Summary
Mre11 and Rad50 are the catalytic components of a highly conserved DNA repair complex that functions in many aspects of DNA metabolism involving double-strand breaks. The ATPase domains in Rad50 are related to the ABC transporter family of ATPases, previously shown to share structural similarities with adenylate kinases. Here we demonstrate that Mre11/Rad50 complexes from three organisms catalyze the reversible adenylate kinase reaction in vitro. Mutation of the conserved signature motif reduces the adenylate kinase activity of Rad50 but does not reduce ATP hydrolysis. This mutant resembles a rad50 null strain with respect to meiosis and telomere maintenance in S. cerevisiae, correlating adenylate kinase activity with in vivo functions. An adenylate kinase inhibitor blocks Mre11/Rad50-dependent DNA tethering in vitro and in cell-free extracts, indicating that adenylate kinase activity by Mre11/Rad50 promotes DNA-DNA associations. We propose a model for Rad50 that incorporates both ATPase and adenylate kinase reactions as critical activities that regulate Rad50 functions.
Introduction
The Mre11/Rad50 (M/R) complex plays an important though still largely undefined role in DNA double-strand break (DSB) repair. Mre11 is an exo-/endonuclease that associates in a tight complex with Rad50, an ATPase with a large coiled-coil domain similar in overall structure to the Structural Maintenance of Chromosomes family of proteins. In eukaryotes, M/R also associates with a nonenzymatic component known as Nbs1 (nibrin) in mammals and in fission yeast and as Xrs2 in budding yeast. Studies in S. cerevisiae clearly show that the Mre11/Rad50/Xrs2 (scM/R/X) complex is required for the repair of DNA DSBs through homologous recombination pathways as well as through nonhomologous end joining (Krogh and Symington, 2004). In vertebrates, each component of the Mre11/Rad50/Nbs1(Xrs2) complex (M/R/N[X]) is essential, and loss of any single component leads to chromosome instability and cell death (Stracker et al., 2004). The M/R/N(X) complex plays an important role in telomere maintenance and is essential for the processing of Spo11-generated DNA breaks during meiotic recombination. In addition, the complex acts as a DSB sensor to initiate DNA damage signaling pathways in eukaryotic cells through the ATM/Tel1 family of serine/threonine protein kinases that are essential for damage-induced checkpoint signaling (Lavin, 2004).
The catalytic motifs in Mre11 and Rad50 are evolutionarily conserved from archaea to humans, indicating the functional importance of their enzymatic activities. In budding yeast, the nuclease activity of Mre11 is required for scM/R/X function during meiosis but is largely dispensable for the other activities of the complex (Moreau et al., 1999; Lee et al., 2002; Zhang and Paull, 2005). In contrast, the ATPase motifs in Rad50 appear to be essential for all known activities of scM/R/X and human Mre11/Rad50/Nbs1 (hsM/R/N), including ATM activation by DNA DSBs in vitro (Alani et al., 1990; Lee and Paull, 2005; Zhang and Paull, 2005).
M/R complexes from several species exhibit DNA binding activity that is stimulated by ATP or nonhydrolyzable ATP analogs (Raymond and Kleckner, 1993; Paull and Gellert, 1999; Hopfner et al., 2000; Lee et al., 2003; Moncalian et al., 2004), although the complexes also show significant ATP-independent DNA binding activity (V.B., T.T.P., unpublished data). ATP-dependent DNA binding is consistent with the model based on the crystal structure of the pfRad50 catalytic domain (Hopfner et al., 2000). This work showed that ATP induces dimerization of the Rad50 catalytic domain and that the surface created by this dimerization interface forms a cleft with the size and electrostatic potential appropriate for DNA binding. We have also shown that ATP (and dATP) promotes unwinding of DNA by hsM/R/N in vitro such that 15–30 base pairs at the DNA end become at least transiently unwound (Paull and Gellert, 1999), and that this activity is important for stimulating ATM activity through hsM/R/N (Lee and Paull, 2005).
In vivo, the scM/R/X complex is one of the first protein complexes to localize at the site of a DNA DSB (Lisby et al., 2004; Shroff et al., 2004) and has been shown to play a major role in holding chromosomal fragments together to prevent missegregation and loss of genomic DNA after DNA breakage (Kaye et al., 2004; Lobachev et al., 2004). Consistent with this biological function, scM/R/X and hsM/R complexes have been shown to bind to and bridge together DNA ends in vitro (Chen et al., 2001; de Jager et al., 2001; Trujillo et al., 2003; Chen et al., 2005). Rad50 catalytic activity has been implicated in this process, but the exact role of the ATPase domain or ATPase activity in end bridging is not yet clear. M/R/N-dependent DNA tethering has also been demonstrated in Xenopus cell-free egg extracts (Costanzo et al., 2004), and the S1202R mutation in the Rad50 catalytic domain abrogates this activity (Dupre et al., 2006), consistent with a catalytic role for Rad50 in DNA tethering.
Rad50 contains Walker A and Walker B motifs at either end of the protein that associate intramolecularly and are responsible for ATP binding and hydrolysis. Rad50 also has a conserved loop in its C-terminal catalytic domain known as the signature motif, which is specific to the ABC transporter family of ATPases. The vast majority of the proteins in this family are integral membrane proteins responsible for the gating of ions or transport of small molecules across a membrane (Holland and Blight, 1999). The role of ATPase activity in this gating process has been controversial, particularly in the case of the Cystic Fibrosis Transmembrane Regulator (CFTR) protein, a well-known member of the ABC transporter family. ATP hydrolysis has not been strongly correlated with ion channel gating by CFTR, although recent evidence suggests that ATP binding rather than ATP hydrolysis may control transport through its effects on the dimerization of the nucleotide binding domains (Higgins and Linton, 2004).
Randak and Welsh have proposed an alternative model for CFTR function by showing that, in addition to its ATPase activity, CFTR catalyzes the reversible adenylate kinase reaction: ATP + AMP ↔ ADP + ADP (Randak and Welsh, 2003). This activity has now been demonstrated for both nucleotide binding domains in CFTR (Randak et al., 1997; Randak and Welsh, 2003; Gross et al., 2005). The 2003 study further showed that CFTR channel activity could be controlled by both ATPase and adenylate kinase reactions and argued that the adenylate kinase activity drives the channel gating cycle under physiological conditions.
Considering the structural similarities between the nucleotide binding domains of CFTR and Rad50, we investigated the ability of M/R complexes to perform the adenylate kinase reaction. Here we show that purified M/R complexes from P. furiosus, S. cerevisiae, and H. sapiens all exhibit adenylate kinase activity. Mutation of the signature motif in the yeast and human Rad50 proteins generates complexes that are specifically impaired in adenylate kinase activity but are proficient in ATPase activity. These mutant complexes show no activity in vivo, suggesting that, like CFTR, the adenylate kinase activity of Rad50 is integral to its biological functions. Tethering of DNA molecules in vitro is blocked by a specific inhibitor of adenylate kinase activity, suggesting a role for this activity in maintaining chromosomal associations in cells. We propose models for Rad50 that incorporate both ATPase and adenylate kinase activities and establish a framework to understand the mechanism of Rad50 catalytic functions.
