Abstract
Dendritic trafficking and translation of BDNF transcripts play a key role in mediating synaptic plasticity. Recently, we demonstrated that siRNA-mediated knockdown of translin, an RNA binding protein, impairs KCl-induced dendritic trafficking of BDNF mRNA in cultured hippocampal neurons. We have now assessed whether translin deletion impairs dendritic trafficking of BDNF mRNA in hippocampal neurons in vivo. We have found that translin and its partner protein, trax, undergo dendritic translocation in response to treatment with pilocarpine, a pro-convulsant muscarinic agonist that increases dendritic trafficking of BDNF mRNA in hippocampal neurons. In translin knockout mice, the basal level of dendritic BDNF mRNA is decreased in CA1 pyramidal neurons. However, translin deletion does not block pilocarpine’s ability to increase dendritic trafficking of BDNF mRNA indicating that the requirement for translin in this process varies with the stimulus employed to drive it. Consistent with this inference, we found that dendritic trafficking of BDNF mRNA induced by bath application of recombinant BDNF in cultured hippocampal neurons, is not blocked by siRNA-mediated knockdown of translin. Taken together, these in vivo and in vitro findings indicate that dendritic trafficking of BDNF mRNA can be mediated by both translin-dependent and -independent mechanisms.
Keywords: trax, hippocampus, pilocarpine, CA1 pyramidal neurons, dendritic translation
The neurotrophin brain-derived neurotrophic factor (BDNF) is a key regulator of neuronal development and is required for various forms of long-term plasticity that depend on protein translation (Huang and Reichardt, 2001; Thoenen, 2000; Poo, 2001). Translation of a subset of dendritically localized mRNAs in close proximity to activated synapses is now emerging as a mechanism underlying long-lasting synaptic changes (Eberwine et al., 2002; Kindler et al., 2005; Sutton and Schuman, 2006; Schuman et al., 2006). As BDNF mRNA is one of these dendritic transcripts, and its localization in dendrites has been implicated in mediating synaptic plasticity (An et al., 2008; Lu et al., 2008; Tongiorgi, 2008), we have conducted studies aimed at defining the molecular mechanisms that orchestrate its trafficking and translation in dendrites. In recent studies, we have obtained evidence that translin, an RNA binding protein that has been localized to dendrites (Finkenstadt et al., 2000; Li et al., 2008), mediates dendritic trafficking of BDNF mRNA in hippocampal neurons grown in vitro (Chiaruttini et al., 2009). In particular, we found that treatment of rat hippocampal cultures with siRNAs targeting translin reduced KCl-induced dendritic trafficking of endogenous BDNF mRNA. Accordingly, we have, in this study, used translin knockout mice to assess its role in dendritic trafficking of BDNF mRNA in vivo.
Dendritic localization of BDNF transcripts has been studied most extensively in the hippocampal formation as the packing and alignment of apical dendrites of pyramidal neurons lends itself well to quantitative analysis. In this region, BDNF mRNA has been detected in apical dendrites of pyramidal neurons under basal conditions (Tongiorgi et al., 2004; An et al., 2008) and its dendritic localization is increased by treatment with chemical convulsants, such as kainate or pilocarpine (Tongiorgi et al., 2004; Chiaruttini et al., 2008). Therefore, to assess translin’s role in mediating dendritic trafficking of BDNF mRNA in vivo, we have checked: 1) whether translin displays increased localization in apical dendrites of hippocampal pyramidal neurons in response to pilocarpine, as found for BDNF mRNA, and 2) whether translin knockout mice show impaired dendritic translocation of BDNF mRNA in hippocampal neurons either under basal conditions or following pilocarpine treatment. Furthermore, since translin forms a heteromeric RNA binding complex with trax (Aoki et al., 1997; Taira et al., 1998; Finkenstadt et al., 2002), we have also compared their localization in neurons.
Experimental Procedures
Animals and reagents
A colony of translin knockout mice was established at Johns Hopkins from the line generated in Dr. M. Kasai’s laboratory (Fukuda et al., 2008). These mice had been backcrossed to C57/BL6 for over 10 generations. The translin knockout mice (Nbio055) were provided by the JCRB Laboratory Animal Resource Bank of the National Institute of Biomedical Innovation (Osaka, Japan). Genotyping of mice was performed on DNA isolated from tail snips. PCR was conducted using the primers and conditions described by Fukuda et al. (2008). Animal housing and procedures adhered to IACUC guidelines.
Rabbit polyclonal translin antiserum had been generated against a fusion protein containing the full-length rat translin sequence fused to glutathione-S-transferase (Finkenstadt et al. 2000). Two trax antibodies were used: one is a rabbit polyclonal antibody provided by Dr. Chern’s laboratory (Sun et al., 2006); the other is a guinea pig polyclonal antibody generated previously (Finkenstadt et al. 2000; Finkenstadt et al. 2001).
A myc-His-translin construct was generated as described in Finkenstadt et al. (2002) except that it encodes the human translin orthologue. An mCherry tagged trax construct was generated by inserting the native rat trax coding region into the HindIII/KpnI restriction sites of Clontech’s pEGFP-C1 vector in which the eGFP region had been replaced with mCherry (gift from M. Meffert). The mCherry tagged Translin construct was also cloned by inserting the rat translin coding sequence into the aforementioned vector using HindIII and SacII restriction sites. Translin GFP and trax GFP constructs were generated by inserting their respective rat coding sequences into the XhoI/EcoRI cloning sites in pEGFP-N1 (Clontech).
Primary Hippocampal Cultures
For immunohistochemical studies, rat and mouse hippocampal cultures were prepared from E18 embryos that were dissected quickly on ice in the following medium: Hank’s balanced salt solution supplemented with 100 U/ml penicillin and 100 U/ml streptomycin (pen/strep: Invitrogen), 1 mM MEM sodium pyruvate (Invitrogen), 30 mM glucose and 10 mM HEPES (Invitrogen). Tissue was then digested with papain (0.67mg/ml; Worthington) and 1% DNase (Sigma) at 37°C for 20 minutes. Following digestion, tissue was rinsed three times followed by trituration in neurobasal medium (Invitrogen) supplemented with 5% fetal bovine serum (FBS) (Hyclone, Logan, UT, USA), 2 mM Gluta-MAX-I (Invitrogen), and 100U/ml pen/strep. The resulting cell suspension was passed through a 70 μm filter. Cells were then plated on polylysine-coated 12 mm coverglasses at a density of 3×104 cells (rat) or 2×105 cells (mouse) in 24 well culture dishes. Neurons were fed glia-conditioned, 1% FBS neuronal medium supplemented with 2% B27 (Invitrogen) every 3–4 days. Neurons were transfected using Lipofectamine 2000 (LF2000; Invitrogen) following manufacturer’s directions.
