Abstract
Ionotropic glutamate receptors (iGluRs) are ligand-gated ion channels that mediate chemical communication between neurons at synapses. A variant iGluR subfamily, the Ionotropic Receptors (IRs), was recently proposed to detect environmental volatile chemicals in olfactory cilia. Here we elucidate how these peripheral chemosensors have evolved mechanistically from their iGluR ancestors. Using a Drosophila model, we demonstrate that IRs act in combinations of up to three subunits, comprising individual odor-specific receptors and one or two broadly expressed co-receptors. Heteromeric IR complex formation is necessary and sufficient for trafficking to cilia and mediating odor-evoked electrophysiological responses in vivo and in vitro. IRs display heterogeneous ion conduction specificities related to their variable pore sequences, and divergent ligand-binding domains function in odor recognition and cilia localization. Our results provide insights into the conserved and distinct architecture of these olfactory and synaptic ion channels and offer perspectives into use of IRs as genetically encoded chemical sensors.
Introduction
Rapid, specific and versatile communication between cells and between individuals relies principally on chemical signals. External molecular cues are usually recognized by dedicated cell surface receptor proteins that can trigger changes in gene expression, physiology or behavior of both cells and organisms. In nervous systems, intercellular communication occurs between neurons at synapses. Diffusible neurotransmitters are released from pre-synaptic cell termini in response to action potentials and recognized by receptor proteins in post-synaptic cell membranes to induce neuronal depolarization and continued propagation of action potentials.
One of the best characterized synaptic communication mechanisms is that mediated by the neurotransmitter glutamate and ionotropic glutamate receptors (iGluRs), which underlies most excitatory neurotransmission in the mammalian central nervous system (Gereau and Swanson, 2008). iGluRs are ligand-gated ion channels, comprising an extracellular “Venus fly-trap” ligand-binding domain (LBD) that undergoes conformational changes upon association with glutamate to open a transmembrane channel pore (Mayer, 2006; Sobolevsky et al., 2009). Several iGluR classes have been defined, including AMPA, Kainate and NMDA receptors, which assemble into subfamily-specific heteromeric complexes with unique signaling properties in post-synaptic membranes (Gereau and Swanson, 2008). iGluRs are structurally and functionally conserved in most animals (Tikhonov and Magazanik, 2009), reflecting their fundamental role in synaptic communication.
Nervous systems are also responsible for detecting myriad volatile chemicals in the environment (Ache and Young, 2005). Odor detection is mediated by large, divergent repertoires of olfactory receptors, which localize to the ciliated dendritic endings of olfactory sensory neurons (OSNs) (Touhara and Vosshall, 2009). In most animals, the vast majority of OSNs express a single odorant receptor (OR) gene, which defines the selectivity of OSN responses to odor stimuli (Fuss and Ray, 2009). Vertebrate ORs are G protein-coupled receptors (GPCRs) and signal through intracellular second messengers to depolarize OSNs (Spehr and Munger, 2009).
We recently described a novel family of olfactory receptors called the Ionotropic Receptors (IRs) (Benton et al., 2009). In contrast to other receptor repertoires, IRs represent a highly divergent subfamily of iGluRs that is present across the protostome branch of the animal kingdom (Croset et al., 2010). Analysis of IR expression in the principal olfactory organ (antenna) of the fruit fly, Drosophila melanogaster, revealed complex combinatorial expression patterns, with individual OSNs expressing 2–5 distinct IR genes (Benton et al., 2009). IRs concentrate in olfactory cilia and not at synapses and misexpression of IRs in other (IR-containing) neurons is sufficient to confer novel odor responsiveness, supporting the hypothesis that they function directly in odor detection (Benton et al., 2009).
The IRs define an intriguing molecular parallel between the chemical communication mechanisms occurring between neurons at synapses and between the external world and olfactory circuits (Shaham, 2010). Our appreciation of the similarities and differences in how these receptors act in these different neural contexts is, however, hampered by our lack of mechanistic knowledge of IRs as olfactory receptors. Here we combine molecular genetic, cellular imaging and electrophysiological approaches to elucidate IR function. Our results provide insights into the transitions that have occurred during evolution of these diverse chemical detectors from their conserved synaptic iGluR ancestors.
Results
Broad sensory neuron expression of two conserved IRs: IR8a and IR25a
Comparative genomic analysis of IR repertoires has defined several classes of receptors (Figure 1A) (Croset et al., 2010). At least 14 “antennal” IRs are conserved across insects and expressed in combinations of up to three different receptors in stereotyped subsets of antennal OSNs in Drosophila(Benton et al., 2009). Forty-five “divergent” IRs are, by contrast, largely specific to drosophilids, and at least some of these are implicated in taste detection (Croset et al., 2010). Finally, two closely related IRs, IR8a and IR25a, are distinguished by their higher sequence identity to iGluRs, the existence of homologous genes across Protostomia and the broad distribution of their transcripts in antennal IR-expressing OSNs (Benton et al., 2009; Croset et al., 2010). These properties suggested that IR8a and IR25a have a conserved and central role in IR function and led us to focus first on these receptors.
Figure 1. Phylogenetic relationships and broad antennal expression of IR8a and IR25a.
(A) Phylogenetic tree of Drosophila iGluRs and IRs. Protein sequences were aligned using MUSCLE, and the tree was calculated with PhyML and visualized in FigTree v1.1.2 (adapted from (Croset et al., 2010)). The scale bar indicates the expected number of substitutions per site. A schematic model of the iGluR/IR structure is shown in the cartoon at the top right.
(B) Schematic of the Drosophila third antennal segment illustrating the different classes of olfactory sensilla and other sensory structures.
(C) Immunostaining on a wildtype antennal section with IR8a (green), IR25a (red) and cilium base marker 21A6 (blue) antibodies. IR8a is detected in ciliated dendritic endings (distal to 21A6) both in coeloconic sensilla (arrowheads) as well as in neurons projecting into the sacculus chamber (asterisk). The scale bar represents 20 µm.
Double immunofluorescence using IR8a- and IR25a-specific antibodies revealed heterogeneous expression in distinct but partially overlapping populations of neurons in the Drosophila antenna (Figure 1B–C). IR25a, but not IR8a, is detected in neurons of the arista, a branched cuticular projection from the antennal surface (Foelix et al., 1989). In the sacculus, an internal multichambered pocket (Shanbhag et al., 1995), IR25a-expressing neurons innervate the first and second chambers, while IR8a is strongly expressed in neurons innervating the third chamber. Throughout the main body of the antenna, many clusters of IR8a- or IR25a-positive neurons are found, corresponding to neurons innervating the coeloconic class of olfactory sensilla (Benton et al., 2009; Stocker, 2001; Yao et al., 2005). These clusters contain 2–3 neurons and often comprise 1–2 cells with stronger IR25a expression and weak or no IR8a expression, and 1–2 cells with stronger IR8a expression and weak or no IR25a expression. As previously described for IR25a (Benton et al., 2009), IR8a is detected in both the cell body and sensory dendritic endings but not in axon termini, consistent with a role in peripheral odor detection (Figure 1C and Figure 3B and data not shown). To confirm the specificity of these antibodies and initiate functional analysis of these receptors, we generated null mutations in IR8a by the same gene-targeting strategy used for IR25a (Benton et al., 2009) (Figure 2A). Homozygous IR8a, IR25a and IR8a/IR25a double mutant animals are viable and fertile, and all corresponding immunoreactivity in the antenna is abolished (Figure 2A).
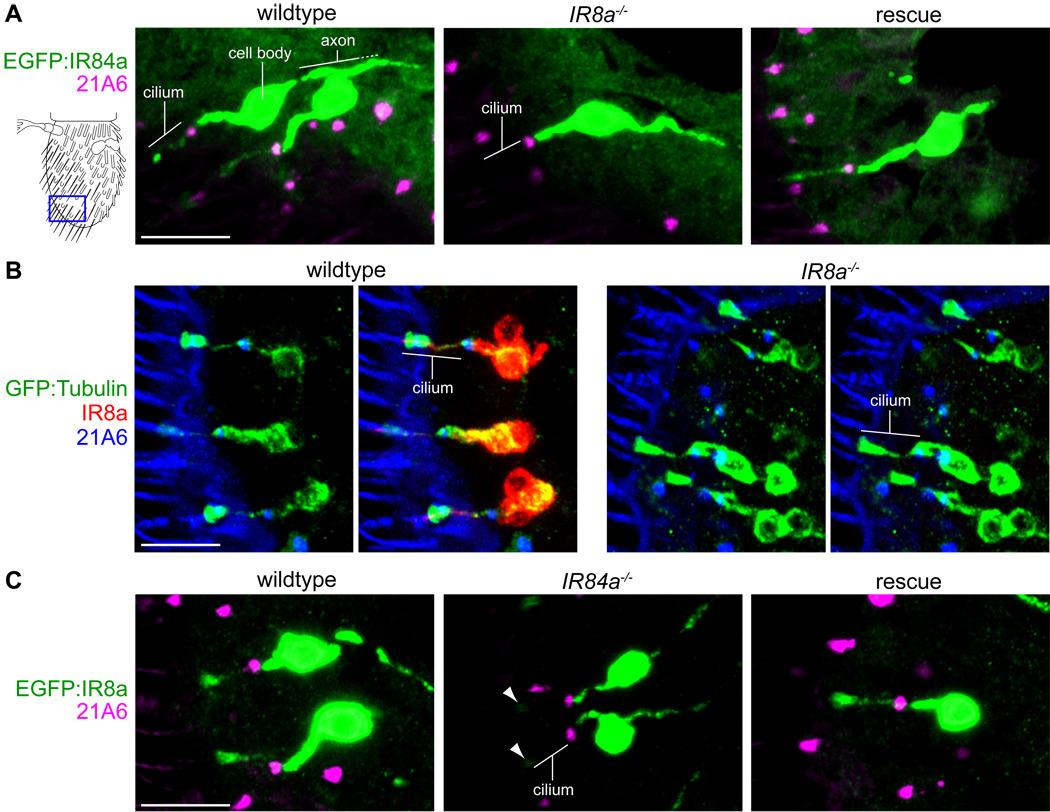
Figure 3. Reciprocal requirement of ligand-specific and co-receptors for sensory cilia localization.
(A) Immunostaining for EGFP:IR84a (anti-GFP, green) and the cilium base (21A6, magenta) in IR84a neurons in wildtype (left), IR8a mutant (middle) and IR8a rescue (right) antennae. The cartoon at the left schematizes the region of the antennae shown in these and subsequent panels. Antennal sections from at least 20 flies, from at least two independent genetic crosses, were examined for each genotype in this and all other immunofluorescence experiments. Cilia localization of EGFP:IR84a was never observed in IR8a mutant neurons. The scale bar represents 10 µm. Genotypes: IR84a-GAL4/UAS-EGFP:IR84a (left), IR8a1;IR84a-GAL4/UAS-EGFP:IR84a (middle), IR8a1;IR84a-GAL4,UAS-IR8a/UAS-EGFP:IR84a (right).
(B) Immunostaining for GFP:Tubulin (anti-GFP, green), IR8a (red) and the cilium base (21A6, blue) in IR8a neurons in wildtype (left images) and IR8a mutant (right images) antennae in anterior-distal coeloconic sensilla where IR84a neurons are located. The scale bar represents 10 µm. Genotypes: IR8a-GAL4/UAS-GFP:α1tub84B (left), IR8a1;IR8a-GAL4/UAS-GFP:α1tub84B (right).
