: 10.1073/pnas.96.6.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, et al. In Vivo Activation of the p53 Pathway by Small-Molecule Antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- Wade M, Rodewald LW, Espinosa JM, Wahl GM. BH3 activation blocks Hdmx suppression of apoptosis and cooperates with Nutlin to induce cell death. Cell Cycle. 2008;7:1973–1982. doi: 10.4161/cc.7.13.6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade M, Wahl GM. Targeting Mdm2 and Mdmx in Cancer Therapy: Better Living through Medicinal Chemistry? Mol Cancer Res. 2009;7:1–11. doi: 10.1158/1541-7786.MCR-08-0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade M, Wong ET, Tang M, Stommel JM, Wahl GM. Hdmx modulates the outcome of p53 activation in human tumor cells. J Biol Chem. 2006;281:33036–33044. doi: 10.1074/jbc.M605405200. [DOI] [PubMed] [Google Scholar]
- Walensky LD, Kung AL, Escher I, Malia TJ, Barbuto S, Wright RD, Wagner G, Verdine GL, Korsmeyer SJ. Activation of apoptosis in vivo by a hydrocarbon-stapled BH3 helix. Science. 2004;305:1466–1470. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walensky LD, Pitter K, Morash J, Oh KJ, Barbuto S, Fisher J, Smith E, Verdine GL, Korsmeyer SJ. A stapled BID BH3 helix directly binds and activates BAX. Mol Cell. 2006;24:199–210. doi: 10.1016/j.molcel.2006.08.020. [DOI] [PubMed] [Google Scholar]
- Wang YV, Wade M, Wong E, Li YC, Rodewald LW, Wahl GM. Quantitative analyses reveal the importance of regulated Hdmx degradation for p53 activation. Proc Natl Acad Sci U S A. 2007;104:12365–12370. doi: 10.1073/pnas.0701497104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia M, Knezevic D, Tovar C, Huang B, Heimbrook DC, Vassilev LT. Elevated MDM2 boosts the apoptotic activity of p53-MDM2 binding inhibitors by facilitating MDMX degradation. Cell Cycle. 2008;7:1604–1612. doi: 10.4161/cc.7.11.5929. [DOI] [PubMed] [Google Scholar]
- Yin H, Lee G-i, Park HS, Payne GA, Rodriguez JM, Sebti SM, Hamilton AD. Terphenyl-based helical mimetics that disrupt the p53/HDM2 interaction. Angew Chem Int Ed Engl. 2005;44:2704–2707. doi: 10.1002/anie.200462316. [DOI] [PubMed] [Google Scholar]