To monitor BDNF mRNA trafficking in vitro, primary hippocampal neurons were prepared from P2 rats as described (Tongiorgi et al., 1997).
Immunoblotting
Forebrains were collected from 3 month old mice and immediately homogenized in cold lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 4 mM EDTA, 1% (v/v) NP-40 and 2X Complete mini EDTA-free protease inhibitor complex (Roche Molecular Biochemicals, Indianapolis, IN, USA). Forebrain homogenates were incubated on ice for 10 minutes then centrifuged at 15000 g for 10 minutes at 4°C. The supernatant was then harvested and Laemmli sample buffer added prior to heating the samples for 5 minutes in a 100°C water bath. Proteins were separated by electrophoresis on 10% polyacrylamide gels and then transferred to a nitrocellulose membrane (Bio-Rad Laboratories, Hercules, CA, USA). Membranes were probed with polyclonal trax antibody (guinea pig: 1:500), polyclonal translin antibody (rabbit: 1:10,000) and mouse monoclonal anti-β-Tubulin type III antibody (T8860 Sigma, St Louis, MO, USA: 1:500) in 1% non-fat dry milk and 0.05% Tween-20 in Tris-buffered saline. After washing the membrane, horseradish peroxidase-conjugated secondary antibodies (anti-rabbit and anti-mouse; Amersham Pharmacia Biotech AB, Piscataway, NJ, USA; goat anti-guinea pig, Chemicon) were used at 1:5000. The bands were visualized by enhanced chemiluminescence (Amersham Pharmacia Biotech AB).
Co-immunoprecipitation
Adult mouse forebrain was collected and immediately homogenized in 1 ml of cold lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2 mM EDTA, 0.2% deoxycholate and 2X Complete mini EDTA-free protease inhibitor complex (Roche Molecular Biochemicals, Indianapolis, IN, USA)). Homogenates were kept on ice for 20 minutes then centrifuged at 15000 g for 10 minutes at 4°C. Supernatant was then harvested and pre-cleared with protein A agarose beads (Pierce) for 30 minutes at 4°C. Supernatant was then collected after a 5 minute spin at 5000 g and 30 μl was kept aside as “offered”. One half of the remaining volume (~485 μl) was mixed with 10 μl of the guinea pig polyclonal trax antibody, while the other half was processed further in the absence of trax antibody. Both tubes were rocked at 4°C for 1.5 hours. Protein A agarose beads were then added to both tubes which were allowed to rock at 4°C for another hour. Beads were then spun down at 5000 g and supernatants removed. Fifty μl of the supernatant from the tube containing trax antibody was kept and designated as “S+”. Beads were washed three times with 500 μl lysis buffer for 5 minutes each at 4°C. After the last wash, 50 μl of 2X Laemmli buffer were added to the beads and S+ sample and 30 μl of 2x Laemmli to the offered sample. All samples were boiled for 5 minutes and then stored at -20°C. Proteins were separated by electrophoresis on 10% polyacrylamide gels and then transferred to a nitrocellulose membrane (Bio-Rad Laboratories, Hercules, CA, USA). Membranes were probed with polyclonal trax antibody (guinea pig: 1:500) and polyclonal translin antibody (rabbit: 1:10,000) as described above.
Electrophoretic Mobility Shift Assays
Gel-shift assays were performed as described previously (Li et al. 2004). In brief, freshly dissected cerebella and hippocampi from translin wt and ko mice were homogenized in a solution containing 20 mM HEPES (pH 7.9), 400 mM NaCl, 20% glycerol, 1.5 mM MgCl2, 20 mM NaF and protease inhibitor cocktail (Roche), incubated on ice for 15 min, and then centrifuged at 15,000×g at 4°C for 15 min. Supernatants were stored at −80 °C for later use.
Radiolabeled probe was prepared by incubating a 39-mer RNA oligo sequence which corresponds to a segment of the 3’UTR of protamine-2 (Li and Baraban, 2004) with [γ-32P] ATP and T4 polynucleotide kinase (New England Biolabs) and purified with a Sephadex G-50 column (GE Healthcare). Five μg of extract protein was incubated with 20,000 cpm of probe in 12 mM HEPES (pH 7.9), 4 mM Tris–HCl (pH 7.9), 50 mM KCl, 50 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 12% glycerol, and 2 μg of poly(dl–dC) in 30 μl total reaction volume for 15 min at room temperature. The reaction was loaded onto a 5% native polyacrylamide gel. After electrophoresis, gels were dried and exposed to Biomax-MR (Kodak) film overnight at −80 °C.
Immunostaining
For immunostaining studies of endogenous translin and trax in brain sections, mice were anesthetized with chloral hydrate (400 mg/kg, i.p.) and then perfused via cardiac puncture with chilled PBS (1 mM KH2P04, 10 mM Na2HP04, 137 mM NaCl, 2.7 mM KCl, pH 7.4), followed by freshly prepared 4% paraformaldehyde (in PBS). Brains were post-fixed in this solution overnight and then cryoprotected by immersion for at least 24 hours in 25% sucrose dissolved in PBS. Thirty μm sections were cut on a sliding microtome. For trax immunostaining, blocking and tissue permeabilization were achieved by incubation of sections in 30mg/ml BSA and 0.1% Triton-X-100 in PBS for one hour. Sections were then incubated overnight at 4°C with polyclonal anti-Trax (rabbit 1:5000) diluted in PBS with 10mg/ml BSA. Extensive washing in PBS was followed by incubation with biotinylated anti-rabbit (1:2000; Vector Labs) overnight at 4°C. The brain sections were then washed in PBS and incubated for 30 minutes with ABC (Vectastain Kit: Vector Labs), and developed for 10 minutes with the tyramide signal amplification solution (1:400; TSA Plus fluorescein, Perkin Elmer). A brief wash period with PBS was followed by 10 minute incubation with DAPI prior to mounting and coverslipping with Permafluor-DABCO (Beckman-Coulter, Marseille, France).