(C) Immunostaining for EGFP:IR8a (anti-GFP, green) and the cilium base (21A6, magenta) in IR84a neurons in wildtype (left), IR84a mutant (middle) and IR84a rescue (right) antennae. Trace levels of EGFP:IR8a are occasionally detected in the cilia tips of IR84a mutant neurons (arrowheads). The scale bar represents 10 µm. Genotypes: UAS-EGFP:IR8a;IR84aGAL4/+ (left), UAS-EGFP:IR8a;IR84aGAL4/IR84aGAL4 (middle), UAS-EGFP:IR8a/UAS-IR84a;IR84aGAL4/IR84aGAL4 (right).
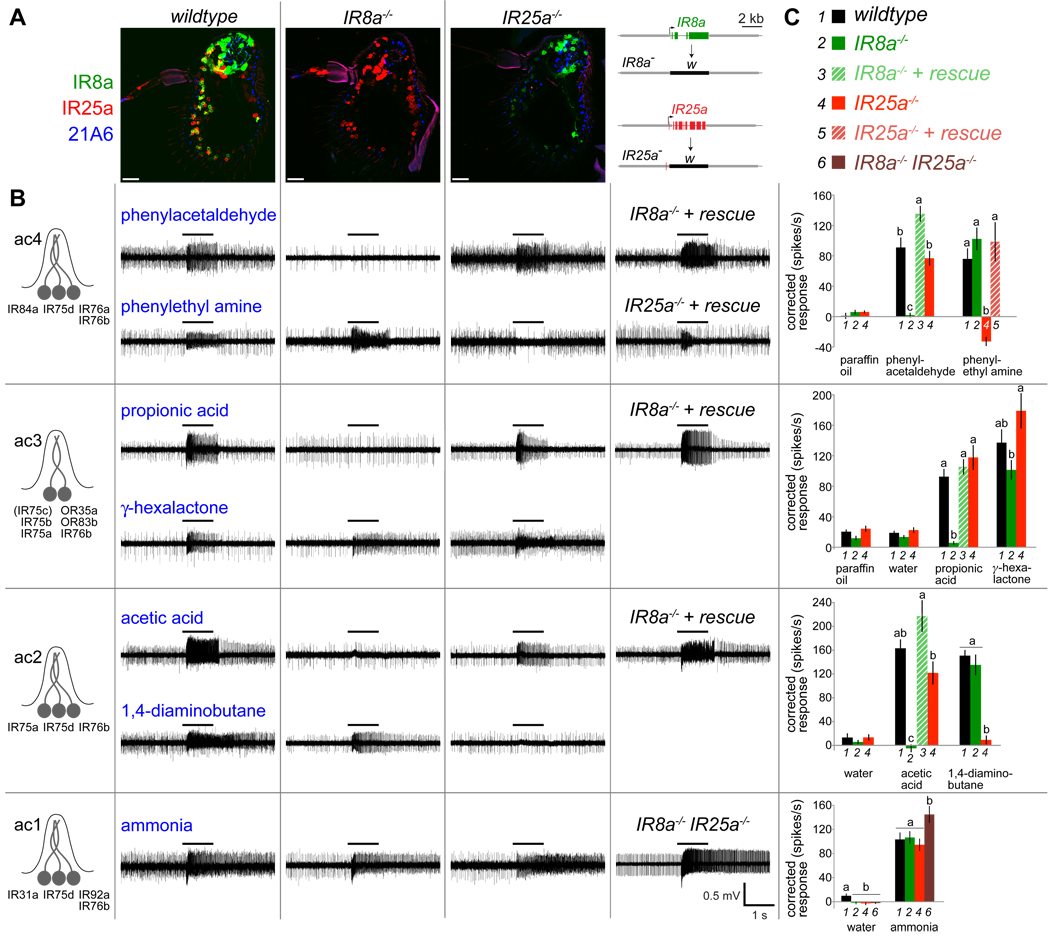
Figure 2. IR8a and IR25a are essential for odor-evoked electrophysiological responses in multiple distinct neuron classes.
(A) Immunostainings on antennal sections from wildtype (left), IR8a1 mutant (middle) and IR25a2 mutant (right) flies with IR8a (green), IR25a (red) and 21A6 (blue) antibodies. The scale bars represent 20 µm. Schematics of gene-targeted IR8a and IR25a null alleles, where the IR coding region is replaced with the white (w) reporter gene, are shown at the far right.
(B) Left: Schematic of IR expression (excluding IR8a and IR25a) in the four classes of coeloconic sensilla (ac1–ac4) (after (Benton et al., 2009)). Right: Representative traces of extracellular recordings of neuronal responses in the four coeloconic sensilla classes in wildtype (first column), IR8a1/Y hemizygous mutant (second column), IR25a2 mutant (third column), and rescue or IR8a1/Y; IR25a2 double mutant (fourth column) flies, stimulated with the indicated odors. Bars above the traces mark stimulus time (1 s). Genotypes for rescue experiments: “IR8a−/− + rescue”: IR8a1/Y;IR8a-GAL4/UAS-IR8a, “IR25a−/− + rescue”: IR25a2,IR25a-GAL4/IR25a2,UAS-IR25a.
(C) Quantification of mean neuronal responses to different odor stimuli in ac1–ac4 sensilla (± s.e.m; n=11–18 (ac4), n=8–16 (ac3), n=5–8 (ac2), n=10–13 (ac1); male flies, ≤3 sensilla/animal) in the genotypes shown in the key at the top and as detailed in (B). Paraffin oil and water are solvent controls. For individual stimuli in each sensilla, bars labeled with different letters are significantly different. ac4: phenylacetaldehyde ANOVA p<0.0001, phenylethyl amine ANOVA p<0.0001; ac3: propionic acid ANOVA p<0.0001, γ-hexalactone ANOVA p>0.011; ac2: acetic acid ANOVA p<0.0001; 1,4-diaminobutane ANOVA p<0.0001; ac1: water ANOVA p>0.0097, ammonia ANOVA p>0.022. Small, but statistically significant, variations in spike responses were observed to some odors in certain mutant backgrounds, for example ac3 γ-hexalactone responses. Although we cannot exclude a modulatory role of IR8a or IR25a in detection of these stimuli, we believe that these effects are likely to be indirect, either because of physiological changes in neighboring neurons in the same sensilla that rely absolutely on these receptors and/or technical difficulties in consistently comparing odor-evoked spike frequencies in sensilla lacking the activity of one or more neurons with those in wildtype sensilla.
IR8a and IR25a are essential for odor-evoked electrophysiological responses in multiple distinct neuron classes
The broad antennal expression and cilia localization of IR8a and IR25a indicated that these receptors might have a widespread role in odor detection in IR neurons. We tested this hypothesis by performing extracellular electrophysiological recordings of odor-evoked neuronal responses in individual coeloconic sensilla in IR8a and IR25a mutants. Four classes of coeloconic sensilla have been defined, named ac1–ac4 (Benton et al., 2009; Yao et al., 2005). These house neurons that express different combinations of IRs and display distinct odor sensitivities (Figure 2B) (Benton et al., 2009; Yao et al., 2005), although matching of specific ligands to receptors awaits in most cases. We tested a panel of seven odors, representing the best sensilla-specific agonists identified in previous or on-going ligand screens (Yao et al., 2005) (R. Rytz and R. B. unpublished), which we assume are recognized by different IRs.
The ac4 sensilla contain three neurons and are stimulated by phenylacetaldehyde and phenylethyl amine (Figure 2B). We observed that IR8a mutant ac4 sensilla lack all responsiveness to phenylacetaldehyde, while phenylethyl amine responses are unchanged (Figure 2B and 2C). Spontaneous activity in IR8a mutant ac4 sensilla (25 ± 4 spikes/sec (mean ± s.e.m); n=13) is also markedly reduced compared to wildtype (82 ± 1 spikes/sec; n=11). These electrophysiological phenotypes resemble those of mutants in IR84a (Y. Grosjean and R. B., unpublished), suggesting a role for IR8a in acting with this receptor in mediating both basal and evoked responses in IR84a neurons. By contrast, mutation of IR25a had no effect on phenylacetaldehyde responses but abolished phenylethyl amine-evoked activity (Figure 2B and 2C), suggesting an essential function in a different IR neuron. Spontaneous activity in IR25a mutant ac4 sensilla was also reduced (43 ± 6 spikes/sec; n=18).
The ac3 sensilla house two neurons, one of which expresses three IRs (IR75a, IR75b and IR75c) and responds to propionic acid (Benton et al., 2009; Yao et al., 2005). Responses to this ligand are abolished in IR8a but not IR25a mutants (Figure 2B). The second neuron is stimulated by many odors, but genetic analysis has ascribed all of these responses to the OR expressed in these cells, OR35a (Yao et al., 2005). OR35a-dependent responses to γ-hexalactone persisted in both IR8a and IR25a mutants (Figure 2B and 2C), indicating independent functioning of this receptor. The ac2 sensilla neurons respond strongly to acetic acid and 1,4-diaminobutane, and these responses are selectively abolished in IR8a and IR25a mutants, respectively (Figure 2B and 2C). Finally, ac1 sensilla contain three IR-expressing neurons, but only one strong agonist, ammonia, has been identified (Yao et al., 2005). Responses to this odor were retained in both IR8a and IR25a mutants, as well as in IR8a/IR25a double mutants (Figure 2B and 2C).
All defects in odor-evoked responses in IR8a and IR25a mutants were rescued by expression of corresponding cDNA transgenes using IR8a or IR25a promoters via the GAL4/UAS system (Figure 2B and 2C and Figure S1) (Brand and Perrimon, 1993). The sole exception was our failure to restore ac2 1,4-diaminobutane responses in IR25a mutants (data not shown). We ascribe this lack of rescue activity to the poor recapitulation of endogenous IR25a expression by our IR25a-GAL4 line (Figure S1B). Expression of IR25a in IR8a mutant neurons did not rescue electrophysiological responses (data not shown), indicating selective functional properties of these two receptors beyond their distinct expression patterns (Figure 1C). Taken together, the loss of multiple, distinct ligand-evoked responses in IR8a and IR25a mutants suggests that these proteins function as co-receptors that act with different subsets of odor-specific IRs.
Reciprocal requirement of odor-specific and co-receptor IRs for sensory cilia targeting
To determine the cellular basis for the loss of electrophysiological responses in these IR co-receptor mutant neurons, we initially focused on the role of IR8a in the correct functioning of the phenylacetaldehyde receptor IR84a (Benton et al., 2009). An EGFP-tagged version of IR84a localizes to the sensory cilium in its endogenous neurons (Figure 3A), defined by the distal distribution relative to the cilium base marker 21A6 (Husain et al., 2006; Zelhof et al., 2006). By contrast, in IR8a mutants, EGFP:IR84a is restricted to the inner dendritic segment (Figure 3A). Restoration of IR8a expression under the control of the IR84a promoter rescues this localization defect, defining a cell autonomous function for IR8a in promoting cilia targeting of IR84a (Figure 3A).
We tested the generality of this requirement for IR8a by examining the cilia localization of a second receptor, IR64a, which is co-expressed with IR8a in morphologically distinct grooved peg sensilla in the third chamber of the sacculus (Ai et al., 2010). EGFP:IR64a is abundant in the outer dendrite of these neurons in wildtype sensilla, and this localization is abolished in IR8a mutants (Figure S2A). We observed more heterogeneous levels of EGFP:IR64a in IR8a mutant neurons, suggesting that this mislocalized protein is destabilized. Cilia targeting is restored by re-expression of IR8a in this mutant background (Figure S2A).