Results
Mre11/Rad50 Complexes Exhibit Adenylate Kinase Activity
Recombinant purified M/R complexes from Homo sapiens (hsM/R), Saccharomyces cerevisiae (scM/R), and Pyrococcus furiosus (pfM/R) (see Figure S1 in the Supplemental Data available with this article online) were tested for adenylate kinase activity: the reversible transfer of the terminal phosphate from ATP to AMP to form two molecules of ADP (Figure 1A). Varying amounts of each M/R complex were incubated with [γ-32P]ATP, either in the absence or the presence of unlabeled AMP, and the reaction products were analyzed by using thin-layer chromatography. All of the M/R complexes hydrolyzed ATP, as shown by the release of [32P]inorganic phosphate (Figures 1B–1D, lanes 2–4), when incubated with ATP only. In the presence of both ATP and AMP, all of the M/R complexes also catalyzed the adenylate kinase reaction, which was evident from the generation of 32P-labeled ADP (Figures 1B–1D, lanes 6–8). Other ATPases unrelated to the ABC transporter family such as RecA and Rad54 do not exhibit adenylate kinase activity (Figure 1E), indicating that the adenylate kinase activity is specific to Rad50 molecules and related ATPases.
Figure 1. Human, Yeast, and Archaeal M/R Complexes Exhibit Adenylate Kinase Activity.

(A) Diagram of adenylate kinase reaction.
(B) hsM/R complexes incubated with [γ-32P]ATP ± AMP as indicated and the reactions separated by TLC. Positions of the [γ-32P]ATP substrate and the [β-32P]ADP and [32Pi] products are shown.
(C) pfM/R complexes assayed as in (B).
(D) scM/R complexes assayed as in (B).
(E) RecA (150 nM), BSA (150 nM), and hsRad54 (100 nM) incubated with [γ-32P]ATP and AMP and characterized as in (B).
Most nucleoside monophosphate kinases require a specific base in the acceptor monophosphate binding site but are not specific to a particular base in the donor triphosphate binding site (Yan and Tsai, 1999). To analyze the requirements for the donor binding site, we used [α-32P]AMP and unlabeled ATP with scM/R and found that the enzyme generated labeled ADP as expected from the adenylate kinase reaction (Figure 2B). We also observed labeled ATP in these reactions (Figure 2B, lanes 3 and 4), indicating that the reverse adenylate kinase reaction occurred (see Figure 2A).
Figure 2. M/R Adenylate Kinase Activity Is Adenine Specific and Reversible.

(A) Diagram of the adenylate kinase reaction with [α-32P]AMP.
(B) scM/R complexes incubated with [α-32P]AMP and ATP and the reactions separated by TLC. Positions of the [α-32P]AMP substrate and the [α-32P]ADP and [α-32P]ATP products are shown.
(C) scM/R complexes assayed as in (B) except that various nucleotides were used in place of ATP as indicated.
(D) scM/R complexes assayed with [γ-32P]ATP and various nucleotides as indicated and characterized as in (B).
(E) scM/R complexes assayed as in (D) with various metal cations (10 mM) as indicated; the amounts of [32Pi] product are shown.
(F) scM/R complexes assayed as in (E); the amounts of [β-32P]ADP product are shown.
Using [α-32P]AMP, we also tested all of the nucleoside and deoxynucleoside triphosphates and found that only ATP and dATP could function as phosphate donors in this reaction (Figure 2C), indicating that the base on the donor nucleotide must be an adenine. Moreover, in the presence of [γ-32P]ATP, we observed that only AMP could function as phosphate acceptor and that the deoxyribonucleoside monophosphate dAMP was not functional in this assay (Figure 2D). The adenine nucleotide specificity has also been observed for human, yeast, and E. coli M/R complexes in DNA binding, nuclease activity, and DNA unwinding (Raymond and Kleckner, 1993; Connelly et al., 1997; Paull and Gellert, 1999; Lee et al., 2003), suggesting that the same active sites are likely responsible for the adenylate kinase reaction.
Kinetic measurements of the ATPase and adenylate kinase activities of hsM/R and scM/R indicate that both enzymes show relatively low affinity for ATP–230 μM for scM/R and 320 μM for hsM/R–and slightly higher affinity for AMP–130 μM for scM/R and 44 μM for hsM/R (Figure S2). We also determined the cation requirements for the adenylate kinase activity of scM/R, which showed that both adenylate kinase and ATPase activities are maximal in the presence of magnesium but that the reaction is also supported by manganese or calcium ions (Figures 2E and 2F).
Ap5A Inhibits the Rad50 Phosphoryl Transfer Reaction but Not ATP Hydrolysis
Adenylate kinases are specifically inhibited by bifunctional molecules containing two adenine nucleotides connected through 5′ linkages to varying numbers of phosphate groups (Lienhard and Secemski, 1973). These molecules mimic the transition state of the phosphoryl transfer reaction between ATP and AMP and are thus very specific to adenylate kinase enzymes. We used four such compounds: Ap3A, Ap4A, Ap5A, and Ap6A, which contain three, four, five, or six phosphates, respectively, to test the sensitivity of M/R adenylate kinase activity. Our results show that none of the inhibitors affected the ATPase activity of scM/R, irrespective of the concentration or the number of phosphates linking the adenine nucleotides (Figures 3A and 3C). In contrast, all the inhibitors with the exception of Ap3A decreased the adenylate kinase activity of scM/R, with Ap5A and Ap6A showing the greatest inhibition (Figures 3B and 3D). We also tested the effects of Ap5A on pfM/R and hsM/R complexes and observed that the inhibitor completely abolished adenylate kinase activity but did not affect ATPase activity (Figures 3E and 3F). With these assays as well as the quantitative data shown in other figures, assays were performed at least three times, and representative data are shown. Collectively, these results show that the adenylate kinase reaction performed by these M/R complexes is structurally similar to the reaction performed by canonical adenylate kinases and that the distance between the two nucleotide binding sites must be ∼16 Å–the length of five phosphate groups.
Figure 3. Adenylate Kinase Inhibitors Block the Adenylate Kinase Activity of M/R Complexes.

(A) scM/R complexes incubated with [γ-32P]ATP and AMP plus Ap4A, Ap5A, or Ap6A as indicated and ATPase activity measured by the release of [32Pi].
(B) scM/R complexes assayed as in (A) and adenylate kinase activity measured by the production of [β-32P]ADP.
(C) scM/R complexes assayed as in (A) plus Ap3A or Ap5A as indicated.
(D) scM/R complexes assayed as in (B) plus Ap3A or Ap5A as indicated.
(E) pfM/R and hsM/R assayed as in (A) with 5 μM Ap5A as indicated.
(F) pfM/R and hsM/R assayed as in (B) with 5 μM Ap5A as indicated.
Mutation of the Rad50 Signature Motif Specifically Disrupts Adenylate Kinase Activity
The signature motif in ABC transporter ATPases is conserved among all of the members of this large family of enzymes and is thought to play an important role in catalysis by binding to the γ phosphate on ATP and thus stabilizing the ATP-bound form of the enzyme (Hopfner et al., 2002). A mutant pfM/R complex containing a serine to arginine mutation in the signature motif (S793R) was found to be deficient in ATP-dependent dimer formation and DNA binding (Hopfner et al., 2002) and also exhibits very low affinity for ATP (Moncalian et al., 2004). This mutation in pfM/R was modeled after an analogous mutation in CFTR (S549R) that results in cystic fibrosis (Kerem et al., 1989). In our assays, this mutant shows lower levels of both ATPase and adenylate kinase activity relative to the wild-type enzyme (Figure 4A), consistent with an overall deficiency in ATP binding.
Figure 4. Mutation of the Rad50 Signature Motif Reduces Adenylate Kinase Activity by hsM/R and scM/R.

(A) pfM/R wild-type and mutant (Rad50 S793R) complexes incubated with [γ-32P]ATP and AMP and the reactions separated by TLC as in Figure 1B.
(B) hsM/R wild-type and mutant (Rad50 S1202R) complexes assayed as in (A).
(C) scM/R wild-type and mutant (Rad50 S1205R) complexes assayed as in (A).
(D) Time course of ATPase activity by hsM/R complexes (300 nM) in the absence of AMP.
(E) Time course of ATPase activity by hsM/R complexes as in (D) but in the presence of AMP.