For translin immunostaining, sections were processed in a similar fashion except that an antigen retrieval step was included after collecting the sections in PBS. In this protocol, sections were processed as described below for the in situ hybridization procedure up to primary antibody step, which included an overnight incubation at 60°C. After washing the sections with PBS, they were incubated overnight in polyclonal rabbit translin antibody (1:6000) and the staining procedure completed as described above for trax staining. As the inclusion or omission of the antigen retrieval step abolished trax and translin staining in brain sections, respectively, double staining could not be performed on brain tissue.
For co-localization studies of recombinant mCherry Translin and Trax GFP in mouse cultures, neurons were transfected with 100ng of each construct at 7 days in vitro (DIV). The following day (18-24 hours later), neurons were rinsed briefly in PBS and then fixed in 4% formaldehyde (in PBS) for 15 minutes. They were then permeabilized in PBS containing 0.1% Triton X-100 for 10 minutes followed by an hour blocking step in 50 mg/ml bovine serum albumin (BSA) in PBS. Cultures were then incubated overnight with mouse anti-MAP2 antibody (1:250, Chemicon International), diluted in PBS with 10mg/ml BSA. After several washes in PBS the following day, the coverslips were incubated for 60 min at 25°C with anti-mouse Alexa-fluor 405 (1:500, Invitrogen) diluted in PBS with 10mg/ml BSA. After several washes in PBS, coverslips were mounted onto slides with Permafluor-DABCO (Beckman-Coulter, Marseille, France). In rat cultures where we stained for both endogenous trax and recombinant translin, neurons were transfected at 9 DIV with a myc-His-tagged translin construct (100 ng) and then processed for staining 18-24 hours later. In this case, cells were incubated simultaneously with both the rabbit trax antibody and a mouse monoclonal anti-myc antibody (1:5000; Invitrogen), followed by incubation with a mixture of anti-rabbit Alexa-fluor 555 and anti-mouse Alexa-fluor 488 (each at 1:2000, Molecular Probes).
The specificity of the polyclonal Trax antibody in rat cultures was determined via co-transfection of rat hippocampal neurons with 100ng eGFP plasmid with siRNA oligos (Dharmacon) at 6 DIV using 30, 50 or 100 nM concentrations. siRNA oligos targeting the following sequences in rat Trax were used: 835: AAGUGGAGAACGCUUGCUA and 218: GGACACAAGACACGACAAA. As control, we also tested a mutant form (835M) of the 835 siRNA with the following sequence: CAGUGGCGAACGAUGAUA (bold indicates nucleotide substitutions). Three days post-transfection, cultures were fixed and processed for staining as described above, using the rabbit Trax polyclonal antibody (1:5000) and the anti-rabbit Alexa-fluor 555 secondary antibody.
Pilocarpine treatment
Mice treated with pilocarpine were 2-3 months old. To reduce mortality from peripheral cholinergic effects, mice were pre-treated with methylscopolamine (1 mg/kg, i.p.,Sigma) dissolved in PBS 20 minutes before receiving pilocarpine (250 mg/kg, i.p., Sigma) dissolved in 0.9% NaCl (Turski et al., 1989). Only mice which developed generalized seizure activity within an hour of pilocarpine administration were used for histological studies. At three hours following pilocarpine administration, mice were anaesthetized with chloral hydrate (400 mg/kg, i.p., Sigma) dissolved in PBS and perfused via intracardiac puncture with freshly made 4% paraformaldehyde (PFA) solution made in DEPC-treated PBS. Control mice were only treated with chloral hydrate prior to perfusion. The brains were then removed and placed overnight in 4% PFA prepared DEPC-treated PBS at 4°C. The next day, brains were transferred to a solution consisting of 25% (w/v) sucrose in DEPC-treated PBS. On the following day, 30-35 μm hippocampal sections were cut on a sliding microtome and collected in DEPC-treated PBS. Sections were then mounted on Superfrost Plus slides (VWR) and processed for in situ hybridization.
In situ hybridization
To monitor the localization of BDNF mRNA in hippocampal sections, hybridization was performed with an antisense probe targeting nucleotides 240-666 of exon IX (NM_007540). The corresponding sense probe was used as a control. To generate these probes, total RNA was isolated from mouse hippocampus and cDNA obtained by reverse transcription. PCR primers targeting this segment contained 5’ HindIII and 3’ XbaI overhangs and the PCR product was cloned into PCR3.1 (Invitrogen). The following primer sequences were used: fwd - 5’CCCAAGCTTGCTGGATGAGGACCAGAAGGT3’ and rev -5’GCTCTAGAGCTTGGGTAGTTCGGCATTGCGA3’. Digoxigenin-labeled RNA probes were generated by using the DIG RNA labeling mix (Roche Applied Sciences) and T7 polymerase. Sense probe was generated by reversing the 5 and 3’ restriction site overhangs and inserting the PCR product in reverse into the plasmid and then continuing with T7 mediated transcription.
Brain sections were post-fixed in 4% paraformaldehyde (PFA) followed by washes in PBS and then permeabilized in PBS with 0.3% Triton-X100. Sections were then rinsed in PBS and treated with proteinase K (1 μg/ml) at 37°C in buffer (100 mM Tris, 50 mM EDTA, pH 8) and treated with acetic anhydride (in 0.1M TEA, pH 8). Slides were washed in PBS and blocked in pre-hybridization buffer (50% deionized formamide, 5X SSC, 1X Denhardt’s, 250 μg/ml yeast tRNA) at 60°C and then hybridized with DIG labeled probe in hybridization buffer (same as pre-hybridization buffer but with 10 mM DTT and 0.5 mg/ml salmon sperm). On the next day, slides were washed sequentially at 60°C in 50:50 2X SSCT (SSC and 0.1% triton-X100): formamide, 2X SSCT, and 0.2X SSCT. Sections were then washed in TBS-T (100 mM Tris, 150 mM NaCl, pH 7.5, 0.1% Tween-20) and blocked in TBS-T containing 5% BSA at room temperature. Sections were then incubated overnight with anti-DIG antibodies that were coupled to peroxidase (1:500; Roche Applied Science) in 1% BSA in TBS-T. On the third day, sections were washed in TBS-T and developed for 10 minutes using the TSA Plus fluorescein kit (1:100; Perkin Elmer). Sections were then washed in PBS and coverslipped.
