Urea has been widely used as a protein denaturant for more than a hundred years; however, its chemical denaturation mechanism still remains controversial, despite the extensive studies from both experimental and theoretical approaches in last several decades.1–19 The denaturing power of urea has been explained by two very different mechanisms: the “indirect” and “direct” mechanisms. The indirect mechanism suggests that urea denatures proteins by disrupting the water structure, which in turn weakens the hydrophobic interaction and makes the protein hydrophobic residues less compact and more readily solvated.4 The direct mechanism, on the other hand, indicates that urea unfolds proteins through direct interactions with protein, either through stronger electrostatic interactions10,11,13,16 with backbone and/or polar residues or through preferential van der Waals attractions14,15,18 with protein residues. Most of the concurrent studies support the direct mechanism involving urea's preferential binding to protein backbone or side chains,10,11,13–15,18 although some recent studies also indicate that the indirect mechanism can also play a role in the urea-induced protein denaturation.8,16,19
In a previous paper, Das and Mukhopadhyay, proposed a consensus view of the urea-induced protein denaturing mechanism.16 In particular, Das et al. found that urea preferentially binds to ubiquitin through its stronger electrostatic interactions with protein than water, and more surprisingly, they found that when a urea molecule moves from the bulk to the protein ubiquitin surface, it experiences more favorable electro-static energy with the surrounding environment by an astounding ~−13 kcal/mol. This contradicts our earlier findings on the protein lysozyme,14,21 where we found that urea denatures the protein through its stronger van der Waals attractions to protein than does water, with about a −2 kcal/mol change in vdW energy when moving from the bulk to the lysozyme surface (versus ~0 kcal/mol for water).14 Das et al. proposed that the difference in mechanism might arise because these are different proteins; i.e., the results might be protein specific. After a careful evaluation, we believe that the differences are not from the different proteins used (see below for more data) and cannot be fully explained with the different cutoffs and temperatures used in both studies.
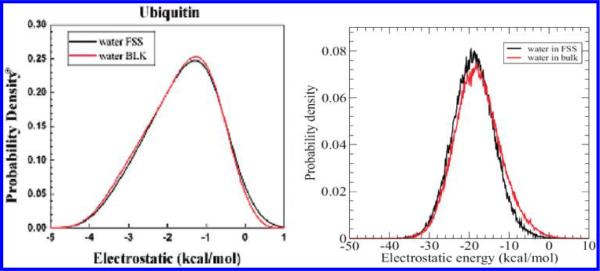
The disagreement between Das et al.'s results and ours can best be seen in the large difference in the water electrostatic interaction energy for both the bulk and protein first solvation shell (FSS). In Figure 1, we reproduce these energy distribution profiles for water. The electrostatic interaction energy for each water molecule with the rest of the system is about −20 kcal/mol in our calculations (with similar values for both the bulk and FSS), while in Das et al.'s paper it is only about −1 kcal/mol. This large a difference (~19 kcal/mol) in bulk water electrostatic interaction energies is clearly beyond normal statistical variance, it is also inconsistent with hydrogen bond strengths. One might argue that the FSS data could be somewhat different for different protein systems (ours is lysozyme, and Das et al.'s is ubiquitin), but the bulk data should be the same since we are using the same force field for water (the CHARMM version of the TIP3P water model20) under similar temperatures. Considering that the water configuration energy is about −10 kcal/mol in TIP3P (all other water models give roughly the same value for this configuration energy),20 and van der Waals energies do not contribute much to the configuration energy (roughly 0–1 kcal/mol in both calculations; see Table 1 below for more details), the pairwise water electrostatic interaction energy should be approximately −20 kcal/mol (the average pairwise interaction energy needs to be divided by 2 to get the average configuration energy, since it is shared by both parties). It is important to note that most of the differences between their and our simulations come from the contribution of the bulk data to the difference in energy of either water or urea in the first solvation shell of the protein from that in the bulk urea–water solution (ΔE = EFSS − EBulk).
Figure 1.
Comparison of the electrostatic energy profiles for water in bulk and FSS: (a) Das et al.16 data; (b) Hua et al.14,21 data. Graphs reproduced from ref 16 and refs 14 and 21 with permission.
Table 1.