To verify that loss of IRs in the outer dendrite in IR8a mutants was not simply due to a failure in formation of this sensory compartment, we expressed a GFP-tagged tubulin isoform (GFP:α1tub84B) in these neurons, which serves as a robust reporter of the outer segment in ciliated sensory neurons in Drosophila (Avidor-Reiss et al., 2004). In wildtype and IR8a mutant neurons, GFP:α1tub84B displayed a similar distribution distal to 21A6 in both coeloconic sensilla in the main body of the antenna and grooved peg sensilla in the sacculus (Figure 3B and Figure S2B). Thus, the outer ciliated segment forms correctly in IR8a mutants, supporting a specific role for this receptor in targeting odor-specific IRs to this cellular compartment.
We investigated whether there was a reciprocal requirement for IR84a for the localization of IR8a by expressing EGFP:IR8a in IR84a mutant neurons (Figure 3C). The normal cilia distribution of this fusion protein is severely impaired in the absence of IR84a (Figure 3C). Similarly, IR8a cilia localization is abolished in IR64a mutant sacculus neurons (Figure S2C). In both mutant backgrounds, IR8a localization is restored by expression of corresponding IR rescue transgenes (Figure 3C and Figure S2C). Thus, efficient cilia targeting of IR8a depends upon the presence of an odor-specific partner.
Functional reconstitution of an IR in OR-expressing sensory neurons
Having shown that phenylacetaldehyde responses in ac4 sensilla require two receptors, IR84a and IR8a (Figure 2B and 2C) (Y. Grosjean and R.B., unpublished), we asked whether these proteins are sufficient for reconstitution of a functional olfactory receptor in heterologous neurons. We previously showed that ectopic expression of IR84a in ac3 neurons is sufficient to confer responsiveness to phenylacetaldehyde (Benton et al., 2009). However, IR8a is also expressed endogenously in these cells (data not shown), raising the possibility that the odor-evoked responses are not due to IR84a alone, but depend on IR84a in combination with IR8a. To resolve this issue, we expressed IR84a in OR22a neurons, which innervate basiconic sensilla and do not express IR8a (Figure 1C).
When expressed alone in these neurons, EGFP:IR84a fails to localize to sensory cilia, where OR22a concentrates (Figure 4A) (Dobritsa et al., 2003). However, when EGFP:IR84a is co-expressed with IR8a, the fusion protein is efficiently transported to the ciliated sensory endings (Figure 4A). As in coeloconic sensilla, we observed a reciprocal requirement for IR84a in the cilia localization of IR8a in OR neurons: alone, EGFP:IR8a was absent from sensory cilia, but co-expression of IR84a was sufficient to promote its re-distribution to the sensory compartment (Figure 4A).
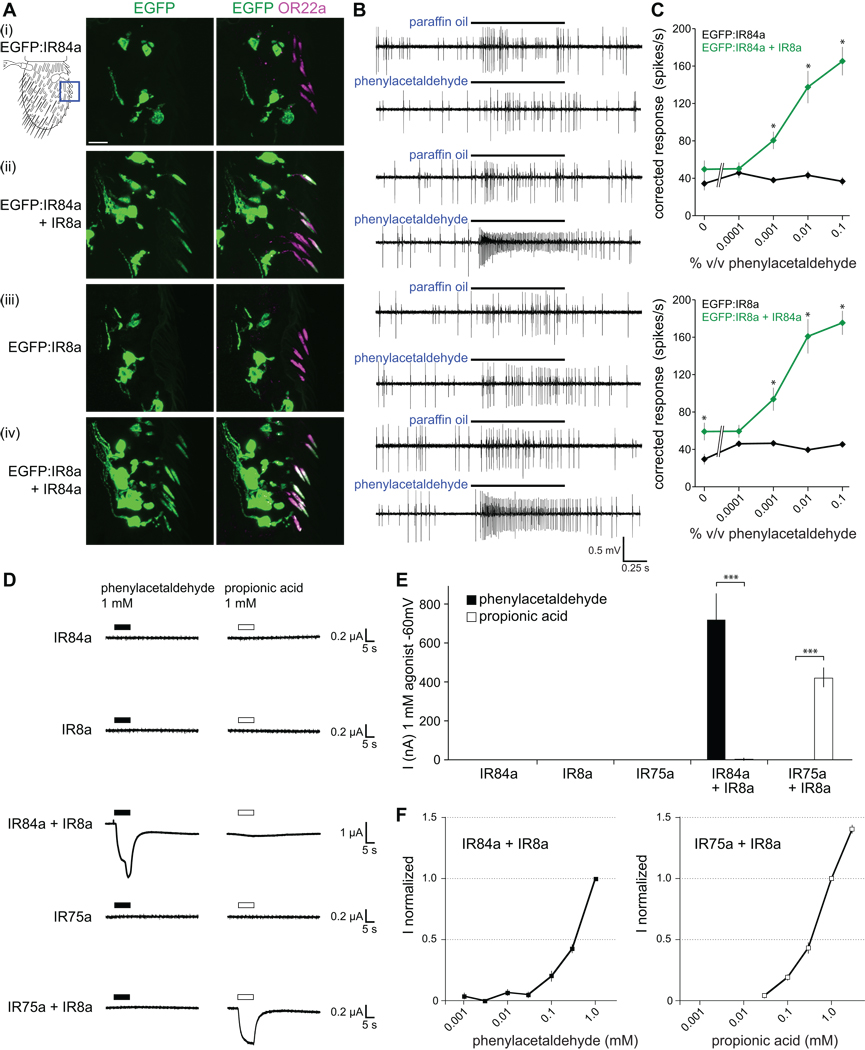
Figure 4. A pair of IRs is sufficient to reconstitute odor responses in OR neurons and Xenopus oocytes.
(A) Immunostaining for EGFP (green) and OR22a (magenta) in OR22a neurons expressing the combinations of IRs shown on the left. Genotypes: (i) UAS-EGFP:IR84a/+;OR22a-GAL4/+, (ii) UAS-EGFP:IR84a/UAS-IR8a;OR22a-GAL4/+, (iii) UAS-EGFP:IR8a/+;OR22a-GAL4/+, (iv) UAS-EGFP:IR8a/UAS-R84a;OR22a-GAL4/+. The cartoon at the top left schematizes the region of the antenna shown in all images. The scale bar represents 10 µm. Approximately 20% of OR22a neurons do not express IR fusion proteins due to incomplete expressivity of the OR22a-GAL4 driver (Dobritsa et al., 2003).
(B) Representative traces of extracellular recordings of neuronal responses in OR22a neurons expressing the combinations of IRs shown to the left in (A) stimulated with paraffin oil (solvent control) or phenylacetaldehyde (0.01% v/v). Bars above the traces mark stimulus time (1 s). OR22a neurons reside in basiconic sensilla with two neurons, visible as two distinct amplitudes of action potentials; the larger amplitude corresponds to OR22a neurons (Dobritsa et al., 2003).
(C) Concentration-response curves for phenylacetaldehyde in the genotypes shown in (A). Mean responses are plotted (± s.e.m; n=12–13 sensilla; ≤4 sensilla/animal, male flies). Asterisks indicate significant differences between responses in sensilla expressing one or both receptors (EGFP:IR84a versus (vs) EGFP:IR84a+IR8a ANOVA p<0.0003; EGFP:IR8a vs EGFP:IR8a+IR84a ANOVA p<0.014 for paraffin oil, p<0.0015 for phenylacetaldehyde-evoked responses).
(D) Representative whole cell current traces recorded at −60 mV with two-electrode voltage-clamp in Xenopus oocytes injected with cRNAs for the combinations of IRs indicated on the left. Currents of IR84a+IR8a-expressing cells often did not return completely to their baseline after phenylacetaldehyde removal, suggesting sustained activation of these receptors after initial agonist exposure or incomplete removal of phenylacetaldehyde from oocyte membranes.
(E) Histogram of current amplitudes of IR84a (n=3–4 oocytes), IR8a (n=3–4), IR75a (n=3–4), IR84a+IR8a (nphenylacetaldehyde=86; npropionic acid=4) and IR75a+IR8a (nphenylacetaldehyde=3; npropionic acid=74) induced by 1 mM phenylacetaldehyde and 1 mM propionic acid. Responses of IR84a+IR8a-expressing oocytes to phenylacetaldehyde and of IR75a+IR8a-expressing oocytes to propionic acid are highly significantly different from a non-cognate odor ligand (unpaired t test, *** p<0.0001).
(F) Concentration-response curves of oocytes expressing IR84a+IR8a for phenylacetaldehyde (left) and IR75a+IR8a for propionic acid (right) recorded at −60 mV and normalized to the current induced by 1 mM of ligand. Mean responses are plotted (± s.e.m; n=4–5 oocytes, 2–3 stimulations per oocyte).
We examined the functionality of these cilia-localized receptors by electrophysiological analysis of phenylacetaldehyde-evoked responses. OR22a neurons expressing EGFP:IR84a or EGFP:IR8a alone do not respond to this odor above basal, solvent-evoked activity. By contrast, when co-expressed with their corresponding partner, robust, concentration-dependent responses to phenylacetaldehyde are observed (Figure 4B and 4C). Thus, IR84a and IR8a are together both necessary and sufficient to reconstitute a cilia-localized and physiologically active olfactory receptor in Drosophila neurons.
Functional reconstitution of IRs in heterologous cells
We extended our investigation of the sufficiency of IR84a and IR8a to form a functional receptor by determining their ability to confer phenylacetaldehyde responsiveness in an ex vivo, non-neuronal system. We chose Xenopus laevis oocytes, which are commonly used for functional expression of iGluRs (Walker et al., 2006). In these cells, single or combinations of IR complementary RNAs (cRNAs) can be injected and odor-evoked current responses across the oocyte membrane measured by two-electrode voltage clamp.
When cRNAs for IR84a or IR8a were injected alone into oocytes, we observed no responses to phenylacetaldehyde (Figure 4D and 4E). By contrast, when IR84a and IR8a cRNAs were co-injected, phenylacetaldehyde induced an inward current of several hundred nA in these cells (Figure 4D and 4E). We further tested the functional properties of a different odor-specific receptor, IR75a, which is expressed in the IR8a-dependent, propionic acid-sensitive neuron in ac3 sensilla (Figure 2B and 2C). Oocytes expressing IR75a and IR8a together, but not either receptor alone, exhibited robust propionic acid-evoked current responses (Figure 4D and 4E). Odor-induced current responses were highly specific to each receptor pair and displayed concentration dependency (Figure 4D–4F). The concentration response curves for both phenylacetaldehyde and propionic acid did not saturate at the highest concentrations obtainable without changing the osmolarity of the solution, preventing our determination of 50% effective concentration (EC50) values. Baseline currents measured in the absence of either agonist were similar between IR84a+IR8a-expressing, IR75a+IR8a-expressing and uninjected oocytes (data not shown), suggesting that these receptors do not have detectable constitutive activity, at least in these cells.