To monitor BDNF mRNA localization in hippocampal cultures, cultures were treated with an RNAi “cocktail” against translin generated as described (Chiaruttini et al., 2009) on DIV 7, and then stimulated with BDNF (50 ng/ml) or KCl (10mM) the following day. After 3h of stimulation, cultures were fixed and processed for in situ hybridization with a probe targeting exon IX (Chiaruttini et al., 2009).
Imaging
To quantify the in situ hybridization signal on brain sections, images were acquired at 20x power using a Zeiss Axiocam MRM camera mounted on an AxioExaminer D1 microscope and then quantified using Image J software (NIH). To measure the intensity of labeling in the stratum radiatum of CA1, we determined the average intensity of staining in rectangles 50 μm wide and 15 μm high positioned 5 to 20 μm, 20 to 35 μm, 35 to 50 μm and 50 to 65 μm away from the cell body layer of CA1. Background measurements taken over adjacent white matter were subtracted from the intensity measurements obtained for cell body or dendritic regions. Dendritic intensity values were divided by cell body labeling intensity to obtain a dendritic trafficking index. Data obtained at each of the dendritic intervals from four experimental groups, wt control (n=5), wt pilocarpine, (n=8), ko control (n=6) and ko pilocarpine (n=8), were then analyzed for statistical significance using 2×2 ANOVA.
Images of BDNF mRNA localization in hippocampal neurons were acquired with a CCD camera and analyzed with Image-ProPlus (Media Cybernetics). The relative dendritic filling (RDF) value was determined as described (Chiaruttini et al., 2009).
Immunocytochemical images of cultured neurons were captured at a resolution of 1024×1024 pixels using the Zeiss Axiovert 200 with 510-Meta confocal module using 63X Plan-apochromat/ 1.4 oil DIC objective and 405 diode laser, 488 argon laser and 542 green HeNe laser and the Zeiss LSM 510 software. Images of Translin and Trax immunostaining and BDNF mRNA in situ hybridization were captured at a resolution of 1024×1024 pixels using the 10X Plan-apochromat objective, the 20X/0.75 Plan-apochromat objective, and the 40X/1.3 Plan-Neofluar oil DIC objective.
To determine the specificity of the rabbit polyclonal Trax antibody, we captured images at 20x power using a Zeiss Axiocam MRM camera mounted on an AxioExaminer D1 microscope, of the EGFP signal and endogenous Trax staining of 9 DIV rat hippocampal cultures at the same exposure across two different conditions: concentration (30, 50, 100 nM) and oligo target (835, 835M, 218). We measured the intensity of the cell body using Image J software (NIH) and normalized the Trax levels to the eGFP levels (took the ratio of the eGFP reading to 255 which is the maximum intensity and multiplied the Trax intensity by that ratio). We averaged all the values per group (n=12, 4 images at each concentration per oligo type).
Results
Dendritic localization of translin and trax
Prior to examining the impact of pilocarpine treatment on translin and trax localization, we first confirmed the specificity of the translin and trax antibodies by immunoblotting forebrain extracts harvested from wild type mice or mice carrying the translin deletion allele. These studies demonstrated that the translin protein band is absent from extracts prepared from translin knockout mice and reduced in heterozygote samples (Figure 1A). Furthermore, since trax protein is degraded in translin knockout mice (Chennathukuzhi et al., 2003), we also checked the specificity of the trax antibody by demonstrating that the trax protein band shows the same profile. Lastly, we also confirmed that the translin/trax gel-shift complex (Finkenstadt et al., 2002) is completely absent in brain extracts prepared from translin knockout mice (Figure 1B).
Figure 1. Specificity of translin and trax immunostaining in brain.
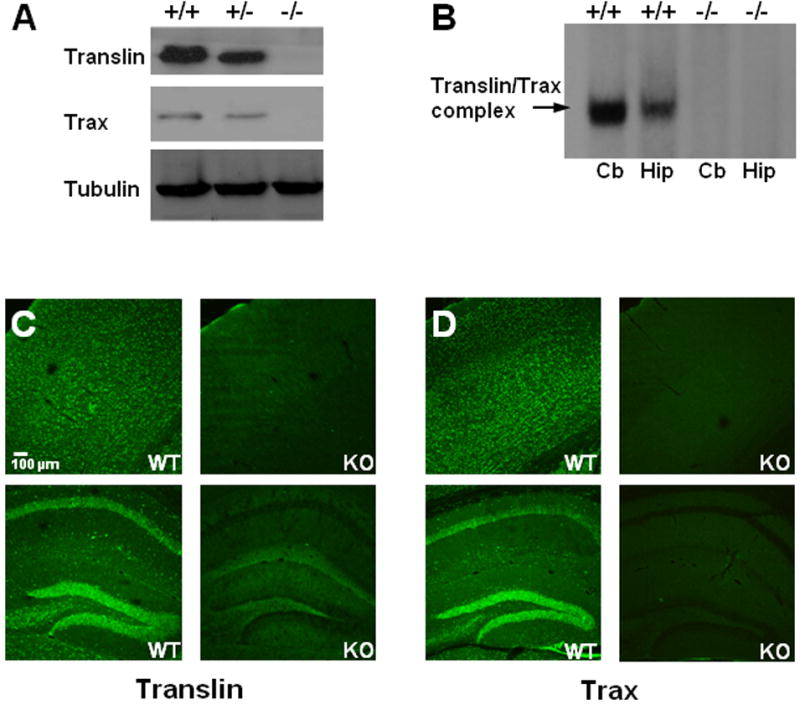
A) Immunoblots of forebrain extracts harvested from adult wild-type (+/+), heterozygous (+/-), or translin knockout (-/-) mice show that translin and trax protein levels are reduced below wild type levels in heterozygous samples and absent in knockout samples. Tubulin blot shown as loading control. B) The translin/trax gel-shift complex (indicated by arrow at left of blot) is absent in cerebellar (Cb) and hippocampal (Hip) extracts harvested from translin knockout mice. C and D) Low power images of cortex (top panels) and hippocampus (bottom panels) show immunostaining for both translin and trax in sections taken from wild type mice (wt). In contrast, only negligible staining is detected in corresponding sections taken from knockout (ko) mice. Note that while some residual translin staining is detected in the dentate gyrus and the overlying stratum lacunosum-moleculare, the CA1 region is devoid of staining.