(F) Time course of adenylate kinase activity by hsM/R complexes in the presence of AMP.
(G–I) The equivalent reactions as in (D)–(F), respectively, except performed with scM/R (2.5 μM).
In contrast to the pfMR mutant, the equivalent mutations in hsM/R and scM/R do not inhibit ATPase activity (Figure 4B, lanes 5–7; Figure 4C, lanes 5–7; Figures 4D, 4E, 4G, and 4H). In fact, the levels of phosphate release from [γ-32P]ATP with hsM/R(S1202R) are 2- to 3-fold higher than with wild-type hsM/R. However, the levels of adenylate kinase activity exhibited by the hsM/R(S1202R) and scM/R(S1205R) mutants are 2- to 10-fold lower than the wild-type complexes (Figures 4B, 4C, 4F, and 4I); thus, these signature motif mutants are specifically impaired in adenylate kinase activity. Inclusion of AMP in the reactions with [γ-32P]ATP lowers the level of ATP hydrolysis (Figure 4D versus Figure 4E, Figure 4G versus Figure 4H), suggesting that AMP may utilize the same binding site as ATP.
Rad50 Signature Motif Mutants Do Not Function In Vivo
We have previously shown that the S1205R mutation in scRad50 is equivalent to a Rad50 deletion in vivo in yeast with respect to MMS and bleomycin resistance (Moncalian et al., 2004). In addition, the mutant fails to complement a deletion strain in assays of nonhomologous end joining in vivo (Zhang and Paull, 2005). Here we also looked at the effects of the S1205R mutation in meiosis and found that the mutant does not support spore viability (Figure 5A). A deletion/S1205R heterozygote did not yield any viable spores, while a deletion/wild-type heterozygote showed spore viability similar to a wild-type homozygote. Interestingly, the S1205R mutant also acts as a dominant negative in meiosis, as evidenced by the complete loss of spore viability in the wild-type/S1205R heterozygote. In contrast, we have not observed any dominant-negative effects of the mutant allele during vegetative growth when we measure MMS resistance (data not shown). We have confirmed that the S1205R Rad50 mutant protein is expressed in yeast and that it forms a complete complex with Mre11 and Xrs2 when coexpressed in insect cells; thus, the mutation does not block Xrs2 association with M/R (data not shown).
Figure 5. Mutation of the Rad50 Signature Motif Inhibits Meiosis and Telomere Maintenance.
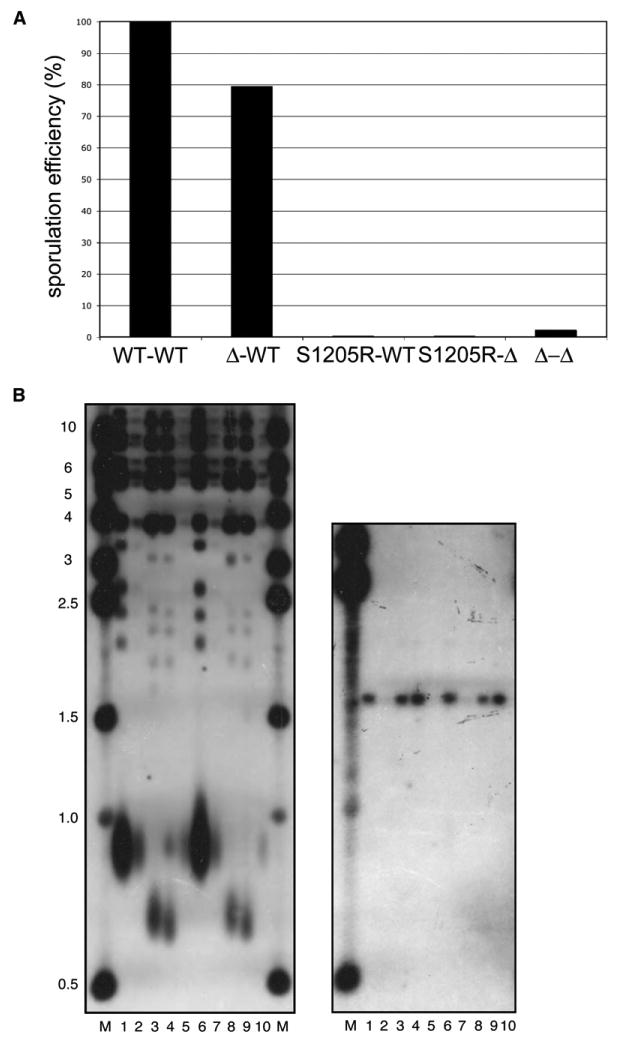
(A) Diploid S. cerevisiae strains with combinations of Rad50 wild-type (WT), rad50 deletion (Δ), or rad50-S1205R (S1205R) alleles were induced to sporulate. The efficiency of sporulation was measured by random spore analysis; values shown are relative to the wild-type diploid.
(B) Telomere length was measured in haploid strains expressing wild-type, rad50, or rad50-S1205R alleles (left panel). Genomic DNA from the wild-type strain was loaded undiluted (lane 1), diluted 1:5 (lane 2), or diluted 1:10 (lane 5). Genomic DNA from the rad50 and rad50-S1205R strains are loaded in lanes 3 and 4, respectively. Lanes 6–10 contain the same pattern of samples from a second group of isolates. The blot was probed with a telomere-specific probe. The same blot (right panel) is shown after stripping and reprobing with CEN4 to confirm equivalent gel loading.
scM/R/X is important for the maintenance of telomere length by telomerase in budding yeast (Moore and Haber, 1996; Boulton and Jackson, 1998; Diede and Gottschling, 2001; Tsukamoto et al., 2001). Genetic analysis has placed the complex in the same epistasis group as telomerase, suggesting that the complex may act on telomere ends to prepare them as a substrate for telomerase or may recruit telomerase to its substrate. Here we analyzed several colonies of the strain expressing Rad50 S1205R and found that the telomeres were as severely shortened as in a rad50 deletion strain (Figure 5B and see also Smith et al. [2005]). The S1205R mutation in Rad50 thus completely abrogates the functions of scM/R/X at telomeres in vivo.
Rad50 Adenylate Kinase Activity Is Required for DNA Tethering
scM/R/X and hsM/R complexes have been shown by atomic force microscopy (AFM) to bridge DNA molecules in vitro (Chen et al., 2001, 2005; de Jager et al., 2001; Trujillo et al., 2003), and Xenopus M/R/N complexes catalyze an analogous DNA tethering reaction in Xenopus egg extracts (Costanzo et al., 2004). Furthermore, scM/R/X has been implicated as an important component of the cellular machinery that prevents broken chromosomal fragments from separating in vivo (Kaye et al., 2004; Lobachev et al., 2004).
To determine the importance of adenylate kinase activity for DNA tethering, we employed the Xenopus egg extract system, which contains functionally active M/R/N complexes. Biotinylated DNA molecules were added to the extracts in the presence of radiolabeled DNA, and the level of DNA tethering was measured by the association of the labeled DNA with streptavidin-coated magnetic beads. A 20-fold increase in DNA-DNA association was observed over background levels, the majority of which has previously been shown to be dependent on M/R/N (Costanzo et al., 2004). The level of DNA tethering was reduced 2-fold and 5-fold by the addition of 0.5 or 1.0 mM Ap5A, respectively, indicating a role for Rad50 adenylate kinase activity in the DNA tethering process (Figure 6A). In contrast, the Ap3A compound, which is structurally similar to Ap5A but does not inhibit Rad50 adenylate kinase activity, did not inhibit DNA tethering in extracts. Note that the high levels of inhibitor used in this assay are necessitated by the high levels of endogenous ATP (estimated 1–4 mM) in Xenopus extracts.