Cutoff Effects on the Electrostatic Interaction Energies for Both Urea and Water in Bulk
| (a) Pure 8 M Urea | ||||
|---|---|---|---|---|
| urea |
water |
|||
| cutoff Å | electrostatic | Vdw | electrostatic | Vdw |
| 8 | −16.7 | −7.7 | −18.9 | 1.31 |
| 10 | −17.4 | −8.3 | −19.6 | 1.20 |
| 12 | −18.7 | −8.5 | −20.1 | 1.15 |
| 20 | −21.3 | −8.7 | −20.2 | 1.10 |
| 80 | −20.1 | −8.8 | −21.4 | 1.10 |
| (b) Lysozyme in 8 M Urea | ||||
|---|---|---|---|---|
| urea |
water |
|||
| cutoff Å | electrostatic | Vdw | electrostatic | Vdw |
| 8 | −13.11 | −6.37 | −15.39 | 0.98 |
| 10 | −15.77 | −6.82 | −17.66 | 0.90 |
| 12 | −16.36 | −6.99 | −18.22 | 0.81 |
| 20 | −20.75 | −7.19 | −18.63 | 0.76 |
| 80 | −20.70 | −7.22 | −20.44 | 0.77 |
We have considered that one possible reason for the discrepancy could be the different cutoffs used, since electrostatic interactions are long-range interactions. It seems Das et al. had been using a 9 Å cutoff for their pairwise electrostatic interaction energy analysis (personal communication; their simulation was run with PME16), while we used an 80 Å cutoff (essentially no cutoff).21 To see if this could explain the observed difference, we addressed this problem by performing four independent calculations, with three different codes, to calculate the electrostatic interaction energies at different cutoffs for both the pure 8 M urea solution and lysozyme in 8 M urea. Table 1 lists the interaction energies with cutoffs of 8, 10, 12, 20, and 80 Å (i.e., for essentially no cutoff so as to be consistent with the PME simulations used). As one can see, with different cutoffs, there can be a 2–3 kcal/mol difference in the electrostatic interactions for the bulk water, and a 3–4 kcal/mol difference for the bulk urea. But these are definitely not in the range of the ~19 kcal/mol difference between our result and Das et al.'s for bulk water! So there must be something else in play. The differences in the bulk urea electrostatic interaction energy in lysozyme in 8 M urea are somewhat larger with various cutoffs (up to 7 kcal/mol), which indicates that too small a cutoff might be troublesome. Once a cutoff of 20 Å or above is used for the electrostatics, however, the results are largely converged. The van der Waals interactions are less sensitive to the cutoffs, as one would expect (we used 80 Å in analysis as well, and Das et al used 7 Å). Ubiquitin in 8 M urea shows similar results (more below). Therefore, we think the different cutoffs used might have contributed to some of the differences, but not all.
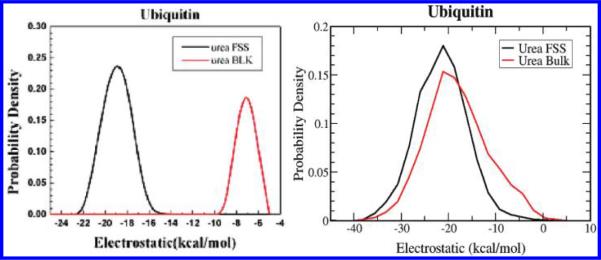
To see if these results are unique to the specific proteins, we repeated our calculations for five different protein systems in 8 M urea, lysozyme, ubiquitin, human γD-drystallin, CI2, and a tetramer of a small Aβ-peptide fragment (NFGAIL), and found similar interaction energy profiles for all of them (data not shown). A quick analysis of the charged residue distributions of each protein shows that they share similar charged residue distributions (see Table 2), suggesting that one should not expect large differences in electrostatic interaction energy profiles among these proteins. Thus presumed differences in charge distributions of different proteins probably cannot explain the discrepancies. Finally, we repeated molecular dynamics simulations on the Das et al. protein system ubiquitin, using two different solvation box sizes (see Methods section for more details): one with their system size and another with a much larger box. We used the various cutoffs, 8, 10, 12, 20, up to 80 Å, for the pairwise interaction energy analysis. Nevertheless, we still could not reproduce their results at any cutoff distance even with their smaller box. Figure 2 shows one representative energy profile of the urea electrostatic interactions for the smaller box with an 80 Å cutoff and found it to agree reasonably well with our previous lysozyme data (the agreement is even better for the larger solvation box). In the current smaller box, the electrostatic interactions are also slightly more favorable in FSS as compared to the bulk, but the van der Waals interactions still dominate the interaction energy change, being 2–3 kcal/mol more favorable than the contribution from the electrostatic interactions (ΔΔE = (EFSSvdw − EBulkvdw) − (EFSSelec − EBulkelec)). Again, various cutoffs can result in some variance (up to ~3 kcal/mol) in the difference between the FSS and bulk urea electrostatic interactions, but definitely not the ~13 kcal/mol difference computed by Das et al. (as shown in Figure 2a). We believe, on the basis of their water bulk data, as discussed above, that there seems to be some question on how the interaction between different parts of the system is computed and reported. The main differences between the two groups are again in the differences in the bulk data, where their data differed significantly from ours (see Figure 2). Finally, it should be noted that the solvation box used by Das et al. seems too small (5 times smaller than our lysozyme box); therefore even if one uses a very large cutoff for the interaction energy analysis, eventually no additional pairs would be included in the pair list because one member of the pair would have to overlap the protein surface.