To assess the specificity of translin and trax immunostaining, we processed sections from wild type and translin knockout mice in parallel. As reported previously, we found robust translin immunostaining in pyramidal cells of the hippocampus and granule cells of the dentate gyrus, as well as in neuronal cell bodies in the overlying cortex (Figure 1C; Finkenstadt et al., 2000). In contrast, translin immunostaining is markedly reduced or absent in sections taken from translin knockout mice. Similarly, trax immunostaining matched that displayed by translin and is also abolished in sections taken from translin knockout mice (Figure 1D).
As pilocarpine treatment has been shown to induce dendritic translocation of BDNF mRNA in pyramidal neurons from cortex (Pattabiraman et al., 2005) and hippocampus (Tongiorgi et al., 2004), we checked whether this treatment also triggers translocation of translin and trax proteins in these regions. In mice, we found that dendritic localization of BDNF mRNA is most prominent in CA1 pyramidal neurons following pilocarpine treatment. Dendritic translocation of both translin and trax showed a similar pattern following pilocarpine treatment (Figure 2A). In contrast to the pattern of translin and trax staining observed in hippocampal pyramidal neurons, dendritic translin and trax staining were readily observed in cortical pyramidal neurons under basal conditions (Figure 2B). Interestingly, BDNF mRNA is also present in dendrites of cortical pyramidal neurons under basal conditions (Figure 2B), consistent with the possibility that the translin/trax complex mediates dendritic translocation of this transcript.
Figure 2. Dendritic localization of translin and trax.
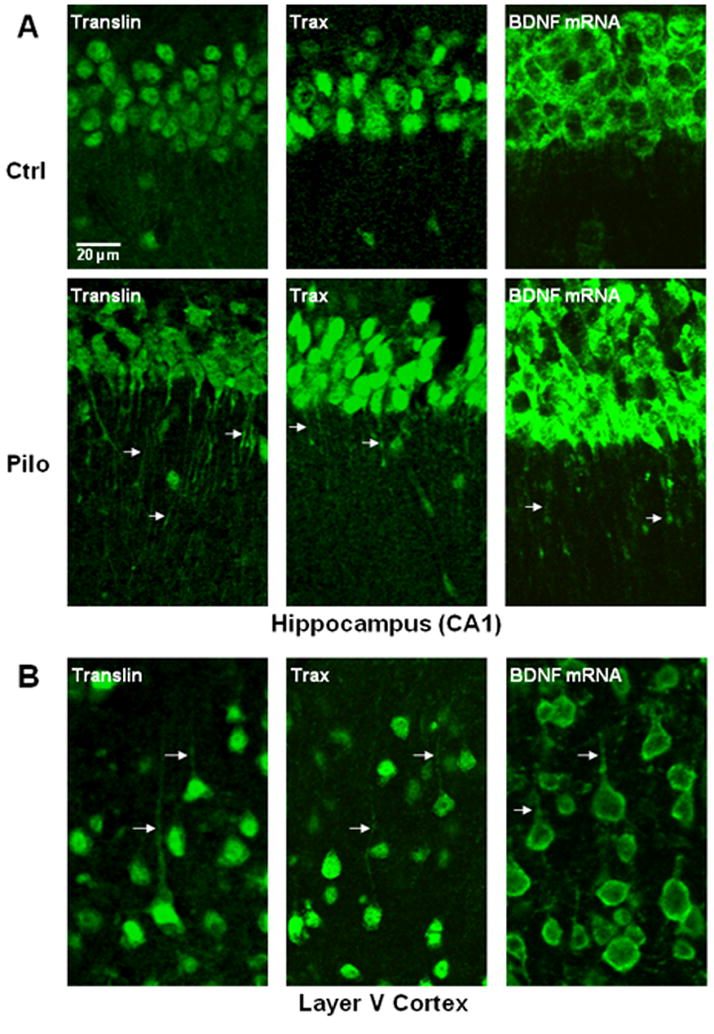
(A) Panels show images taken of the hippocampal CA1 region. In control mice (Ctrl), translin and trax staining is restricted to the cell bodies of CA1 pyramidal neurons but translocate out into apical dendrites in the stratum radiatum in mice treated with pilocarpine (Pilo) 3 hours prior to perfusion. As shown in the right hand panels, BDNF mRNA is restricted to the cell bodies of pyramidal neurons of CA1 in control mice but undergoes translocation into apical dendrites following pilocarpine treatment. (B) In contrast to their localization pattern in hippocampus, both translin and trax, as well as BDNF mRNA, are present in dendrites of cortical pyramidal neurons in control mice (arrows). These panels show layer V pyramidal neurons in the barrel field of the somatosensory cortex (S1BF).
Although these immunostaining studies indicate that translin and trax display similar patterns of localization, we were unable to identify conditions which allowed us to perform double staining to assess whether they co-localize. Detection of translin immunostaining required processing of tissue sections with relatively harsh antigen retrieval conditions that abolished trax immunostaining. Given these technical limitations, we conducted co-immunoprecipitation studies to check whether these proteins form a heteromeric complex in mouse brain, as previously reported in studies conducted on rat brain (Finkenstadt et al., 2000). As expected, immunoprecipitation of trax from extracts prepared from adult mouse forebrain also pulled down translin (Figure 3A).
Figure 3. Association of translin and trax.