Figure 6. Adenylate Kinase Activity Is Required for M/R-Mediated DNA Tethering.
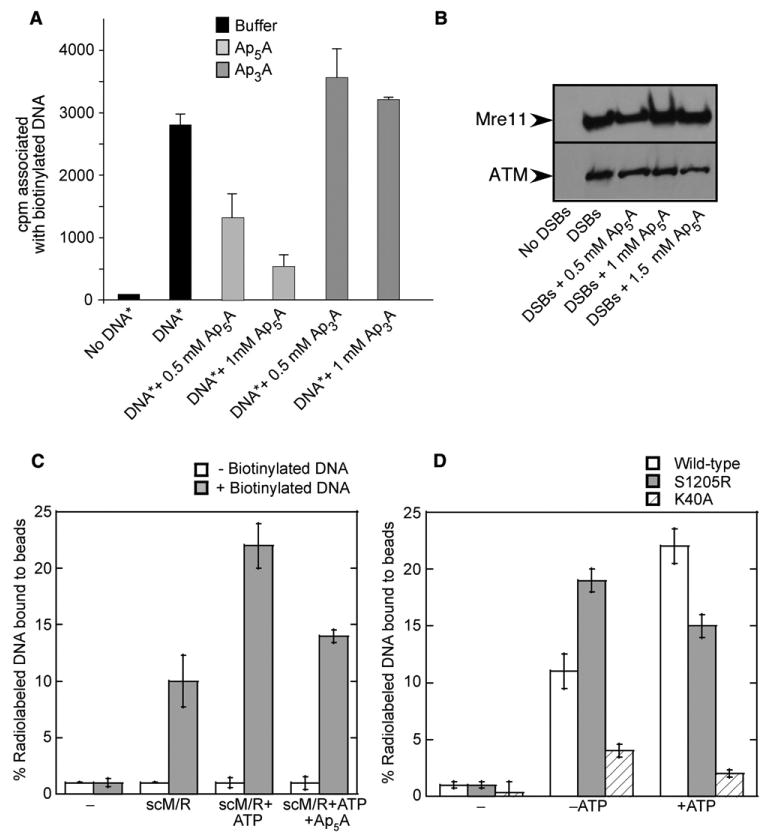
(A) Xenopus egg extracts incubated for 20 min with buffer, Ap5A, or Ap3A as indicated. Biotinylated DNA (0.6 × 1011 ends/μl) and nonbiotinylated radioactive DNA (DNA*; 3 × 1011 ends/μl) were added, and then biotinylated DNA was isolated and the associated radioactivity counted by scintillation.
(B) ATM and Mre11 association with DNA was monitored by western blot in DNA-bound fractions using specific antibodies directed against ATM or Mre11. The equivalent of 100 ng of DNA was loaded on the gel.
(C) scM/R complexes (30 nM) incubated with 32P-labeled linear DNA (2.5 kb, 1 nM) in the presence of an unlabeled, biotinylated DNA fragment attached to streptavidin-coated magnetic beads. The percentage of labeled DNA associated with the beads is shown as an average of two experiments, with standard deviation as shown.
(D) Reactions performed as in (C) except with scM/R(S1205R) and scM/R(K40A) mutant complexes as indicated.
The effect of Ap5A in Xenopus extracts is specific to the DNA tethering reaction, as the binding of M/R/N and ATM to DNA was not affected by Ap5A (Figure 6B). This result is consistent with our observations of purified M/R complexes, which have not shown any effect of adenylate kinase activity on the association of M/R or M/R/N(X) complexes with DNA molecules in binding assays in vitro (data not shown).
To examine the effects of Rad50 adenylate kinase activity on DNA tethering in a purified system, we used a similar assay that measures association of a 32P-labeled 2.5 kb DNA molecule with an unlabeled, biotinylated DNA molecule attached to streptavidin-coated magnetic beads. After incubation of the DNA-bound magnetic beads with scM/R, ∼10% of the labeled DNA was associated with the biotinylated DNA on the beads, and this level increased to ∼22.5% in the presence of ATP (Figure 6C). The addition of AMP to these reactions did not have any effect (data not shown), although the Ap5A inhibitor reduced the level of DNA tethering to ∼14%, close to the level observed in the absence of ATP. No association of the labeled DNA was seen with scM/R in the absence of the biotinylated DNA. The adenylate kinase activity of Rad50 therefore does contribute to DNA tethering as measured in this in vitro assay.
To further test if the DNA tethering activity requires the adenylate kinase function of Rad50, we used a mutant scM/R complex containing Rad50 S1205R. Surprisingly, this mutant scM/R complex was able to bridge DNA molecules more efficiently than the wild-type, although the presence of ATP did not increase the DNA tethering any further (Figure 6D). We also used a mutant scM/R complex that contained a mutation in the Rad50 Walker A domain (K40A). This mutant complex completely lacked DNA tethering ability, both in the presence and in the absence of ATP (Figure 6D).
Rad50 Catalyzes the Reverse Adenylate Kinase Reaction
As shown in Figure 6, we have observed inhibitory effects of the Ap5A adenylate kinase inhibitor in reactions that contain only ATP and no AMP. This suggests that the presence of AMP is not essential for adenylate kinase activity by Rad50. We have also consistently failed to observe any effect of AMP on the ATP-dependent activities of hsM/R, hsM/R/N, pfM/R, scM/R, or scM/R/X in vitro (data not shown). Considering these results, we hypothesized that Rad50 could enter the adenylate kinase cycle through the reverse reaction, starting with ATP hydrolysis (see Figure 7A).
Figure 7. Human and Yeast M/R Complexes Exhibit Reverse Adenylate Kinase Activity.
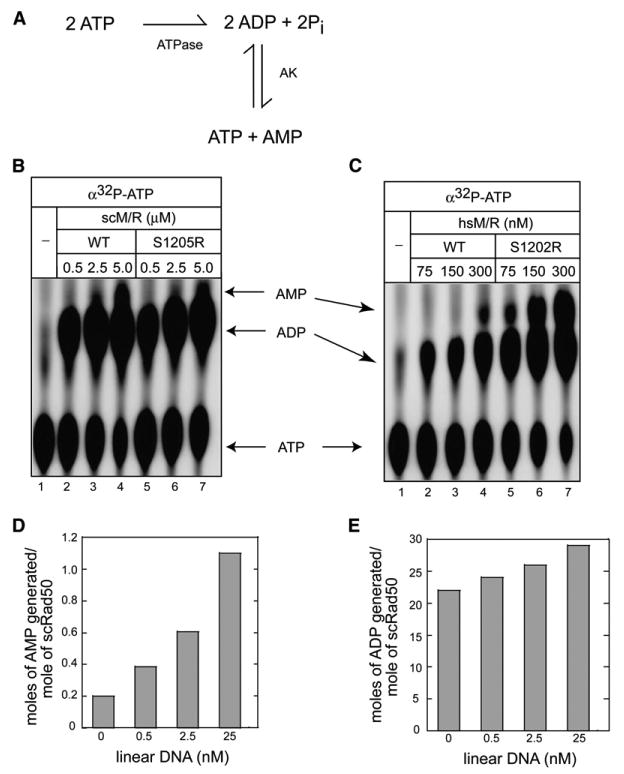
(A) Diagram of adenylate kinase reaction starting with ATP hydrolysis (ATPase) and the reverse adenylate kinase reaction (AK).
(B) Wild-type or scM/R(S1205R) complexes incubated with [α-32P]ATP as indicated and analyzed by TLC. The positions of the [α-32P]ATP substrate and [α-32P]ADP and [α-32P]AMP products are shown.
(C) Wild-type or hsM/R(S1202R) complexes assayed as in (B).