Table 2.
Charged Residue Distributions in Different Proteinsa
| positive charge | negative charge | |||
|---|---|---|---|---|
| proteins (#AA) | number | percentage | number | percentage |
| lysozyme (129) | 17 | 13.1% | 9 | 7.0% |
| ubiquitin (76) | 11 | 14.5% | 11 | 14.5% |
| CI2 (64) | 10 | 15.6% | 8 | 12.5% |
| γD-crystallin (173) | 22 | 12.7% | 22 | 12.7% |
These proteins share a similar charged residue distributions, so one might not expect a large difference in their electrostatic interaction energies with urea or water.
Figure 2.
Comparison of the electrostatic energy profiles for urea in bulk and FSS for protein ubiquitin. (a) Das et al.16 data; (b) our current data with the same size box as that of Das et al. (the smaller box). Graph (a) reproduced from ref 16 with permission. It should be noted that there are still some slight differences between the electrostatic interaction energies we computed for urea in ubiquitin with this smaller solvation box and our previous lysozyme data. For the larger solvation box, the agreement of the ubiquitin data with our former lysozyme calculations is much better.
To summarize, we could not reproduce Das et al.'s results and we believe there might be some question on how the interaction between different parts of the system is computed and reported, on top of the too-small cutoffs used in the calculation of the pairwise interaction energy profiles. Meanwhile, our simulations support a direct mechanism in urea-induced lysozyme unfolding.21 These studies show that urea denaturizes protein though a two-stage kinetic process: first by forming a “dry molten globule” and then unfolding with both urea and water exposure. The dry globule has been observed directly by FRET and far-UV CD spectra recently.22 The dominant driving force is through the preferential binding of the protein to urea, with a stronger van der Waals dispersion interaction with urea than water.21 Interestingly, the preferential binding through stronger van der Waals interactions mechanism has now been accepted by researchers who were previously in the electrostatic interaction camp where the mechanism was believed to be based on urea interacting with the protein through stronger hydrogen bonds.15,21
Methods
The ubiquitin in 8 M urea solution is set up in a similar way to the previous hen lysozyme system, which was reported in detail elsewhere.21,23 The simulation was performed using the NAMD2 molecular dynamics program,24,25 with the all-atom CHARMM force field26 for proteins (lysozyme and ubiqutin) and solvent urea using PME. A slightly simplified TIP3P water model20 was used for water. Lysozyme was solvated in a 8Murea box of size 73.1 Å × 73.1 Å × 73.1 Å (129 residues, 7799 water molecules, 1811 urea molecules, and 8 Cl− counterions), and ubiquitin in two different sizes of 8 M urea boxes, one the same size as Das et al., 47.1 Å × 44.7 Å × 44.9 Å (76 residues, 1739 water molecules, and 438 urea molecules), and the other much larger, 68.5 Å × 69.4 Å × 68.1 Å (76 residues, 6722 water molecules, and 1575 urea molecules). Molecular dynamics simulations of 1000 ns for the lysozyme system and 100+ ns each for the two ubiquitin systems have been performed for data collection in this study.
ACKNOWLEDGMENT
We thank Steven Plotkin for bringing the discrepancy to our attention. We also thank Dave Thirumalai, Payel Das, and Peng Xiu for helpful discussions. R.Z. and B.J.B. acknowledge the support from the IBM BlueGene Science Program. B.J.B. also acknowledges support from NIH (GM44330).