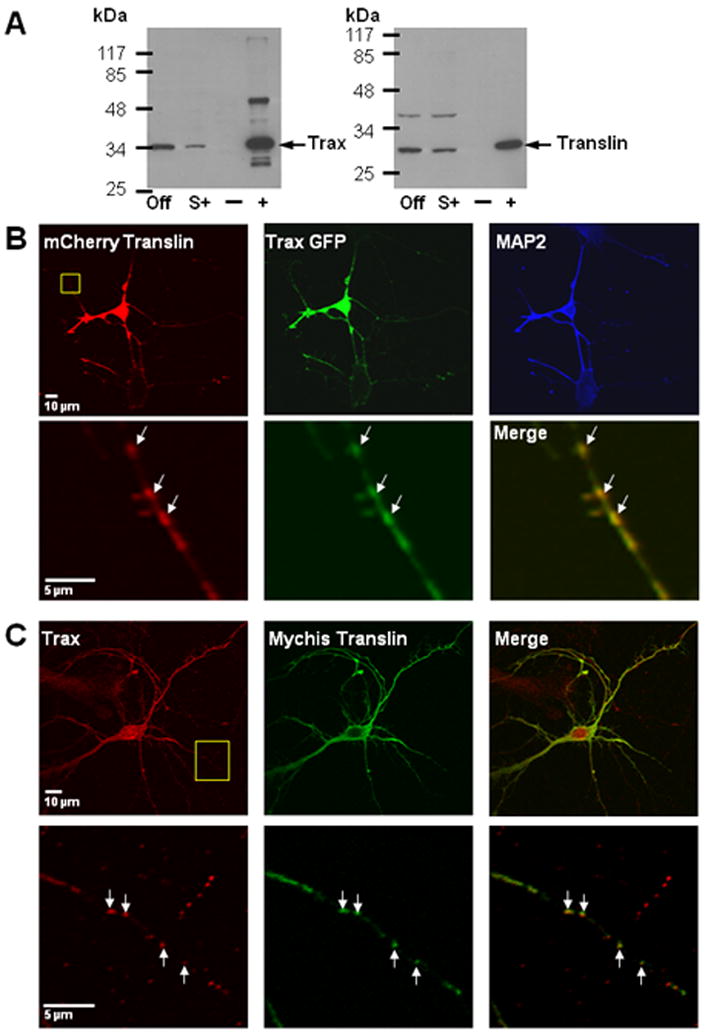
(A) Co-immunoprecipitation of translin and trax from mouse forebrain extracts. Extracts were incubated with or without trax antibody and then aliquots of the starting lysate, supernatant or pellet were processed for immunoblotting with either trax (left blot) or translin (right blot). The lanes located at the right of each blot contain the pellets obtained without (-) or with (+) trax antibody. (The additional bands present in the rightmost lane of the trax blot reflect heavy and light chains of the trax antibody, since the same guinea pig antibody was used for both the i.p. and blotting steps.) The lanes located at the extreme left of both blots contain aliquots of the starting lysate or “offered” (Off). The lanes labeled (S+) contain the supernatant from the tubes that were incubated with trax antibody and show depletion of both trax and translin relative to the levels found in the offered sample. B) Co-localization of recombinant translin and trax in mouse hippocampal neurons. Mouse cultures were transfected at 7 DIV with both mCherry Translin and Trax GFP plasmids and then fixed the next day. Low power images shown in top row illustrate co-localization of these constructs throughout the cell. Staining with MAP2 antibody indicates that both proteins localize to MAP2-positive, dendritic processes as well as a MAP2-negative process that extends from the top right corner of the cell body, which appears to be an axonal process. Bottom panels show high power images of boxed area in top left panel. We have also detected endogenous trax puncta in MAP2-negative, presumed axonal processes indicating that localization to axons is not an artifact of overexpression of these recombinant constructs (data not shown). Co-localization of recombinant translin and trax was also observed following co-transfection of rat hippocampal cultures. C) Co-localization of endogenous trax puncta with recombinant translin. Top panels show low power images of a rat hippocampal neuron (8DIV) that was transfected with a myc-His translin construct (green) and stained for endogenous trax (red). Bottom panels show high power images of boxed area in top left panel. Arrowheads indicate discrete puncta that are stained for both myc-His translin and endogenous trax.
Co-localization of translin and trax in vitro
Consistent with these co-immunoprecipitation findings, we also found that recombinant translin and trax co-localized in mouse or rat hippocampal cultures co-transfected with these expression plasmids (Figure 3B). To assess whether endogenous translin and trax also co-localize in rat hippocampal cultures, we also performed immunostaining studies on rat hippocampal cultures with the translin and trax antibodies employed successfully for tissue staining. For trax, we found a punctate staining pattern in neuronal processes. We confirmed the specificity of this staining by demonstrating that transfection with siRNA oligos targeting trax reduced the observed staining (Supplementary figure 1). Unfortunately, we were unsuccessful in identifying staining conditions that yielded specific translin immunostaining in neuronal cultures, as the conditions needed for antigen retrieval damaged the cells. Accordingly, we compared the localization of a transfected translin expression construct with endogenous trax. These studies revealed a high degree of co-localization of these proteins in puncta in dendrites (Fig 3C). To quantify the level of co-localization, we selected clearly defined translin positive puncta and then assessed whether they were also trax-positive. Using this approach, approximately 90% of translin puncta were also positive for trax (88± 4%, mean ± SEM).
Effect of translin deletion on dendritic trafficking of BDNF mRNA
Prior to assessing whether translin deletion impairs dendritic trafficking of BDNF mRNA, we checked the specificity of the BDNF mRNA in situ hybridization procedure by confirming that the hybridization signal was abolished in adjacent sections processed with the corresponding sense probe (Figure 4A, right hand panels). Furthermore, we checked whether translin deletion alters the basal level of expression of BDNF mRNA in cell body layers of the hippocampus (Figure 4, bottom panel). Qualitatively, the pattern of BDNF mRNA localization in cell body layers appeared similar in wild type and translin ko mice. Densitometric analysis of the CA1 pyramidal cell layers showed that the average intensity of BDNF mRNA labeling was reduced by approximately 20% in the sections from translin ko mice compared to control mice. However, this difference did not reach significance (112 +/- 18 vs. 88 +/- 17; wt vs. ko; mean +/- SEM; p>0.3).