Heteromeric complex formation between IRs
The co-dependency of IR84a and IR8a for cilia localization and odor-evoked responses suggested that these proteins might form a complex. We tested this possibility through optical imaging of fluorescent protein-tagged receptors. We first generated an mCherry-tagged IR8a fusion protein and confirmed that this promotes cilia targeting of EGFP:IR84a in OR22a neurons (Figure 5A). In these cells, we observed precise colocalization and consistent relative intensities of EGFP and mCherry fluorescence throughout the cell bodies, inner dendrites and cilia (Figure 5A). Odor-evoked responses conferred on these neurons by the fluorescent protein-tagged IRs were indistinguishable from those generated by untagged receptors (Figure 5B), indicating that the fluorescent tags do not interfere with their function.
Figure 5. Heteromeric IR complex formation.
(A) Colocalization of endogenous fluorescence of EGFP:IR84a (green) and mCherry:IR8a (false-colored magenta) in OR22a neurons. Genotype: UAS-EGFP:IR84a/+;UAS-mCherry:IR8a/OR22a-GAL4. Autofluorescence of sensilla cuticle is visible in the magenta (but not green) channel. The scale bar represents 10 µm.
(B) Concentration-response curves for phenylacetaldehyde in OR22a neurons expressing tagged (UAS-EGFP:IR84a/+;UAS-mCherry:IR8a/OR22a-GAL4, red symbols) or untagged (UAS-IR84a/+;UAS-IR8a/OR22a-GAL4, black symbols) combinations of IR84a and IR8a. Mean responses are plotted (± s.e.m; n=11–16 sensilla; ≤4 sensilla/animal, male flies). Responses are not statistically different at any stimulus concentration (ANOVA p>0.18).
(C) Single molecule fluorescence colocalization in membranes of live Xenopus oocytes visualized with total internal reflection fluorescence microscopy. Molecules of EGFP:IR84a and mCherry:IR8a (left) or EGFP:IR84a and mCherry:IR25a (right) were detected as bright fluorescent spots (top), and positions were extracted from intensity peaks (bottom) (see Experimental Procedures); colocalizing EGFP and mCherry spots are displayed as filled circles. The scale bars represent 1 µm.
(D) Quantification of colocalization of EGFP and mCherry signals in oocytes expressing the indicated combinations of tagged IRs. Each circle represents the percentage colocalizing fluorescent spots within a fresh, unbleached membrane patch, calcluated as: [number of spots with EGFP and mCherry fluorescence/(number of EGFP spots + number of mCherry spots)] × 100. Each membrane patch contains ~100–800 fluorescent spots and 7–18 patches were analyzed for each combination of receptors. Statistical differences between IR84a+IR8a and IR84a+IR25a colocalization frequency were determined by Student’s t test, *** p<0.0001.
(E–F) Example fluorescence intensity traces of colocalizing spots from cells expressing (E) mCherry:IR8a+EGFP:IR84a or (F) mCherry:IR84a+EGFP:IR8a. The magenta and green bars indicate the collection periods of fluorescence emitted (arbitrary units (a.u.)) originating from mCherry and EGFP, respectively. One (top traces) or two (bottom traces) EGFP tags in one spot result in the stepwise bleaching process (black arrows).
We next used these tagged IRs in a single molecule optical approach in Xenopus oocytes (Ulbrich and Isacoff, 2007, 2008). Here, low expression density of receptors permits simultaneous detection and spatial resolution of many individual fluorescent molecules. Total internal reflection fluorescence microscopy was used to restrict illumination to the plasma membrane, thereby excluding fluorescence from the intracellular space and focusing on receptors that have passed through the quality control process of cell surface targeting. In contrast to the mutual requirement for IR84a and IR8a in cilia membrane targeting in vivo (Figure 3 and Figure 4A), these receptors can localize independently to the oocyte plasma membrane, albeit less efficiently than when co-expressed (Figure S3A).
When EGFP:IR84a and mCherry:IR8a were expressed in oocytes at low concentrations, these fusion proteins appeared as bright fluorescent spots of relatively uniform fluorescence intensity in the plasma membrane (Figure 5C). A large fraction (~40%) of spots showed fluorescence from both EGFP and mCherry, consistent with assembly of EGFP:IR84a and mCherry:IR8a into a protein complex (Figure 5C and 5D and Figure S3B; see Experimental Procedures). By contrast, when EGFP:IR84a was co-expressed with mCherry:IR25a, with which it does not function in vivo (Figure 2B and 2C), fluorescence overlap was detected in <5% of the spots (Figure 5C and 5D). This value is consistent with the expected random colocalization (~4%; see Experimental Procedures) of mCherry and EGFP spots at the tested receptor density. Similar observations were made for both receptor pairs in which the fluorescent protein tags were exchanged (Figure 5D). These results indicate that IR84a forms a specific complex with IR8a.
To determine the number of IR8a and IR84a subunits within individual complexes, we analyzed the intensity traces from the EGFP-tagged partner in the spots where the mCherry and EGFP signal colocalized. EGFP photobleaches within a short time under high intensity illumination (as achieved in the single molecule observations), permitting deduction of the number of EGFP-tagged subunits by counting the bleaching steps (Ulbrich and Isacoff, 2007, 2008); mCherry photobleaches too rapidly to be analyzed in this way. Unfortunately, the intensities of most spots (>75%) were too noisy to be evaluated (Figure S3C), likely due to a high mobility of the proteins in the plasma membrane. However, in the fraction of spots where distinct bleaching steps were discernible, we observed either one or two bleaching steps, but never more (Figure 5E and 5F), suggesting a stoichiometry of up to 2 IR8a:2 IR84a subunits in these complexes.
To test this interpretation with an alternative analysis that included all spots regardless of the noise in their intensity traces, we integrated the fluorescence intensity from the start of EGFP illumination until complete photobleaching for all spots. These values were then normalized by the integrated intensities for the spots that we were confident contained only one EGFP molecule (i.e. had a single, clear bleaching step). This analysis revealed that the entire population of spots had an average of 1.57 EGFP molecules in EGFP:IR8a+mCherry:IR84a complexes and 1.49 EGFP molecules in EGFP:IR84a+mCherry:IR8a complexes (Figure S3D and S3E). Because only 80% of EGFP tags are fluorescent (Ulbrich and Isacoff, 2007), some complexes with two EGFP-tagged subunits would have only one visible EGFP. Therefore, from the normalized integrated EGFP intensities, we calculated that 83% of EGFP:IR8a+mCherry:IR84a complexes and 70% of EGFP:IR84a+mCherry:IR8a complexes had two EGFP-tagged subunits per spot (see Experimental Procedures). Together, these analyses are consistent with IR complexes containing up to two subunits each of IR8a and IR84a, similar to the dimer-of-dimers structure of many types of iGluR (Gereau and Swanson, 2008).
Ion conduction and pharmacological properties of IRs
The similarity in the domain organization of IRs and iGluRs led to the hypothesis that these olfactory receptors function as odor-gated ion channels (Benton et al., 2009). Our functional reconstitution of IRs in heterologous cells - in the absence of other Drosophila proteins - supports this model of signaling. We wished, however, to test the ionotropic activity of IRs.
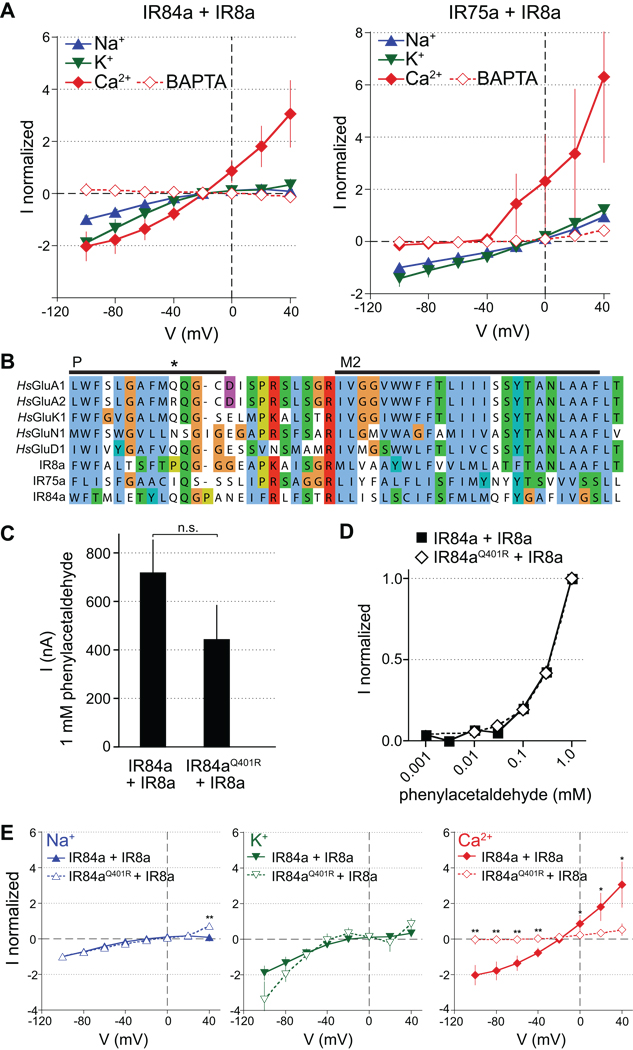
We first examined the ion conduction properties of phenylacetaldehyde- or propionic acid-evoked currents induced by IR84a+IR8a and IR75a+IR8a, respectively. This was performed in oocytes by measuring current/voltage (IV) relationships when the extracellular solution contained primarily Na+, K+ or Ca2+ as the cationic charge carrier (Figure 6A). For both receptor combinations, the reversal potential with Na+ or K+ solutions was slightly negative and not different between the two cations (IR84a+IR8a: −8 ± 7 mV (Na+) and −11 ± 5 mV (K+); IR75a+IR8a: −6 ± 2 mV (Na+) and −4 ± 1 mV (K+); n=8–3; paired t test, p>0.05). Current amplitudes were similar for K+ and Na+ for both IR84a+IR8a and IR75a+IR8a (Figure 6A), except when measured at −100 mV where they were higher for K+ than for Na+ for IR84a+IR8a (paired t test, p=0.03), but indistinguishable for IR75a+IR8a (paired t test, p=0.20) (Figure 6A). In the presence of extracellular Ca2+ and absence of extracellular Na+ and K+, activation of IR84a+IR8a, but not IR75a+IR8a, also induced inward currents (Figure 6A). These currents were, however, abolished by pre-injection of IR84a+IR8a-expressing oocytes with the Ca2+ chelator BAPTA, which prevents indirect activation of endogenous Ca2+-dependent chloride channels (Kuruma and Hartzell, 1999). Together, these experiments indicate that odor-evoked IR-dependent currents are carried principally by monovalent cations, but that IR84a+IR8a-dependent activation also leads to low Ca2+ entry, which is amplified by oocyte Ca2+-dependent channels to produce a measurable current.
Figure 6. Ion conduction properties of IRs.
(A) Current/voltage (I/V) relationships of oocytes expressing IR84a+IR8a stimulated with 1 mM phenylacetaldehyde (left) and IR75a+IR8a stimulated with 1 mM propionic acid (right) in extracellular Na+ (blue), K+ (green) or Ca2+ (red) solutions. Currents were measured as the amplitudes at the end of 1500 ms voltage steps from −100 to +40 mV at 20 mV increments. Currents recorded in the absence of agonist were subtracted from the current amplitudes recorded in the presence of 1 mM agonist. For each oocyte, currents were normalized to the current amplitude measured at −100 mV in the Na+ solution. Mean normalized currents are plotted (± s.e.m; n=9–35). Open symbols represent currents in oocytes (in Ca2+ solution) injected with BAPTA prior to the electrophysiological measurements to chelate the entering Ca2+.