REFERENCES
- (1).Nozaki Y, Tanford C. J. Biol. Chem. 1963;238:4074–4081. [PubMed] [Google Scholar]
- (2).Wetlaufer DB, Malik SK, Stoller L, Coffin RL. J. Am. Chem. Soc. 1964;86:508–514. [Google Scholar]
- (3).Robinson DR, Jencks WP. J. Am. Chem. Soc. 1965;87:2462–2470. doi: 10.1021/ja01089a028. [DOI] [PubMed] [Google Scholar]
- (4).Frank HS, Franks F. J. Chem. Phys. 1968;48:4746–4757. [Google Scholar]
- (5).Finer EG, Franks F, Tait MJ. J. Am. Chem. Soc. 1972;94:4424–4429. [Google Scholar]
- (6).Wallqvist A, Covell DG, Thirumalai D. J. Am. Chem. Soc. 1998;120:427–428. [Google Scholar]
- (7).Caflisch A, Karplus M. Structure Fold. Des. 1999;7:477–488. doi: 10.1016/s0969-2126(99)80064-1. [DOI] [PubMed] [Google Scholar]
- (8).Bennion BJ, Daggett V. Proc. Natl. Acad. Sci. U.S.A. 2003;100:5142–5147. doi: 10.1073/pnas.0930122100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Mountain RD, Thirumalai DJ. Am. Chem. Soc. 2003;125:1950–1957. doi: 10.1021/ja020496f. [DOI] [PubMed] [Google Scholar]
- (10).Tobi D, Elber R, Thirumalai D. Biopolymers. 2003;68:359–369. doi: 10.1002/bip.10290. [DOI] [PubMed] [Google Scholar]
- (11).O'Brien EP, Dima RI, Brooks B, Thirumalai D. J. Am. Chem. Soc. 2007;129:7346–7353. doi: 10.1021/ja069232+. [DOI] [PubMed] [Google Scholar]
- (12).Stumpe MC, Grubm€uller H. J. Phys. Chem. B. 2007;111:6220–6228. doi: 10.1021/jp066474n. [DOI] [PubMed] [Google Scholar]
- (13).Stumpe MC, Grubm€uller H. J. Am. Chem. Soc. 2007;129:16126–16131. doi: 10.1021/ja076216j. [DOI] [PubMed] [Google Scholar]
- (14).Hua L, Zhou R, Thirumalai D, Berne BJ. Proc. Natl. Acad. Sci. U.S.A. 2008;105:16928–16933. doi: 10.1073/pnas.0808427105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Stumpe MC, Grubm€uller H. PLoS Comput. Biol. 2008;4:e1000221. doi: 10.1371/journal.pcbi.1000221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Das A, Mukhopadhyay C. J. Phys. Chem. B. 2009;113:12816–12824. doi: 10.1021/jp906350s. [DOI] [PubMed] [Google Scholar]
- (17).Stumpe MC, Grubm€uller H. Biophys. J. 2009;96:3744–3752. doi: 10.1016/j.bpj.2009.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Das P, Zhou R. J. Phys. Chem. B. 2010;114:5427–5430. doi: 10.1021/jp911444q. [DOI] [PubMed] [Google Scholar]
- (19).Yang L, Gao YQ. J. Am. Chem. Soc. 2010;132:842–848. doi: 10.1021/ja9091825. [DOI] [PubMed] [Google Scholar]
- (20).Neria E, Fischer S, Karplus M. J. Chem. Phys. 1996;105:1902–1921. [Google Scholar]
- (21).Hua L, Zhou RH, Thirumalai D, Berne BJ. Proc. Natl. Acad. Sci. U.S.A. 2008;105:16928–16933. doi: 10.1073/pnas.0808427105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Jha SK, Udgaonkar JB. Proc. Natl. Acad. Sci. U.S.A. 2009;106:12289–12294. doi: 10.1073/pnas.0905744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Zhou RH, Eleftheriou M, Royyuru AK, Berne BJ. Proc. Natl. Acad. Sci. U.S.A. 2007;104:5824–5829. doi: 10.1073/pnas.0701249104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Kumar S, Huang C, Zheng G, Bohm E, Bhatele A, Phillips JC, Yu H, Kale LV. IBM J. Res. Dev. 2008;52:177–188. [Google Scholar]
- (26).MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]