Figure 4. Effect of translin deletion on the pattern of BDNF mRNA localization.
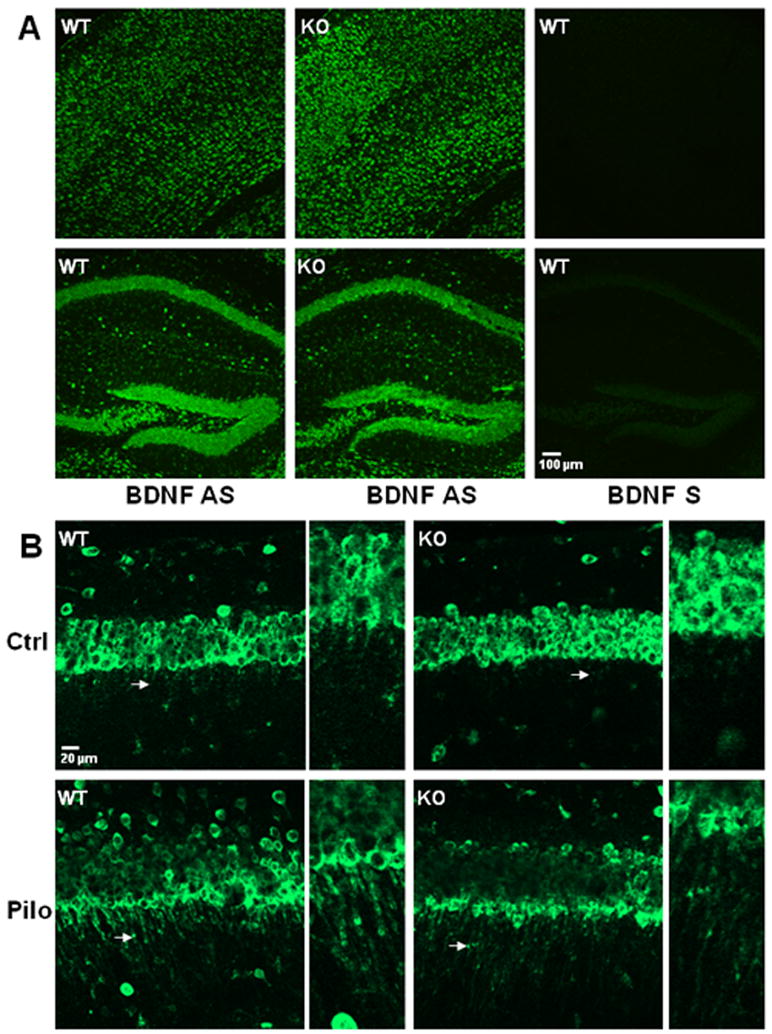
A) Low power images of cortex (top panels) or hippocampus (bottom panels) show staining obtained by processing brain sections from wild type (wt) and translin knockout (ko) mice for in situ hybridization with a BDNF mRNA probe targeting the coding region. The overall hybridization pattern displayed by wt and ko mice with the anti-sense probe is similar. The hybridization signal was absent in adjacent sections from wt mice that were processed with the corresponding sense probe (right panels). B) Top panels show BDNF mRNA hybridization signal in the CA1 region of control wild type (left panel) and translin knockout (right panel) mice. Bottom panels show increased dendritic localization (arrows) following pilocarpine treatment. High magnification images correspond to areas indicated by arrows in the panels directly to their left.
To assess whether translin deletion blocked the ability of pilocarpine treatment to increase dendritic trafficking of BDNF mRNA, we measured the intensity of in situ staining in the CA1 stratum radiatum at different distances from the cell body layer, as this subregion of the hippocampus displayed the most prominent dendritic localization of BDNF mRNA following pilocarpine treatment (Figures 4B and 5). Furthermore, to ensure that any decreases in dendritic trafficking detected in translin ko mice are not due to decreased expression of BDNF mRNA at the cell body level, we divided each of the dendritic values by the intensity measured at the CA1 cell body layer. Analysis of these quantitative data by ANOVA revealed that both translin deletion and pilocarpine produced significant main effects, without a significant interaction effect (Figure 5). We obtained the same results if the dendritic values were analyzed without normalizing them to the cell body layer (supplementary figure 2). Thus, these findings indicate that translin deletion decreases the basal level of BDNF mRNA trafficking, but does not impair the ability of pilocarpine to increase dendritic trafficking of BDNF mRNA.
Figure 5. Effect of translin deletion on dendritic localization of BDNF mRNA in CA1 region in control and pilocarpine-treated mice.
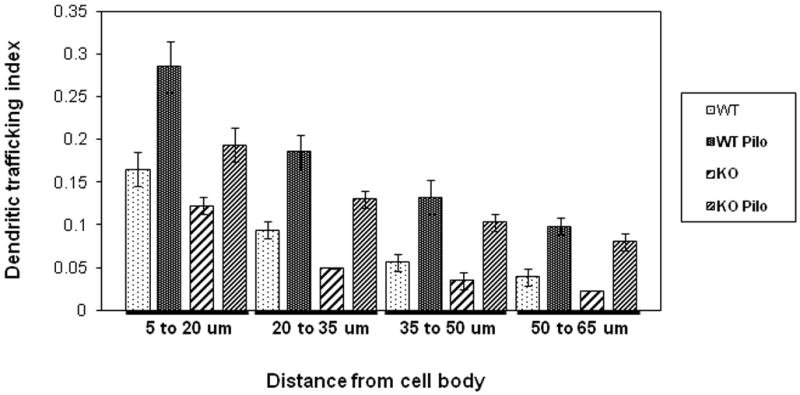
Bar graph shown presents quantification of hybridization signal in dendrites by treatment (control and pilocarpine) and genotype (wt and ko). For each of the distances shown from the edge of the CA1 cell body layer, the dendritic trafficking index was determined by dividing the average signal intensity for that 15 μm interval in the stratum radiatum by the average staining intensity of the overlying cell body layer. Error bars indicate standard errors. Two-way ANOVA analysis at each interval, except the 50 to 65 μm distance, revealed significant main effects of both genotype and treatment (p<0.005). At the longest distance, 50 to 65 μm, there is a significant effect of pilocarpine (p<0.0005) but not of genotype. There was no significant interaction effect at any of these intervals (p>0.5).