(B) Sequence alignment of the pore selectivity filter (P) and M2 transmembrane domain for selected human (Hs) AMPA (GluA1, GluA2), Kainate (GluK1), NMDA (GluN1) and Delta (GluD1) subfamily iGluRs and Drosophila IRs. iGluR names follow new nomenclature conventions (Collingridge et al., 2009). The asterisk highlights the Q/R editing site that regulates calcium permeability in GluA2 (Liu and Zukin, 2007).
(C) Histogram of mean Na+ current amplitudes (± s.e.m) at −60 mV in oocytes expressing IR84a+IR8a (n=86) or IR84aQ401R+IR8a (n=7) induced by 1 mM phenylacetaldehyde. Responses are not statistically different (unpaired t test, p=0.16).
(D) Concentration-response curves for phenylacetaldehdyde of oocytes expressing either IR84a+IR8a or IR84aQ401R+IR8a recorded at −60 mV in extracellular Na+ solution. Currents were normalized to that measured by stimulation with 1 mM agonist. Responses are not statistically different at any stimulus concentration (unpaired t test, p=0.14).
(E) I/V relationships of IR84a+IR8a (solid symbols and lines; data reproduced from (A)) and IR84aQ401R+IR8a (open symbols and dotted lines) in extracellular Na+ (blue), K+ (green) or Ca2+ (red) bath solutions. Currents were measured and corrected as described as in (A). Mean normalized currents are plotted (± s.e.m; IR84a+IR8a n=35; IR84aQ401R+IR8a n=8). Statistically significant differences between responses of wildtype and mutant receptors are indicated by asterisks (unpaired Student’s t test, ** p<0.01, * p<0.05).
To further characterize these putative IR ion channels, we compared their pharmacological properties to those of iGluRs. NMDA receptors display a characteristic inhibition by extracellular Mg2+ (Nowak et al., 1984). Addition of 2 mM Mg2+ had, however, no effect on IR84a+IR8a or IR75a+IR8a currents measured when the primary charge carrier was Na+ (Figure S4A). We also tested several iGluR antagonists for their influence on IR-dependent currents, including two NMDA pore blockers, memantine and MK-801 (Kashiwagi et al., 2002), and an AMPA and Kainate receptor blocker, philanthotoxin (Jones et al., 1990; Ragsdale et al., 1989). None of these had effects on either IR84a+IR8a or IR75a+IR8a currents, except for memantine, which inhibited phenylacetaldehyde-induced IR84a+IR8a currents with a half maximal inhibitory concentration (IC50) of 39 ± 9 µM (Figure S4B), a value that is ~40 times the IC50 of memantine for NMDA receptors (Parsons et al., 2008). Antagonists for several other classes of ion channel, including amiloride, Cd2+, tetraethylammonium (TEA) and ruthenium red had mostly only modest effects on IR84a+IR8a or IR75a+IR8a currents, even at high concentrations (Figure S4B). Notably, while ruthenium red slightly inhibited IR84a+IR8a currents, it enhanced IR75a+IR8a current amplitudes (Figure S4B). Together, these experiments distinguish IRs pharmacologically from both iGluRs and other classes of ion channel, and further highlight the physiological differences between different IR complexes.
To understand the molecular basis for the functional heterogeneity of IR84a+IR8a and IR75a+IR8a, we compared the sequence of the putative ion channel pore domain of IR84a, IR75a and IR8a with those of iGluRs. While this region is highly conserved in iGluRs, individual IRs bear a large number of amino acid substitutions (Figure 6B). This sequence divergence may account for the observed insensitivity of IRs to iGluR pore blockers as well as the pharmacological differences between IR84a+IR8a and IR75a+IR8a (Figure 6A and Figure S4A–B). We focused on residues aligned with a glutamine that controls Ca2+ permeability in iGluRs (Dingledine et al., 1992) (Figure 6B). In GluA2, RNA editing-regulated substitution of this glutamine to arginine renders channels Ca2+-impermeable (Hume et al., 1991; Liu and Zukin, 2007). While IR75a contains an isoleucine (I388) in this position, IR84a retains a glutamine (Q401) (Figure 6B). We hypothesized that this residue might account for the difference in Ca2+ conductance mediated by IR75a+IR8a and IR84a+IR8a channels (Figure 6A). To test this, we generated an IR84aQ401R mutant receptor, which we predicted to lack Ca2+ permeability. IR84aQ401R+IR8a expressing oocytes showed similar Na+ current amplitudes (Figure 6C) and phenylacetaldehyde concentration responses as the wildtype receptors (Figure 6D). Importantly, IV curve measurements revealed that IR84aQ401R+IR8a-dependent conductance of monovalent cations was unchanged compared to the wildtype receptors, but that Ca2+-dependent conductance was abolished (Figure 6E). The selective alteration of ion permeability by a single mutation in the putative IR84a pore filter supports the notion that IRs function as ion channels.
IR8a contains a proline (P576) at the equivalent position in the pore sequence (Figure 6B). Expression of an IR8aP576R mutant, together with wildtype IR84a, markedly decreased phenylacetaldehyde-evoked currents (9.5% of that in oocytes expressing wildtype receptors at −60 mV), suggesting either a global effect on protein structure, plasma membrane expression and/or ion conductance. We were, however, able to establish IV curves for the remaining small IR84a+IR8aP576R–dependent current (Figure S4C). These revealed small but significant differences in the normalized conductance of monovalent cations between oocytes expressing wildtype and mutant channels, and abolishment of the Ca2+-dependent conductance in the mutant channel-expressing oocytes (Figure S4C). These observations suggest that IR8a also contributes to ion conduction and selectivity within a heteromeric IR complex.
IR ligand-binding domains function in both odor recognition and receptor localization
To define the domains contributing to the localization and odor-recognition properties of IRs in vivo, we generated a series of transgenic flies expressing mutant versions of EGFP:IR84a or EGFP:IR8a in combination with a wildtype partner in OR22a neurons (Figure 7 and Figures S5 and S6). We examined both the cilia targeting properties of these receptors and their ability to confer concentration-dependent responses to phenylacetaldehyde (Figure 7).
Figure 7. IR ligand-binding domains function in cilia localization and odor responses.
(A–K) Left: Immunostaining for EGFP (green) and OR22a (magenta) in OR22a neurons expressing an EGFP-tagged deletion or site-directed mutant IR in combination with an untagged wildtype IR8a or IR84a partner, as illustrated in the cartoons on the far left. Genotypes are of the form (A–D) UAS-EGFP:IR84aX/UAS-IR8a;OR22a-GAL4/+ or (E–K) UAS-EGFP:IR8aX/UAS-IR84a;OR22a-GAL4/+, where “X” denotes a wildtype or mutant version of the IR fusion protein. The arrowhead in (G) marks the very weak cilia localization of EGFP:IR8a ΔC+IR84a. The scale bar represents 10 µm for all panels. Right: Concentration-response curves for phenylacetaldehyde in OR22a neurons expressing the combination of receptors shown on the left. Mean responses are plotted (± s.e.m; n=12–29 sensilla; ≤4 sensilla/animal, male flies). The responses of corresponding wildtype receptor combinations (panels (A) and (E)) are shown as blue lines in each graph for comparison with mutant receptor responses. Responses were corrected for small, but slightly variable, baseline solvent responses to permit direct comparison of phenylacetaldehyde-evoked activity. Statistically significant differences between responses of wildtype and mutant receptors are indicated by asterisks (Student’s t test, *** p<0.001, ** p<0.01, * p<0.05).
IR84a, like most odor-specific IRs, lacks the large amino-terminal domain (ATD) present in iGluRs, IR8a and IR25a (Croset et al., 2010), and retains only a short, ~200 amino acid N-terminal region before the S1 region. Although this region does not bear obvious homology to known protein domains and is highly divergent in IRs, its deletion abolished the normal cilia targeting and phenylacetaldehyde responsiveness of the wildtype receptor (Figure 7A and 7B), suggesting it is important for folding, complex assembly and/or localization of this receptor. By contrast, deletion of the C-terminal cytoplasmic tail had no effect on either localization or function (Figure 7C).
Odor-specific IR LBDs are highly divergent in primary structure from both iGluRs and among each other (Benton et al., 2009). While this sequence variability is consistent with their predicted diverse ligand-binding properties, it complicates analysis of their putative role in IR odor recognition. However, IR84a has an arginine residue (R317) that aligns with the conserved arginine in iGluR LBDs that contacts the α-carboxyl group of glutamate or artificial agonists (Figure S5) (Armstrong et al., 1998). We substituted this residue in IR84a with alanine (IR84aR317A). Strikingly, this mutation had no effect on receptor targeting to cilia, but completely eliminated phenylacetaldehyde responses (Figure 7D). This observation supports a direct role for the IR LBD in odor recognition.
Deletion of the ATD in the IR8a co-receptor also abolished localization and function (Figure 7E and 7F), supporting a role for this domain in protein folding, IR complex assembly or cilia targeting. IR8a bears a much longer C-terminal tail than odor-specific IRs (Figures S5 and S6), and, in contrast to the dispensability of the IR84a C-terminus, deletion of this domain strongly reduced cilia targeting efficiency and phenylacetaldehyde responsiveness (Figure 7G).
We were particularly interested in defining the role of the IR8a LBD, given the apparent function of this protein as a co-receptor rather than in defining odor specificity. Intriguingly, the IR8a LBD is more similar in primary sequence to those of iGluRs than other IR LBDs and preserves the triad of three principal glutamate-binding residues: arginine (described above), threonine (which contacts the γ-carboxyl group of glutamate) and aspartate (which contacts the α-amino group of glutamate) (Benton et al., 2009; Mayer, 2006) (Figure S6). Mutation of the conserved arginine to alanine (IR8aR481A) had little observable effect on IR8a localization or function (Figure 7H), in contrast to the equivalent mutation in IR84a (Figure 7D). However, a more drastic charge reversal substitution with glutamate at this position, IR8aR481E, reduced the efficiency of cilia targeting and resulted in modest but significant reduction in phenylacetaldehyde responses (Figure 7I). Mutation of the threonine (IR8aT645A) had no effect on either localization or function (Figure 7J). This lack of phenotype is consistent with the fact that this residue is not conserved in IR8a orthologs in several species (Figure S6). By contrast, mutation of the conserved aspartate, IR8aD724A, completely abolished cilia localization and phenylacetaldehyde responses (Figure 7K). These observations reveal a role for the IR8a LBD in receptor localization.