(D and E) scM/R complexes (2.5 μM) assayed with [α-32P]ATP in the presence of varying amounts of linear DNA as indicated. The moles of [α-32P]AMP produced, indicating adenylate kinase activity, are shown in (D) and the moles of [α-32P]ADP produced, indicating ATP hydrolysis, are shown in (E).
To test this model, we used [α-32P]ATP in the absence of AMP and found that both scM/R and hsM/R generated labeled [32P]AMP (Figures 7B and 7C), indicating that the enzymes can initiate the cycle with ATP hydrolysis followed by the reverse adenylate kinase reaction. Surprisingly, the signature motif mutants scM/R(S1205R) and hsM/R(S1202R) also generated labeled AMP in this reaction; thus, the mutations do not inhibit the reverse adenylate kinase reaction (2ADP → ATP + AMP), yet they are specifically deficient in the forward adenylate kinase reaction (ATP + AMP → 2 ADP). Moreover, the hsM/R(S1202R) complex generated 7.5- to 10-fold more labeled AMP compared with the wild-type enzyme (Figure 7C). The overproduction of labeled AMP by the hsM/R(S1202R) mutant compared with the wild-type was also observed when we used [14C]ADP to directly measure the reverse adenylate kinase reaction independently from ATP hydrolysis (Figure S3).
The ATPase and adenylate kinase assays shown above were performed in the absence of DNA, thus DNA is not essential for either of these catalytic activities. However, we did observe that the presence of linear DNA stimulated the adenylate kinase activity of scM/R by 5.5-fold, as measured by the production of labeled AMP (Figure 7D), while there was only a 1.5-fold stimulation of the ATPase activity under the same conditions (Figure 7E). In contrast, supercoiled DNA did not have any effect on either the ATPase or the adenylate kinase activities of Rad50 (data not shown).
Discussion
We show in this study that M/R complexes from three different species catalyze the reversible adenylate kinase reaction: the transfer of a phosphate from ATP to AMP to generate two molecules of ADP. The relationship between adenylate kinases and the ATPase domains in ABC transporter enzymes was first noted in a sequence comparison analysis (Hyde et al., 1990). The nucleotide binding domains of the CFTR ion channel, also a member of the ABC transporter family, were subsequently shown to catalyze adenylate kinase activity in vitro (Randak et al., 1997; Randak and Welsh, 2003; Gross et al., 2005). Furthermore, the gating of the channel was inhibited by Ap5A, an inhibitor specific to adenylate kinases because of its similarity to the transition state of the reaction, demonstrating that adenylate kinase activity is important for the function of CFTR (Randak and Welsh, 2003).
Here we demonstrate that M/R complexes catalyze the “forward” adenylate kinase reaction (AMP + ATP → ADP + ADP) as well as the “reverse” reaction (ADP + ADP → AMP + ATP). Furthermore, Rad50 can enter the adenylate kinase reaction through ATP hydrolysis, because incubation of M/R complexes with α-32P-ATP generates α-32P-AMP in addition to α-32P-ADP. Both the forward and reverse reactions are blocked by Ap5A (Figure 3 and data not shown), an indication that a bona fide adenylate kinase reaction is responsible for these products.
We have observed that, in the presence of equimolar AMP and ATP, M/R complexes catalyze both ATP hydrolysis and the forward adenylate kinase reaction. The presence of AMP reduces the level of ATP hydrolysis by ∼2- to 4-fold depending on the species of M/R, indicating that AMP acts competitively with ATP. Depending on the intracellular concentrations of AMP, which have been estimated at ∼1%–2% of the ATP pool (Dean and Perrett, 1976; Menze et al., 2005), it is possible that Rad50 molecules in vivo will catalyze both ATP hydrolysis and the forward adenylate kinase reaction, considering that the KM values of hsM/R and scM/R for AMP are 44 μM and 130 μM, respectively (Figure S2). Alternatively, it is possible that Rad50 complexes enter the adenylate kinase reaction through ATP hydrolysis as shown in Figure 7 and therefore are independent of the cellular AMP pool. This is consistent with our observation of Ap5A inhibition of DNA tethering in the absence of AMP and the fact that none of the activities of the M/R complexes appear to be affected by AMP in vitro (data not shown). We hypothesize that the Rad50 in a cell may be cycling back and forth in the adenylate kinase reaction between the ADP/ADP and the AMP/ATP-bound states. This is logical energetically because Rad50 ATPase activity is not dependent on DNA and would consume ATP unnecessarily if it were not engaged in the energetically neutral adenylate kinase reaction.
Rad50 adenylate kinase activity exhibits a strict adenine specificity for both the donor triphosphate nucleotide and the acceptor monophosphate nucleotide, identical to the specificity observed for Rad50 ATPase activity. The simplest explanation for these data is that the original ATP binding sites visualized in the structural analysis of pfRad50 (Hopfner et al., 2000) are also the sites that catalyze adenylate kinase activity. One complication with this model, however, is the fact that the ATP molecules present in the two active sites in the crystal structure are too far apart (∼35 Å) to accommodate the adenylate kinase reaction. The specificity of the Ap5A inhibitor strongly suggests that the distance between the nucleotides in the adenylate kinase reaction is ∼16 Å. To explain this discrepancy, we propose two alternative models. In the first model, Rad50 enters the adenylate kinase cycle through ATP hydrolysis but then undergoes a significant conformational change after ATP hydrolysis such that the ATP binding sites move closer together. Such a large conformational change would likely also affect the coiled-coil domains of Rad50, which are connected to the ATP binding domains. This type of conformational switch may be analogous to the transport cycle proposed for ABC transporters in which ATP-dependent changes in the nucleotide binding domains induce a conformational change in the orientation of the transmembrane domains that open or close the channel (Higgins and Linton, 2004).
In the second model, Rad50 would bind AMP at a site separate from the known ATP binding sites. Considering that ATP-bound Rad50 catalytic domains form dimers of dimers (Hopfner et al., 2000), this may allow one dimer to catalyze ATP hydrolysis while the other catalyzes the adenylate kinase reaction. Direct labeling of the AMP binding site will be necessary to distinguish between these models.
The Biological Importance of Rad50 Adenylate Kinase Activity
The signature motif mutation used in this study was originally made on the basis of a mutation found in CFTR that causes severe cystic fibrosis (Kerem et al., 1989). This mutant protein that carries a serine to arginine change in NBD1 of CFTR has not been characterized biochemically as far as we know but leads to a significant loss of gating function in vivo. The same mutation made in P. furiosus Rad50(S793R) causes a substantial decrease in ATP binding, ATP-induced dimerization, and ATP hydrolysis (Hopfner et al., 2000; Moncalian et al., 2004).
Mutation of the conserved serine to arginine in human Rad50 generates a complex that behaves like wild-type with respect to complex formation but is completely deficient in all ATP-dependent activities in vitro, including AMP-PNP-dependent DNA binding and ATP-dependent effects on Mre11 nuclease activity (Lee et al., 2003). Furthermore, the hsM/R/N S1202R mutant complex fails to stimulate ATM kinase activity in vitro, a deficiency that stems at least in part from its inability to perform ATP-dependent DNA unwinding (Lee and Paull, 2005). These results led us to initially assume that the analogous mutations in yeast and human Rad50 reduce ATP binding and hydrolysis as the S793R mutation does in pfM/R; however, it is clear that both scRad50(S1205R) and hsRad50(S1202R) mutant proteins retain the ability to hydrolyze ATP. We further show in this work that the yeast and human Rad50 mutants are specifically impaired in the forward adenylate kinase reaction.