A three subunit IR olfactory receptor
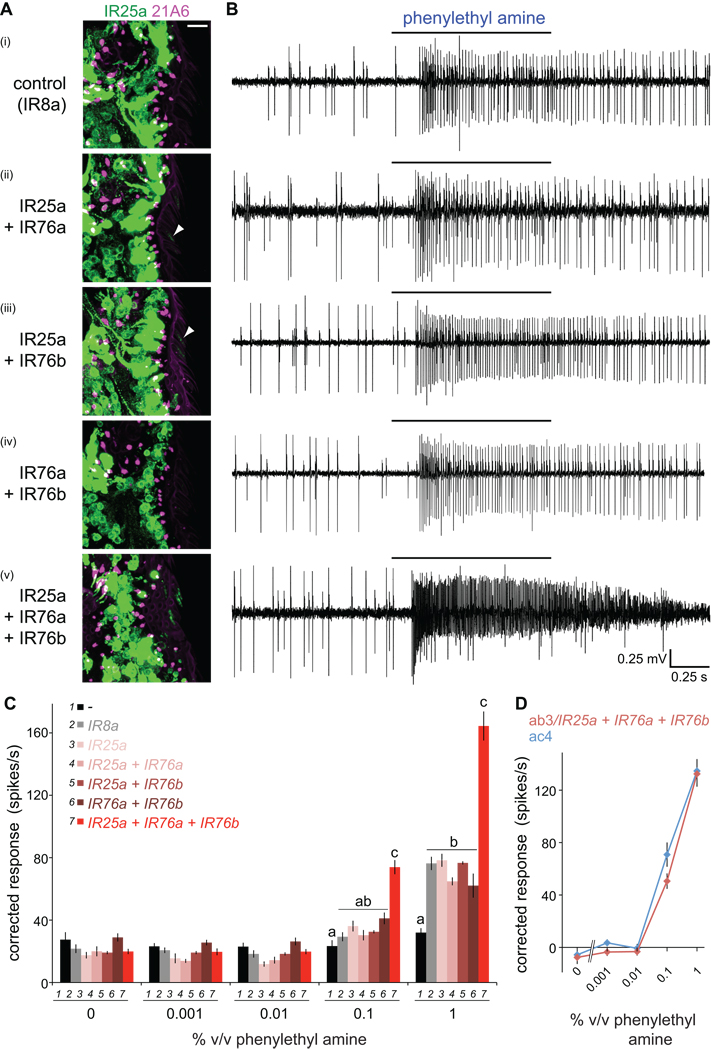
We asked whether the second IR co-receptor, IR25a, also functions together with a single odor-specific IR. As shown above (Figure 2B and 2C), IR25a is essential for ac4-specific electrophysiological responses to phenylethyl amine. Analysis of the IR expression map suggested that IR76a could be the odor-specific receptor for this stimulus, as this is the only IR whose expression is confined to ac4. We therefore attempted to reconstitute phenylethyl amine responses in OR22a neurons by misexpression of IR76a together with IR25a. We used the IR25a antibody to detect cilia localization of this putative receptor complex, but observed only very weak or no staining within the sensory compartment of these cells (Figure 8A). Electrophysiological analysis revealed only low basal responses to phenylethyl amine that were indistinguishable from control sensilla misexpressing IR8a (Figure 8B and 8C). IR76a is co-expressed with IR76b, a receptor that is also found in one neuron in each of the three other coeloconic sensilla classes (Benton et al., 2009), suggesting that this IR may also function as a co-receptor. We tested this possibility by expressing combinations of IR76a, IR76b and IR25a in OR22a neurons. All possible pairs of IRs resulted in either no or very weak localization of IR25a to cilia and basal phenylethyl amine responses (Figure 8A–8C). By contrast, upon co-expression of all three IRs, we observed consistent localization of IR25a to sensory cilia and robust, concentration-dependent odor-evoked responses (Figure 8A–8C). Moreover, the magnitude of these responses was highly comparable to phenylethyl amine-evoked activity in endogenous ac4 sensilla neurons (Figure 8D). These results reveal a thus-far unique case where three distinct subunits form a functional olfactory receptor.
Figure 8. An olfactory receptor of three IR subunits.
(A) Immunostaining for IR25a (green) and the cilia base (21A6, magenta) in OR22a neurons expressing the indicated combinations of IRs. Weak cilia localization of IR25a in neurons co-expressing IR25a+IR76a or IR25a+IR76b is indicated by arrowheads. Genotypes: (i) UAS-IR8a/+;OR22a-GAL4/+, (ii) UAS-IR76a/UAS-IR25a; OR22a-GAL4/+, (iii) UAS-IR25a/+;UAS-IR76b/OR22a-GAL4, (iv) UAS-IR76a/+;UAS-IR76b/OR22a-GAL4, (v) UAS-IR76a/UAS-IR25a;UAS-IR76b/OR22a-GAL4.
(B) Representative traces of extracellular recordings of neuronal responses in OR22a neurons expressing the combinations of IRs shown to the left in (A) stimulated with phenylethyl amine (1% v/v). Bars above the traces mark stimulus time (1 s). Misexpression of a control receptor, IR8a, confers weak responsiveness to phenylethyl amine, which may reflect non-specific sensitization of these neurons to this odor, as IR8a does not localize to sensory cilia in the absence of IR84a (Figure 3A).
(C) Concentration-responses for phenylethyl amine in OR22a neurons expressing the combinations of IRs shown in the key. Genotypes are as in (A) as well as a no IR control (“-“) (OR22a-GAL4/OR22a-GAL4) and IR25a misexpression alone (UAS-IR25a/+;OR22a-GAL4/+). Mean responses are plotted (± s.e.m; n=12–28 sensilla; ≤4 sensilla/animal, male flies). For responses to 0.1% and 1% phenylethyl amine, bars labeled with different letters are significantly different (ANOVA p<0.0001).
(D) Comparison of concentration-responses for phenylethyl amine in OR22a neurons ectopically expressing IR25a+IR76a+IR76b (red) to those in endogenous ac4 sensilla (blue). Responses in OR22a neurons were corrected for background endogenous neural responses (black bars in (C)). Responses to phenylethyl amine in ac4 were measured in an IR8a1 mutant background to facilitate quantification of odor-evoked spikes in the absence of IR84a neuron activity (Figure 2B).
Discussion
Conservation and divergence in the molecular architecture of olfactory IRs and synaptic iGluRs
“Chemosensory synapses” between the environment and sensory neurons have been proposed as novel models to characterize mechanisms of neuronal activation and regulation by external stimuli (Shaham, 2010). The IRs provide an intriguing example of molecular homology between peripheral sensory and post-synaptic receptors and motivated us to define the conserved and divergent properties of these olfactory receptors compared to their iGluR ancestors.
Cross-species analyses have demonstrated that IR25a is the “ancestral” IR, as orthologs of this gene are expressed in chemosensory neurons in insects, nematode worms and mollusks (Croset et al., 2010). By contrast, IR8a is a recently evolved, insect-specific duplicate of IR25a, although it retains a similar domain organization and sequence identity to iGluRs (Croset et al., 2010). The chemosensory role of IR25a in the common protostome ancestor is unknown, but it is attractive to suggest that it initially retained function as a glutamate-sensing receptor in the distal dendritic membranes of peripheral sensory neurons, analogous to the role of iGluRs in post-synaptic membranes of interneurons. Subsequent expansion of the IR repertoire may have allowed specialization of IR8a and IR25a as co-receptors acting in conjunction with more divergent odor-specific IRs. The dedication of these relatively slowly evolving members of the IR repertoire as a structural core of heteromeric IR complexes may help maintain the central function of these receptors as ligand-gated cation channels.
Our analysis of IR8a suggests that one specific function of the co-receptors may be to link IR complexes to the cilia transport pathway through their intracellular cytoplasmic tail, similar to the role of this region in coupling iGluRs to the post-synaptic transport machinery (Groc and Choquet, 2006). Conserved motifs for subcellular targeting are not apparent between iGluRs and IR8a or IR25a (data not shown), perhaps reflecting the novel signals required to localize IRs to specialized sensory cilia membranes. The maintenance of LBDs in co-receptor IRs raises the possibility that these proteins still bind ligands. Our mutational analysis of IR8a argues that glutamate is very unlikely to be recognized by its LBD and suggests that this domain serves in complex localization rather than peripheral ligand responses. Notably, LBD mutations in certain Kainate receptors also reduce cell surface expression in cultured cells (Valluru et al., 2005). While IR8a (and IR25a) may associate with unknown ligands important for trafficking, we favor instead a model in which the conformation of co-receptor LBDs contributes to a scaffold for correct assembly of an IR complex to ensure only functional heteromers reach sensory cilia.
In contrast to IR8a and IR25a, evolution of odor-specific IRs, such as IR84a and IR75a, was accompanied by significant reduction in structural complexity, as these proteins lack an ATD and bear only short, and apparently dispensable, cytosolic C-termini. Divergent LBDs and pore filters in these proteins appear to confer specificity of odor recognition and ion conduction properties of IR receptor complexes, respectively. Traces of ancestral glutamate-binding mechanisms are detectable, however, as we show that a glutamate-conjugating arginine is conserved and essential in IR84a for recognition of its odor ligand, phenylacetaldehyde. Odor-specific IR sequences may provide a valuable source of natural (and functional) “site-directed mutants” to understand how the ion conduction and other properties of these ligand-gated ion channels are specified at the molecular level.
Our reconstitution of olfactory responses using a combination of three distinct IRs (IR25a, IR76a and IR76b) highlights a further level of sophistication in how these proteins assemble into functional odor-sensing complexes. While IR76a is very likely to define ligand-specificity, the precise contributions of IR25a and a second putative co-receptor, IR76b, have not yet been resolved. It is possible that IR76b - which is more closely related to odor-specific IRs than to IR25a or IR8a - recognizes an unknown chemical ligand, whose co-presence with phenylethyl amine in an odor blend could lead to synergistic or diminished neuronal responsiveness. Further variations in IR complexes are apparent. For example, IR25a is likely to have IR76b-independent roles as a co-receptor for sacculus and aristal odor-specific IRs, as the latter receptor is not expressed in these structures (Benton et al., 2009). Moreover, the ammonia receptor in ac1 is independent of both IR8a and IR25a. Thus, while OR-expressing neurons in vertebrates and insects encode odor stimuli through the activity of singularly-expressed odor-specific receptors (Touhara and Vosshall, 2009), the IRs appear to function in “combinatorial codes” within individual OSNs. These may define unique ligand sensitivities and signaling dynamics, akin to the heteromer-specific properties of iGluRs in synaptic localization and signaling (Coussen, 2009; Greger et al., 2007; Kohr, 2006). In contrast to iGluRs (Walker et al., 2006), however, IRs do not appear to depend upon additional accessory proteins, such as TARPs (Tomita, 2010), for cell surface expression or function.
IRs as a novel model to understand olfactory receptor function and evolution
Olfactory receptor repertoires have long attracted the attention of molecular, structural and evolutionary biologists interested in the outstanding problems of odor recognition specificity and functional adaptability of these rapidly evolving proteins (Kaupp, 2010; Nei et al., 2008). While ever-expanding numbers of OR genes are being identified in genome sequences (Nei et al., 2008), progress in our understanding of the functional properties of the corresponding proteins has been relatively slow. Vertebrate ORs are notoriously difficult to express in experimentally-amenable heterologous systems (McClintock and Sammeta, 2003; Mombaerts, 2004), although recent identification in mammals of accessory factors that enhance their expression and/or function have begun facilitating matching of odors to receptors (Saito et al., 2009; Saito et al., 2004; Von Dannecker et al., 2006; Yoshikawa and Touhara, 2009). More challengingly, their seven transmembrane domain organization has eluded crystallization, obliging experimental probing of the odor-binding site to be guided by bioinformatic and modeling approaches (Katada et al., 2005; Schmiedeberg et al., 2007).
In insects, in vivo analyses of ORs have assigned ligands to a large fraction of this repertoire (Hallem and Carlson, 2006). Similar to IRs, odor-specific ORs function with a common co-receptor OR83b, which has an essential role in cilia targeting in vivo (Benton et al., 2006; Larsson et al., 2004; Neuhaus et al., 2005). Detailed understanding of insect ORs has, however, been hampered by the lack of homology of these polytopic membrane proteins to known receptors (Benton et al., 2006). Although initially assumed to be GPCRs (Hill et al., 2002), more recent analyses suggest these receptors function at least in part as odor-gated ion channels (Sato et al., 2008; Smart et al., 2008; Wicher et al., 2008).