In budding yeast, the Rad50 S1205R mutant fails to complement a rad50 deletion strain for MMS or bleomycin resistance and is equivalent to a null strain for nonhomologous end joining of cohesive or mismatched DNA ends (Moncalian et al., 2004; Zhang and Paull, 2005). In addition, the S1205R allele of Rad50 generates gross chromosomal rearrangements at a rate similar to a null allele of Rad50 (Smith et al., 2005). Here we show that the mutant also does not support telomere maintenance or spore viability in meiosis. Interestingly, the Rad50 S1205R mutation also acts as a dominant negative in meiosis. This mutation may block Spo11 processing similarly to the rad50S mutants that were previously identified (Alani et al., 1990), though this question has not been specifically investigated. It is clear that S1205R is not a separation of function mutant, however, because it shows a severe phenotype in both vegetative cells and those undergoing meiosis.
The in vitro DNA tethering assays described in this study show that M/R complexes utilize adenylate kinase activity in DNA tethering reactions. The Ap5A adenylate kinase inhibitor blocks both DNA tethering (Figure 6) and ATM activation (data not shown) in the Xenopus extracts, consistent with Rad50 adenylate kinase activity functioning upstream of ATM. It is not yet clear what step in DNA tethering is dependent on Rad50 adenylate kinase activity, but it must be a step that is distinct from DNA binding, which is unaffected by Ap5A either in Xenopus extracts or in DNA binding assays with purified proteins. Earlier studies demonstrated that the majority of DNA tethering activity in Xenopus extracts is MRN dependent (Costanzo et al., 2004) and that a mutant complex containing the Rad50 S1202R mutation was unable to complement extracts immunodepleted for Xenopus MRN (Dupre et al., 2006).
Using recombinant proteins, we also show that the wild-type scM/R complex catalyzes ATP-stimulated association of two DNA molecules. A mutation in a conserved lysine of the Walker A motif (K40A) completely blocks all DNA tethering, consistent with a previous report that analyzed end bridging by AFM (Chen et al., 2005). In contrast, the S1205R mutant complex shows high levels of DNA tethering, but it is not stimulated by ATP. It is not yet clear why the S1205R complex behaves differently in the purified system versus the Xenopus extract. One possibility is that DNA tethering in the extract requires multiple catalytic cycles while the purified system does not. Alternatively, the mutant may exhibit a higher level of nonspecific DNA binding, which could be more pronounced in the absence of other cellular factors.
The Role of Adenylate Kinase Activity in Rad50 Functions
What is the functional relationship between Rad50 ATPase and adenylate kinase activities? One possibility is that the forward adenylate kinase reaction is the mechanistically important reaction carried out by M/R complexes independent of ATP hydrolysis, as proposed for CFTR (Randak and Welsh, 2003). While the similarities with CFTR are compelling, several observations argue against this hypothesis for Rad50. First, unlike CFTR, the addition of AMP to in vitro reactions with M/R or M/R/N(X) complexes does not have any effect even though ATP is required in these reactions (data not shown). While we demonstrate here that M/R can enter the adenylate kinase reaction through ATP hydrolysis, one would expect that the addition of AMP would increase the functional activity of the enzyme if the forward adenylate kinase reaction was the critical catalytic step. Second, the addition of Ap5A to in vitro assays with M/R or M/R/N(X) complexes has no effect on the exonuclease activity of pfM/R or on the endonuclease activity of hsM/R/N on 3′ overhangs (data not shown), even though these activities are clearly ATP dependent. This suggests that ATP binding and ATP hydrolysis are functionally important apart from any relationship to adenylate kinase activity.
As an alternative model, we propose that Rad50 enters the adenylate kinase reaction through ATP hydrolysis but that different functions of the enzyme may require different nucleotide-bound states. For instance, there may be at least three different classes of Rad50 functions: ones that only require ATP binding, others that require binding and hydrolysis, and others that require binding, hydrolysis, and adenylate kinase activity.
DNA binding by M/R complexes is stimulated by ATP binding, not by hydrolysis, as shown with scRad50, pfRad50, hsM/R, and hsM/R/N (Raymond and Kleckner, 1993; Hopfner et al., 2000; de Jager et al., 2002; Lee et al., 2003), since nonhydrolyzable ATP analogs stimulate DNA binding as efficiently or more efficiently than ATP. In contrast, the partial unwinding of DNA duplexes by hsM/R/N requires ATP hydrolysis (Paull and Gellert, 1999), and hydrolysis is also required for scM/R/X to induce changes in DNA topology (Trujillo et al., 2003). Lastly, ATP stimulation of DNA tethering by scM/R and Xenopus M/R/N requires both ATP hydrolysis and adenylate kinase activity, as this activity is sensitive to Ap5A. In vivo, M/R/N(X) complexes are very likely engaged in all of these activities–DNA binding, unwinding, and DNA tethering–which are likely to be coordinated by Rad50 molecules in different catalytic states. The difficult challenge for future studies is to determine how each nucleotide-bound state alters the conformation of the Rad50 catalytic domains and coiled coils and how these changes translate into changes in DNA structure.
The evidence shown here and previously with CFTR suggests that adenylate kinase activity may be common to all of the enzymes in the ABC transporter ATPase family. It is clear that the relationship between ABC ATPases and adenylate kinases is not merely a structural similarity but is a functional and mechanistic connection that may govern how these enzymes respond to their respective substrates and may also control their activities in response to alterations in nucleotide pools that occur with changes in the intracellular environment.
Experimental Procedures
Plasmid Expression Constructs
See the Supplemental Data.
Protein Purification
See the Supplemental Data.
ATPase and Adenylate Kinase Assays
For ATPase assays, M/R complexes were incubated in 10 μl reactions containing buffer A (20 mM Tris [pH 8.0], 100 mM NaCl, 10% glycerol, 1 mM DTT) with 1 mM MgCl2 and 50 μM [γ-32P]ATP. Adenylate kinase assays also contained 50 μM cold AMP. Reactions were incubated at 37°C (human and archaebacteria) or 30°C (yeast) for 2, 8, or 16 hr and were stopped with the addition of 1% SDS and 10 mM EDTA. The reaction (1 μl) was then spotted on a polyethyleneimide (PEI) plate (EMD Biosciences) and separated by TLC for ATP, ADP, and Pi by using 0.75 M KH2PO4 (pH 3.4). The plates were dried and analyzed by phosphorimager (Bio-Rad). The levels of Pi generated were used as a measure of ATPase activity, and the levels of [β-32P]ADP were used as a measure of adenylate kinase activity. Adenylate kinase activity was also measured in some cases by using 50 μM [α-32P]AMP and unlabeled ATP (Figure 2) or with 50 μM [α-32P]ATP in the absence of AMP (Figure 7).
Xenopus Egg Extracts
Membrane-free egg cytosols were prepared as described (Smythe and Newport, 1991). All extract incubations were performed at 21°C. Extracts were incubated for 20 min with buffer, Ap5A, or Ap3A prior to DNA binding or tethering assays. For DNA binding assays, Xenopus egg extracts supplemented with buffer, Ap5A, or Ap3A were incubated with 150 bp biotinylated DNA fragments bound to streptavidin beads (10 ng/μl). Biotinylated DNA was generated by PCR by using M13-ssDNA as a template (Costanzo et al., 2004). After incubation, DNA-bound streptavidin beads were separated per the manufacturer's instructions (Dynabeads, M280 Streptavidin, Dynal Biotech) and washed in ELB buffer (10 mM HEPES [pH 7.7], 2.5 mM MgCl2, 0.05 mM KCl, and 250 mM sucrose) containing 0.1% Triton X-100. Samples were separated on 3%–8% NuPAGE gels and analyzed by Western blot using antibodies targeting Xenopus Mre11 or ATM (Robertson et al., 1999; Dupre et al., 2006). The DNA tethering assay was performed as described previously (Dupre et al., 2006). Extracts were incubated with buffer, Ap5A, or Ap3A and then supplemented with cold streptavidin-bound DNA (10 ng/μl) and free radioactive DNA fragments generated by PCR in presence of α32P-dCTP (50 ng/μl). After DNA removal from extracts, the radioactivity associated with biotinylated DNA was counted by scintillation.