In the face of these challenges, we propose that our comprehensive functional analysis of the IRs now establishes these proteins as an attractive new model olfactory receptor repertoire to determine how diverse molecular recognition and signaling properties have evolved and contribute to odor perception in vivo. The clear modular organization of the IRs offers the possibility to selectively manipulate the localization, ligand-recognition and signaling properties of these receptors. Perhaps most significantly, the amenability of the iGluR LBD to crystallographic analysis (Armstrong and Gouaux, 2000; Armstrong et al., 1998; Nanao et al., 2005), suggests that atomic-resolution visualization of odor/IR interactions will also be feasible, which would provide important insights into how olfactory receptors achieve their diverse ligand specificity.
IRs as genetically encoded chemical sensors
Finally, our definition of the molecular constituents of functional IR complexes in heterologous cells lays the foundation for use of these receptors as new types of genetically encoded chemical sensors. Although the LBDs of iGluRs are well-described and their potential as targets for directed modifications already demonstrated through generation of a light-activated iGluR (Szobota et al., 2007; Volgraf et al., 2006), this class of ion channel has been surprisingly underexploited as a tool to couple recognition of different types of chemicals with cellular physiological responses. The existence of many hundreds of divergent IRs of presumed distinct specificity reveals a natural exploitation of this ligand-gated ion channel for chemical sensing (Croset et al., 2010; Liu et al., 2010). The molecular properties of IRs uncovered here provide a basis for their rational modification to generate custom-designed chemoreceptors of desired specificity. Such sensors could offer invaluable tools as genetically encoded neuronal activators or inhibitors as well as have broad practical applications, for example, in environmental pollutant detection or clinical diagnosis.
Experimental Procedures
Drosophila genetics
Standard methods were used for Drosophila genetics, as described, together with a list of strains used, in the Supplemental Experimental Procedures.
Molecular biology
Standard methods were used in construction of all plasmids; details are provided in the Supplemental Experimental Procedures.
Histology
Standard methods were employed for immunofluorescence as described, together with all antibodies used, in the Supplemental Experimental Procedures.
Electrophysiology
Drosophila sensory neurons: extracellular recordings in single sensilla of 2–14 day old flies were performed and quantified essentially as described (Benton et al., 2009; Benton et al., 2007); details are provided, together with odor sources, in the Supplemental Experimental Procedures.
Xenopus oocytes: oocyte preparation and injection was carried out essentially as described (Vukicevic et al., 2006); details are provided in the Supplemental Experimental Procedures. Solutions containing agonists were applied once every minute for 10 s; between applications, the recording chamber was perfused with standard bath solution (110 mM NaCl, 2 mM BaCl2, 10 mM Hepes-NaOH, pH adjusted to 7.4 with NaOH) without agonist. For current/voltage (IV) curves in the presence of different ions, NaCl was replaced by 110 mM KCl or 40 mM CaCl2 and the osmolarity was adjusted with sucrose. The Na+ and K+ solutions contained 2 mM Ba2+ as divalent cation. Kaleidagraph (Synergy software) was used to fit the inhibition curves to the Hill equation: I = I0/[1+([inh]/IC50)nH], where I0 is the current in the absence of inhibitor (inh), IC50 is the inhibitor concentration that induces 50% inhibition and nH is the Hill coefficient. For IV curve measurements in high extracellular Ca2+, we injected 50 nl of 40 mM BAPTA 1–2 h prior to the electrophysiological measurements to test the contribution of the Ca2+ currents by endogenous Ca2+-dependent chloride currents. Phenylacetaldehyde and propionic acid were prepared as 1 M stock solutions in DMSO and diluted in bath solution to the desired final concentration. Philanthotoxin 433 tris(trifluoroacetate) (Sigma) was diluted to 1 mM in standard bath solution containing 0.3% BSA; (+)-MK-801 (Sigma) was diluted to 100 mM in water; memantine (Sigma) was diluted to 10 mM in water; amiloride (Sigma), TEA (Merck) and ruthenium red (Applichem) were directly added to bath solution. Statistical analyses are described in the figure legends.
Single molecule imaging
Imaging of individual IR protein complexes in Xenopus oocyte membranes by total internal reflection fluorescence microscopy was performed essentially as described (Sonnleitner et al., 2002; Ulbrich and Isacoff, 2007); details are provided in the Supplemental Experimental Procedures. Red (mCherry) and green (EGFP) spots were considered to be colocalized when their center positions were closer than 3 pixels (150 nm). However, most colocalization events reflect much shorter separation (Figure S3B). The expectation value for colocalization of red and green spots from random spot distributions was calculated by the formula: f = a*dg*dr/(dg+dr), where a=π*r2 is the area of the disk around a spot with r = 150 nm, dg and dr are the green and red spot densities, respectively, and f is the resulting fraction of overlapping spots. For spot densities as observed in the EGFP:IR84a+mCherry:IR25a co-expression experiment, the resulting expectation values for red/green colocalization was 1.4–6.9% (mean 3.6 ±0.7%).
To analyze the influence of co-expression of the partner subunit on plasma membrane expression density (Figure S3A), we injected a total volume of 50 nl per cell, with 0.1 µg/µl cRNA for the EGFP-tagged subunit and - where included - 0.25 µg/µl cRNA for the mCherry-tagged subunit. For each condition, we counted surface-localized spots of EGFP in two randomly selected 13 × 13 µm plasma membrane areas in each of eight different cells.
To deduce the fraction, f, of dimers from the average integrated intensity, x, we assumed that a fraction, p = 0.8, of the EGFP tags were fluorescent (Ulbrich and Isacoff, 2007). The relation between x, f and p is:
where the numerator is the total fluorescence from all complexes and the denominator the fraction that is fluorescent. Not all complexes are fluorescent because some monomers have a non-fluorescent EGFP and some dimers have two non-fluorescent EGFPs. Solving for f results in:
which yields f = 0.70 for x = 1.49 and f = 0.83 for x = 1.57.
Supplementary Material
Acknowledgements
We thank Yael Grosjean for sharing the IR84a mutant prior to publication, Raphael Rytz for generating the tree in Figure 1A and Michael Saina for analyzing IR8a expression in axon termini. We acknowledge Kazushige Touhara for use of pXpress, Roger Tsien for use of mCherry, the Bloomington Stock Center for Drosophila strains and the Developmental Studies Hybridoma Bank for monoclonal antibodies. We are grateful to Sophie Martin, Chun Tang and members of the Benton group for discussions and comments on the manuscript. Research in S.K.’s laboratory is supported by the Swiss National Science Foundation (31003A-117717). Research in E.Y.I.’s laboratory is supported by the NIH (R01NS035549) and NSF (FIBR 0623527). This work was funded by a European Research Council Starting Independent Researcher Grant to R.B.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ache BW, Young JM. Olfaction: diverse species, conserved principles. Neuron. 2005;48:417–430. doi: 10.1016/j.neuron.2005.10.022. [DOI] [PubMed] [Google Scholar]
- Ai M, Min S, Grosjean Y, Leblanc C, Bell R, Benton R, Suh GSB. Acid sensing by the Drosophila olfactory system. Nature. 2010 doi: 10.1038/nature09537. 10.1038/nature09537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong N, Gouaux E. Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron. 2000;28:165–181. doi: 10.1016/s0896-6273(00)00094-5. [DOI] [PubMed] [Google Scholar]
- Armstrong N, Sun Y, Chen GQ, Gouaux E. Structure of a glutamate-receptor ligand-binding core in complex with kainate. Nature. 1998;395:913–917. doi: 10.1038/27692. [DOI] [PubMed] [Google Scholar]
- Avidor-Reiss T, Maer AM, Koundakjian E, Polyanovsky A, Keil T, Subramaniam S, Zuker CS. Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell. 2004;117:527–539. doi: 10.1016/s0092-8674(04)00412-x. [DOI] [PubMed] [Google Scholar]
- Benton R, Sachse S, Michnick SW, Vosshall LB. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 2006;4:e20. doi: 10.1371/journal.pbio.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell. 2009;136:149–162. doi: 10.1016/j.cell.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R, Vannice KS, Vosshall LB. An essential role for a CD36-related receptor in pheromone detection in Drosophila. Nature. 2007;450:289–293. doi: 10.1038/nature06328. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Collingridge GL, Olsen RW, Peters J, Spedding M. A nomenclature for ligand-gated ion channels. Neuropharmacology. 2009;56:2–5. doi: 10.1016/j.neuropharm.2008.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussen F. Molecular determinants of kainate receptor trafficking. Neuroscience. 2009;158:25–35. doi: 10.1016/j.neuroscience.2007.12.052. [DOI] [PubMed] [Google Scholar]
- Croset V, Rytz R, Cummins SF, Budd A, Brawand D, Kaessmann H, Gibson TJ, Benton R. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. 2010;6:e1001064. doi: 10.1371/journal.pgen.1001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingledine R, Hume RI, Heinemann SF. Structural determinants of barium permeation and rectification in non-NMDA glutamate receptor channels. J Neurosci. 1992;12:4080–4087. doi: 10.1523/JNEUROSCI.12-10-04080.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobritsa AA, van der Goes van Naters W, Warr CG, Steinbrecht RA, Carlson JR. Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron. 2003;37:827–841. doi: 10.1016/s0896-6273(03)00094-1. [DOI] [PubMed] [Google Scholar]
- Foelix RF, Stocker RF, Steinbrecht RA. Fine structure of a sensory organ in the arista of Drosophila melanogaster and some other dipterans. Cell Tissue Res. 1989;258:277–287. doi: 10.1007/BF00239448. [DOI] [PubMed] [Google Scholar]
- Fuss SH, Ray A. Mechanisms of odorant receptor gene choice in Drosophila and vertebrates. Mol Cell Neurosci. 2009;41:101–112. doi: 10.1016/j.mcn.2009.02.014. [DOI] [PubMed] [Google Scholar]
- Gereau RW, Swanson GT. The Glutamate Receptors. Totowa, N.J: Humana Press; 2008. [Google Scholar]
- Greger IH, Ziff EB, Penn AC. Molecular determinants of AMPA receptor subunit assembly. Trends Neurosci. 2007;30:407–416. doi: 10.1016/j.tins.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Groc L, Choquet D. AMPA and NMDA glutamate receptor trafficking: multiple roads for reaching and leaving the synapse. Cell Tissue Res. 2006;326:423–438. doi: 10.1007/s00441-006-0254-9. [DOI] [PubMed] [Google Scholar]