DNA Tethering Assay with Purified Components
Biotinylated DNA was prepared by the PCR amplification of a 2.5 kb DNA fragment, separated on a 1% agarose gel, excised in the absence of ethidium bromide, and electroeluted into 2 ml of T.E. (10 mM Tris [pH 8.0], 1 mM EDTA). The electroeluted DNA was dialyzed against water, concentrated to 50 μl, and bound to magnetic beads (Dynal Biotech). The radiolabeled DNA was prepared by PCR amplification of an unrelated 2.5 kb fragment in the presence of [α-32P]dATP. The PCR product was purified as described above and stored in Tris-EDTA. DNA tethering was performed in binding buffer (25 mM MOPS [pH 8.0], 2 mM dithiothreitol [DTT], 5 mM MgCl2, 0.1% CHAPS, 0.1 mg/ml bovine serum albumin) with 12.5 mM NaCl and 1 nM 32P-labeled linear dsDNA. These components were combined with scM/R complex (30 nM) and DNA-bound magnetic beads (1 nM) in 9 μl. Reactions were incubated at 37°C for 30 min ± 100 μM ATP followed by three washes with binding buffer for 10 min each. Beads were resuspended in 20 μl binding buffer and were analyzed by liquid scintillation counting versus the total counts used in the reaction.
Determination of Kinetic Parameters
See the Supplemental Data.
Yeast Constructs and Strains
See the Supplemental Data.
Random Spore Analysis
Diploid strains were sporulated by incubation in 1% KOAc and 0.025% glucose for 10 days, then analyzed for viable spores as previously described (Adams et al., 1997). Percent sporulation is calculated in comparison to the wild-type homozygote TP2219, which exhibited 1.4% ± 0.3% sporulation. Strains expressing the rad50-S1205R allele showed no viable spores on any of the dilution plates; thus, the sporulation rate was at least 1000-fold reduced compared with the wild-type strain.
Telomere Length Assays
Haploid strains TP1218 (wild-type), TP1219 (rad50), and TP1991 (rad50-S1205R) were streaked onto YEPD for single colonies. Five colonies from each strain were used to study telomere length. In each strain, the five isolates exhibited identical telomere phenotypes (two are shown in Figure 4B). The strains were grown to log phase in YEPD at 30°C, and telomere length was analyzed by Southern blot of genomic DNA digested with XhoI and PstI run on a 1% gel. DNA from approximately the same number of cells was loaded in each lane. The blot was hybridized with a C1–3A telomere probe (Alexander and Zakian, 2003), which detects all telomeric fragments. This blot was stripped and reprobed with a CEN4 probe (Conrad et al., 1990) to verify loading. In all cases, telomere length in TP1219 and TP1991 strains was very short; reproducibly, telomeres in TP1991 were even modestly shorter than in TP1219.
Supplementary Material
Acknowledgments
We are grateful to Jim Carney and John Tainer for expression constructs and reagents; to Tom Petes, Lorraine Symington, and Hannah Klein for yeast strains; and to members of the Paull lab and Marty Gellert for critical comments about this project. This work was supported by National Institutes of Health grant R01 CA094008 to T.T.P and RO1 CA92245 to J.G.
Footnotes
Supplemental Data: Supplemental Data include three figures, Supplemental Experimental Procedures, and Supplemental References and can be found with this article online at http://www.molecule.org/cgi/content/full/25/5/647/DC1/.
References
- Adams A, Gottschling DE, Kaiser CA, Stearns T. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- Alani E, Padmore R, Kleckner N. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990;61:419–436. doi: 10.1016/0092-8674(90)90524-i. [DOI] [PubMed] [Google Scholar]
- Alexander MK, Zakian VA. Rap1p telomere association is not required for mitotic stability of a C(3)TA(2) telomere in yeast. EMBO J. 2003;22:1688–1696. doi: 10.1093/emboj/cdg154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton SJ, Jackson SP. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 1998;17:1819–1828. doi: 10.1093/emboj/17.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Trujillo K, Ramos W, Sung P, Tomkinson AE. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol Cell. 2001;8:1105–1115. doi: 10.1016/s1097-2765(01)00388-4. [DOI] [PubMed] [Google Scholar]
- Chen L, Trujillo KM, Van Komen S, Roh DH, Krejci L, Lewis LK, Resnick MA, Sung P, Tomkinson AE. Effect of amino acid substitutions in the rad50 ATP binding domain on DNA double strand break repair in yeast. J Biol Chem. 2005;280:2620–2627. doi: 10.1074/jbc.M410192200. [DOI] [PubMed] [Google Scholar]
- Connelly JC, de Leau ES, Okely EA, Leach DR. Overexpression, purification, and characterization of the SbcCD protein from Escherichia coli. J Biol Chem. 1997;272:19819–19826. doi: 10.1074/jbc.272.32.19819. [DOI] [PubMed] [Google Scholar]
- Conrad MN, Wright JH, Wolf AJ, Zakian VA. RAP1 protein interacts with yeast telomeres in vivo: overproduction alters telomere structure and decreases chromosome stability. Cell. 1990;63:739–750. doi: 10.1016/0092-8674(90)90140-a. [DOI] [PubMed] [Google Scholar]
- Costanzo V, Paull T, Gottesman M, Gautier J. Mre11 assembles linear DNA fragments into DNA damage signaling complexes. PLoS Biol. 2004;2:e110. doi: 10.1371/journal.pbio.0020110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean BM, Perrett D. Studies on adenine and adenosine metabolism by intact human erythrocytes using high performance liquid chromatography. Biochim Biophys Acta. 1976;437:1–5. doi: 10.1016/0304-4165(76)90342-1. [DOI] [PubMed] [Google Scholar]
- de Jager M, van Noort J, van Gent DC, Dekker C, Kanaar R, Wyman C. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol Cell. 2001;8:1129–1135. doi: 10.1016/s1097-2765(01)00381-1. [DOI] [PubMed] [Google Scholar]
- de Jager M, Wyman C, van Gent DC, Kanaar R. DNA end-binding specificity of human Rad50/Mre11 is influenced by ATP. Nucleic Acids Res. 2002;30:4425–4431. doi: 10.1093/nar/gkf574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diede SJ, Gottschling DE. Exonuclease activity is required for sequence addition and Cdc13p loading at a de novo telomere. Curr Biol. 2001;11:1336–1340. doi: 10.1016/s0960-9822(01)00400-6. [DOI] [PubMed] [Google Scholar]
- Dupre A, Boyer-Chatenet L, Gautier J. Two-step activation of ATM by DNA and the Mre11-Rad50-Nbs1 complex. Nat Struct Mol Biol. 2006 doi: 10.1038/nsmb1090. in press. [DOI] [PubMed] [Google Scholar]
- Gross CH, Abdul-Manan N, Fulghum J, Lippke J, Liu X, Prabhakar P, Brennan D, Willis MS, Faerman C, Connelly P, et al. Nucleotide-binding domains of cystic fibrosis transmembrane conductance regulator, an ABC transporter, catalyze adenylate kinase activity but not ATP hydrolysis. J Biol Chem. 2005;281:4058–4068. doi: 10.1074/jbc.M511113200. Published online December 16, 2005. [DOI] [PubMed] [Google Scholar]
- Higgins CF, Linton KJ. The ATP switch model for ABC transporters. Nat Struct Mol Biol. 2004;11:918–926. doi: 10.1038/nsmb836. [DOI] [PubMed] [Google Scholar]