- Hallem EA, Carlson JR. Coding of odors by a receptor repertoire. Cell. 2006;125:143–160. doi: 10.1016/j.cell.2006.01.050. [DOI] [PubMed] [Google Scholar]
- Hill CA, Fox AN, Pitts RJ, Kent LB, Tan PL, Chrystal MA, Cravchik A, Collins FH, Robertson HM, Zwiebel LJ. G protein-coupled receptors in Anopheles gambiae. Science. 2002;298:176–178. doi: 10.1126/science.1076196. [DOI] [PubMed] [Google Scholar]
- Hume RI, Dingledine R, Heinemann SF. Identification of a site in glutamate receptor subunits that controls calcium permeability. Science. 1991;253:1028–1031. doi: 10.1126/science.1653450. [DOI] [PubMed] [Google Scholar]
- Husain N, Pellikka M, Hong H, Klimentova T, Choe KM, Clandinin TR, Tepass U. The agrin/perlecan-related protein eyes shut is essential for epithelial lumen formation in the Drosophila retina. Dev Cell. 2006;11:483–493. doi: 10.1016/j.devcel.2006.08.012. [DOI] [PubMed] [Google Scholar]
- Jones MG, Anis NA, Lodge D. Philanthotoxin blocks quisqualate-, AMPA- and kainate-, but not NMDA-, induced excitation of rat brainstem neurones in vivo. Br J Pharmacol. 1990;101:968–970. doi: 10.1111/j.1476-5381.1990.tb14189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwagi K, Masuko T, Nguyen CD, Kuno T, Tanaka I, Igarashi K, Williams K. Channel blockers acting at N-methyl-D-aspartate receptors: differential effects of mutations in the vestibule and ion channel pore. Mol Pharmacol. 2002;61:533–545. doi: 10.1124/mol.61.3.533. [DOI] [PubMed] [Google Scholar]
- Katada S, Hirokawa T, Oka Y, Suwa M, Touhara K. Structural basis for a broad but selective ligand spectrum of a mouse olfactory receptor: mapping the odorant-binding site. J Neurosci. 2005;25:1806–1815. doi: 10.1523/JNEUROSCI.4723-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaupp UB. Olfactory signalling in vertebrates and insects: differences and commonalities. Nat Rev Neurosci. 2010;11:188–200. doi: 10.1038/nrn2789. [DOI] [PubMed] [Google Scholar]
- Kohr G. NMDA receptor function: subunit composition versus spatial distribution. Cell Tissue Res. 2006;326:439–446. doi: 10.1007/s00441-006-0273-6. [DOI] [PubMed] [Google Scholar]
- Kuruma A, Hartzell HC. Dynamics of calcium regulation of chloride currents in Xenopus oocytes. Am J Physiol. 1999;276:C161–C175. doi: 10.1152/ajpcell.1999.276.1.C161. [DOI] [PubMed] [Google Scholar]
- Larsson MC, Domingos AI, Jones WD, Chiappe ME, Amrein H, Vosshall LB. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron. 2004;43:703–714. doi: 10.1016/j.neuron.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Liu C, Pitts RJ, Bohbot JD, Jones PL, Wang G, Zwiebel LJ. Distinct olfactory signaling mechanisms in the malaria vector mosquito Anopheles gambiae. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SJ, Zukin RS. Ca2+-permeable AMPA receptors in synaptic plasticity and neuronal death. Trends Neurosci. 2007;30:126–134. doi: 10.1016/j.tins.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Mayer ML. Glutamate receptors at atomic resolution. Nature. 2006;440:456–462. doi: 10.1038/nature04709. [DOI] [PubMed] [Google Scholar]
- McClintock TS, Sammeta N. Trafficking prerogatives of olfactory receptors. Neuroreport. 2003;14:1547–1552. doi: 10.1097/00001756-200308260-00001. [DOI] [PubMed] [Google Scholar]
- Mombaerts P. Genes and ligands for odorant, vomeronasal and taste receptors. Nat Rev Neurosci. 2004;5:263–278. doi: 10.1038/nrn1365. [DOI] [PubMed] [Google Scholar]
- Nanao MH, Green T, Stern-Bach Y, Heinemann SF, Choe S. Structure of the kainate receptor subunit GluR6 agonist-binding domain complexed with domoic acid. Proc Natl Acad Sci U S A. 2005;102:1708–1713. doi: 10.1073/pnas.0409573102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Niimura Y, Nozawa M. The evolution of animal chemosensory receptor gene repertoires: roles of chance and necessity. Nat Rev Genet. 2008;9:951–963. doi: 10.1038/nrg2480. [DOI] [PubMed] [Google Scholar]
- Neuhaus EM, Gisselmann G, Zhang W, Dooley R, Stortkuhl K, Hatt H. Odorant receptor heterodimerization in the olfactory system of Drosophila melanogaster. Nat Neurosci. 2005;8:15–17. doi: 10.1038/nn1371. [DOI] [PubMed] [Google Scholar]
- Nowak L, Bregestovski P, Ascher P, Herbet A, Prochiantz A. Magnesium gates glutamate-activated channels in mouse central neurones. Nature. 1984;307:462–465. doi: 10.1038/307462a0. [DOI] [PubMed] [Google Scholar]
- Parsons CG, Gilling KE, Jatzke C. Memantine does not show intracellular block of the NMDA receptor channel. Eur J Pharmacol. 2008;587:99–103. doi: 10.1016/j.ejphar.2008.03.053. [DOI] [PubMed] [Google Scholar]
- Ragsdale D, Gant DB, Anis NA, Eldefrawi AT, Eldefrawi ME, Konno K, Miledi R. Inhibition of rat brain glutamate receptors by philanthotoxin. J Pharmacol Exp Ther. 1989;251:156–163. [PubMed] [Google Scholar]
- Saito H, Chi Q, Zhuang H, Matsunami H, Mainland JD. Odor coding by a mammalian receptor repertoire. Sci Signal. 2009;2:ra9. doi: 10.1126/scisignal.2000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H, Kubota M, Roberts RW, Chi Q, Matsunami H. RTP family members induce functional expression of mammalian odorant receptors. Cell. 2004;119:679–691. doi: 10.1016/j.cell.2004.11.021. [DOI] [PubMed] [Google Scholar]
- Sato K, Pellegrino M, Nakagawa T, Nakagawa T, Vosshall LB, Touhara K. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature. 2008;452:1002–1006. doi: 10.1038/nature06850. [DOI] [PubMed] [Google Scholar]
- Schmiedeberg K, Shirokova E, Weber HP, Schilling B, Meyerhof W, Krautwurst D. Structural determinants of odorant recognition by the human olfactory receptors OR1A1 and OR1A2. J Struct Biol. 2007;159:400–412. doi: 10.1016/j.jsb.2007.04.013. [DOI] [PubMed] [Google Scholar]
- Shaham S. Chemosensory organs as models of neuronal synapses. Nat Rev Neurosci. 2010;11:212–217. doi: 10.1038/nrn2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanbhag SR, Singh K, Singh RN. Fine structure and primary sensory projections of sensilla located in the sacculus of the antenna of Drosophila melanogaster. Cell Tissue Res. 1995;282:237–249. doi: 10.1007/BF00319115. [DOI] [PubMed] [Google Scholar]
- Smart R, Kiely A, Beale M, Vargas E, Carraher C, Kralicek AV, Christie DL, Chen C, Newcomb RD, Warr CG. Drosophila odorant receptors are novel seven transmembrane domain proteins that can signal independently of heterotrimeric G proteins. Insect Biochem Mol Biol. 2008;38:770–780. doi: 10.1016/j.ibmb.2008.05.002. [DOI] [PubMed] [Google Scholar]
- Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnleitner A, Mannuzzu LM, Terakawa S, Isacoff EY. Structural rearrangements in single ion channels detected optically in living cells. Proc Natl Acad Sci U S A. 2002;99:12759–12764. doi: 10.1073/pnas.192261499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spehr M, Munger SD. Olfactory receptors: G protein-coupled receptors and beyond. J Neurochem. 2009;109:1570–1583. doi: 10.1111/j.1471-4159.2009.06085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker RF. Drosophila as a focus in olfactory research: mapping of olfactory sensilla by fine structure, odor specificity, odorant receptor expression, and central connectivity. Microsc Res Tech. 2001;55:284–296. doi: 10.1002/jemt.1178. [DOI] [PubMed] [Google Scholar]
- Szobota S, Gorostiza P, Del Bene F, Wyart C, Fortin DL, Kolstad KD, Tulyathan O, Volgraf M, Numano R, Aaron HL, et al. Remote control of neuronal activity with a light-gated glutamate receptor. Neuron. 2007;54:535–545. doi: 10.1016/j.neuron.2007.05.010. [DOI] [PubMed] [Google Scholar]
- Tikhonov DB, Magazanik LG. Origin and molecular evolution of ionotropic glutamate receptors. Neurosci Behav Physiol. 2009;39:763–773. doi: 10.1007/s11055-009-9195-6. [DOI] [PubMed] [Google Scholar]
- Tomita S. Regulation of ionotropic glutamate receptors by their auxiliary subunits. Physiology (Bethesda) 2010;25:41–49. doi: 10.1152/physiol.00033.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touhara K, Vosshall LB. Sensing odorants and pheromones with chemosensory receptors. Annu Rev Physiol. 2009;71:307–332. doi: 10.1146/annurev.physiol.010908.163209. [DOI] [PubMed] [Google Scholar]
- Ulbrich MH, Isacoff EY. Subunit counting in membrane-bound proteins. Nat Methods. 2007;4:319–321. doi: 10.1038/NMETH1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbrich MH, Isacoff EY. Rules of engagement for NMDA receptor subunits. Proc Natl Acad Sci U S A. 2008;105:14163–14168. doi: 10.1073/pnas.0802075105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valluru L, Xu J, Zhu Y, Yan S, Contractor A, Swanson GT. Ligand binding is a critical requirement for plasma membrane expression of heteromeric kainate receptors. J Biol Chem. 2005;280:6085–6093. doi: 10.1074/jbc.M411549200. [DOI] [PubMed] [Google Scholar]
- Volgraf M, Gorostiza P, Numano R, Kramer RH, Isacoff EY, Trauner D. Allosteric control of an ionotropic glutamate receptor with an optical switch. Nat Chem Biol. 2006;2:47–52. doi: 10.1038/nchembio756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Dannecker LE, Mercadante AF, Malnic B. Ric-8B promotes functional expression of odorant receptors. Proc Natl Acad Sci U S A. 2006;103:9310–9314. doi: 10.1073/pnas.0600697103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vukicevic M, Weder G, Boillat A, Boesch A, Kellenberger S. Trypsin cleaves acid-sensing ion channel 1a in a domain that is critical for channel gating. J Biol Chem. 2006;281:714–722. doi: 10.1074/jbc.M510472200. [DOI] [PubMed] [Google Scholar]
- Walker CS, Brockie PJ, Madsen DM, Francis MM, Zheng Y, Koduri S, Mellem JE, Strutz-Seebohm N, Maricq AV. Reconstitution of invertebrate glutamate receptor function depends on stargazin-like proteins. Proc Natl Acad Sci U S A. 2006;103:10781–10786. doi: 10.1073/pnas.0604482103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicher D, Schafer R, Bauernfeind R, Stensmyr MC, Heller R, Heinemann SH, Hansson BS. Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature. 2008;452:1007–1011. doi: 10.1038/nature06861. [DOI] [PubMed] [Google Scholar]
- Yao CA, Ignell R, Carlson JR. Chemosensory coding by neurons in the coeloconic sensilla of the Drosophila antenna. J Neurosci. 2005;25:8359–8367. doi: 10.1523/JNEUROSCI.2432-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa K, Touhara K. Myr-Ric-8A enhances G(alpha15)-mediated Ca2+ response of vertebrate olfactory receptors. Chem Senses. 2009;34:15–23. doi: 10.1093/chemse/bjn047. [DOI] [PubMed] [Google Scholar]
- Zelhof AC, Hardy RW, Becker A, Zuker CS. Transforming the architecture of compound eyes. Nature. 2006;443:696–699. doi: 10.1038/nature05128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.