- Holland IB, Blight MA. ABC-ATPases, adaptable energy generators fuelling transmembrane movement of a variety of molecules in organisms from bacteria to humans. J Mol Biol. 1999;293:381–399. doi: 10.1006/jmbi.1999.2993. [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Karcher A, Shin DS, Craig L, Arthur LM, Carney JP, Tainer JA. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Craig L, Moncalian G, Zinkel RA, Usui T, Owen BA, Karcher A, Henderson B, Bodmer JL, McMurray CT, et al. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002;418:562–566. doi: 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- Hyde SC, Emsley P, Hartshorn MJ, Mimmack MM, Gileadi U, Pearce SR, Gallagher MP, Gill DR, Hubbard RE, Higgins CF. Structural model of ATP-binding proteins associated with cystic fibrosis, multidrug resistance and bacterial transport. Nature. 1990;346:362–365. doi: 10.1038/346362a0. [DOI] [PubMed] [Google Scholar]
- Kaye JA, Melo JA, Cheung SK, Vaze MB, Haber JE, Toczyski DP. DNA breaks promote genomic instability by impeding proper chromosome segregation. Curr Biol. 2004;14:2096–2106. doi: 10.1016/j.cub.2004.10.051. [DOI] [PubMed] [Google Scholar]
- Kerem B, Rommens JM, Buchanan JA, Markiewicz D, Cox TK, Chakravarti A, Buchwald M, Tsui LC. Identification of the cystic fibrosis gene: genetic analysis. Science. 1989;245:1073–1080. doi: 10.1126/science.2570460. [DOI] [PubMed] [Google Scholar]
- Krogh BO, Symington LS. Recombination proteins in yeast. Annu Rev Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- Lavin MF. The Mre11 complex and ATM: a two-way functional interaction in recognising and signaling DNA double strand breaks. DNA Repair (Amst) 2004;3:1515–1520. doi: 10.1016/j.dnarep.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- Lee SE, Bressan DA, Petrini JH, Haber JE. Complementation between N-terminal Saccharomyces cerevisiae mre11 alleles in DNA repair and telomere length maintenance. DNA Repair (Amst) 2002;1:27–40. doi: 10.1016/s1568-7864(01)00003-9. [DOI] [PubMed] [Google Scholar]
- Lee JH, Ghirlando R, Bhaskara V, Hoffmeyer MR, Gu J, Paull TT. Regulation of Mre11/Rad50 by Nbs1: effects on nucleotide-dependent DNA binding and association with ATLD mutant complexes. J Biol Chem. 2003;278:45171–45181. doi: 10.1074/jbc.M308705200. [DOI] [PubMed] [Google Scholar]
- Lienhard GE, Secemski II. P 1, P 5 -Di(adenosine-5′)pentaphosphate, a potent multisubstrate inhibitor of adenylate kinase. J Biol Chem. 1973;248:1121–1123. [PubMed] [Google Scholar]
- Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- Lobachev K, Vitriol E, Stemple J, Resnick MA, Bloom K. Chromosome fragmentation after induction of a double-strand break is an active process prevented by the RMX repair complex. Curr Biol. 2004;14:2107–2112. doi: 10.1016/j.cub.2004.11.051. [DOI] [PubMed] [Google Scholar]
- Menze MA, Clavenna MJ, Hand SC. Depression of cell metabolism and proliferation by membrane-permeable and -impermeable modulators: role for AMP-to-ATP ratio. Am J Physiol Regul Integr Comp Physiol. 2005;288:R501–R510. doi: 10.1152/ajpregu.00490.2004. [DOI] [PubMed] [Google Scholar]
- Moncalian G, Lengsfeld B, Bhaskara V, Hopfner KP, Karcher A, Alden E, Tainer JA, Paull TT. The rad50 signature motif: essential to ATP binding and biological function. J Mol Biol. 2004;335:937–951. doi: 10.1016/j.jmb.2003.11.026. [DOI] [PubMed] [Google Scholar]
- Moore JK, Haber JE. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau S, Ferguson JR, Symington LS. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol Cell Biol. 1999;19:556–566. doi: 10.1128/mcb.19.1.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull TT, Gellert M. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 1999;13:1276–1288. doi: 10.1101/gad.13.10.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randak C, Welsh MJ. An intrinsic adenylate kinase activity regulates gating of the ABC transporter CFTR. Cell. 2003;115:837–850. doi: 10.1016/s0092-8674(03)00983-8. [DOI] [PubMed] [Google Scholar]
- Randak C, Neth P, Auerswald EA, Eckerskorn C, Assfalg-Machleidt I, Machleidt W. A recombinant polypeptide model of the second nucleotide-binding fold of the cystic fibrosis transmembrane conductance regulator functions as an active ATPase, GTPase and adenylate kinase. FEBS Lett. 1997;410:180–186. doi: 10.1016/s0014-5793(97)00574-7. [DOI] [PubMed] [Google Scholar]
- Raymond WE, Kleckner N. RAD50 protein of S. cerevisiae exhibits ATP-dependent DNA binding. Nucleic Acids Res. 1993;21:3851–3856. doi: 10.1093/nar/21.16.3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson K, Hensey C, Gautier J. Isolation and characterization of Xenopus ATM (X-ATM): expression, localization, and complex formation during oogenesis and early development. Oncogene. 1999;18:7070–7079. doi: 10.1038/sj.onc.1203194. [DOI] [PubMed] [Google Scholar]
- Shroff R, Arbel-Eden A, Pilch D, Ira G, Bonner WM, Petrini JH, Haber JE, Lichten M. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr Biol. 2004;14:1703–1711. doi: 10.1016/j.cub.2004.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, Gupta A, Kolodner RD, Myung K. Suppression of gross chromosomal rearrangements by the multiple functions of the Mre11-Rad50-Xrs2 complex in Saccharomyces cerevisiae. DNA Repair (Amst) 2005;4:606–617. doi: 10.1016/j.dnarep.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Smythe C, Newport JW. Systems for the study of nuclear assembly, DNA replication, and nuclear breakdown in Xenopus laevis egg extracts. Methods Cell Biol. 1991;35:449–468. doi: 10.1016/s0091-679x(08)60583-x. [DOI] [PubMed] [Google Scholar]
- Stracker TH, Theunissen JW, Morales M, Petrini JH. The Mre11 complex and the metabolism of chromosome breaks: the importance of communicating and holding things together. DNA Repair (Amst) 2004;3:845–854. doi: 10.1016/j.dnarep.2004.03.014. [DOI] [PubMed] [Google Scholar]
- Trujillo KM, Roh DH, Chen L, Van Komen S, Tomkinson A, Sung P. Yeast xrs2 binds DNA and helps target rad50 and mre11 to DNA ends. J Biol Chem. 2003;278:48957–48964. doi: 10.1074/jbc.M309877200. [DOI] [PubMed] [Google Scholar]
- Tsukamoto Y, Taggart AK, Zakian VA. The role of the Mre11-Rad50-Xrs2 complex in telomerase- mediated lengthening of Saccharomyces cerevisiae telomeres. Curr Biol. 2001;11:1328–1335. doi: 10.1016/s0960-9822(01)00372-4. [DOI] [PubMed] [Google Scholar]
- Yan H, Tsai MD. Nucleoside monophosphate kinases: structure, mechanism, and substrate specificity. Adv Enzymol Relat Areas Mol Biol. 1999;73:103–134. doi: 10.1002/9780470123195.ch4. [DOI] [PubMed] [Google Scholar]
- Zhang X, Paull TT. The Mre11/Rad50/Xrs2 complex and non-homologous end-joining of incompatible ends in S. cerevisiae. DNA Repair (Amst) 2005;4:1281–1294. doi: 10.1016/j.dnarep.2005.06.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