Stimulus selective effects of translin siRNA on BDNF mRNA trafficking in vitro
As these results suggest that translin deletion impairs BDNF mRNA trafficking into dendrites under basal conditions but not following pilocarpine administration, we wondered whether this may reflect the ability of BDNF mRNA to undergo dendritic translocation via both translin-dependent and translin-independent targeting mechanisms under different stimulation conditions. Previous studies indicate that pilocarpine treatment in vivo activates BDNF/ TrkB signaling in hippocampal neurons (Roberts et al., 2006; He et al., 2010). Accordingly, we compared the role of translin in mediating dendritic trafficking of BDNF mRNA in cultured hippocampal neurons in response to two stimuli, KCl-induced depolarization and BDNF, which have been shown previously to trigger this process in hippocampal cultures (Tongiorgi et al., 1997; Righi et al., 2000). We found that treatment with translin siRNA markedly reduces trafficking induced by KCl, as reported previously (Chiaruttini et al., 2009), but does not impair trafficking induced by BDNF (Figure 6).
Figure 6. Differential effect of translin siRNA on dendritic trafficking of BDNF mRNA induced by BDNF or KCl.
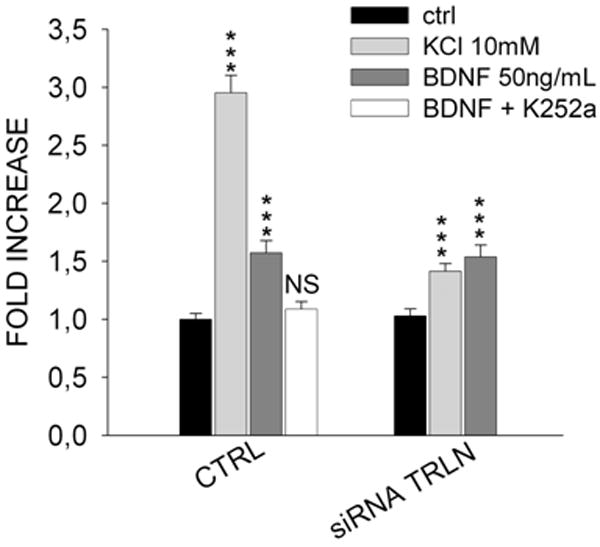
Treatment of rat hippocampal cultures with translin siRNA markedly decreases the ability of KCl to stimulate dendritic trafficking of BDNF mRNA, but does not impair the response to BDNF (50ng/ml). The effects of these stimuli are plotted as the fold increase in the relative dendritic filling value determined for each experimental condition. The specificity of the BDNF response is confirmed by its blockade by pre-treatment with K252a (30nM, 30 minutes), a selective antagonist of TrkB tyrosine kinase activity. Asterisks indicate significant difference (p<0.001) compared to control values.
Discussion
These studies indicate that the dependence of dendritic trafficking of BDNF mRNA on translin varies with the stimulus used to activate this process. Our prior studies demonstrated that siRNA-mediated knockdown of translin produced a marked reduction in dendritic trafficking of BDNF mRNA in cultured hippocampal neurons (Chiaruttini et al., 2009). However, that study was restricted to examining dendritic trafficking triggered by KCl-induced depolarization. In this study, we wanted to check whether translin deletion also impaired dendritic trafficking of BDNF mRNA in hippocampal neurons in vivo. Consistent with this hypothesis, we found that the basal level of dendritic BDNF mRNA in CA1 pyramidal neurons is decreased in sections obtained from translin KO mice. However, contrary to this hypothesis, we found that the ability of pilocarpine treatment to induce dendritic trafficking of BDNF mRNA in CA1 pyramidal neurons persists in translin knockout mice. Thus, these in vivo results suggest that the requirement for translin to mediate dendritic trafficking of BDNF mRNA depends on the stimulus driving this process. To test this inference, we have compared the impact of siRNA translin on dendritic trafficking elicited by KCl and BDNF in hippocampal cultures. These in vitro studies revealed that translin siRNA blocks dendritic trafficking of BDNF mRNA induced by KCl but not by BDNF. Taken together, these in vivo and in vitro studies imply that dendritic trafficking of BDNF mRNA, and presumably other dendritic transcripts, is mediated by multiple signaling pathways that respond to different patterns of extracellular stimuli.
These studies also provide direct evidence that the translin/trax complex undergoes activity-dependent translocation into dendrites. Under basal conditions, translin and trax immunostaining is barely detectable in apical dendrites of CA1 pyramidal neurons. However, dendritic staining is readily apparent following pilocarpine treatment. Nevertheless, it is important to note that the observed translocation of translin and trax into dendrites in response to pilocarpine is not required to mediate dendritic trafficking of BDNF mRNA, as the latter persists in translin KO mice.
In a previous study, we showed that both translin siRNA and introduction of a well-studied single nucleotide polymorphism (SNP), G196A, located in the coding region of BDNF mRNA blocked dendritic translocation of this transcript in vitro (Chiaruttini et al., 2009). Furthermore, the ability of the translin/trax complex to bind to a short segment of the coding region containing this SNP was reduced by introduction of this mutation. Accordingly, we proposed that this segment of RNA might contain a binding site for the translin/trax complex. In addition, we found that knock-in mice with the G196A SNP showed impaired dendritic trafficking of BDNF mRNA in response to pilocarpine. Thus, the results of this study indicating that translin deletion does not block the ability of pilocarpine to induce dendritic translocation of BDNF mRNA appear, at first glance, to be at odds with the previously proposed model which states that the translin/trax complex mediates dendritic trafficking of BDNF mRNA by binding to a cis element containing the G196A site. However, it would be premature to reject this model at this point in light of the following considerations. First, it is possible that translin acts at the G196A site to mediate dendritic trafficking mediated by basal neuronal activity in vivo or KCl treatment in vitro, but that it can be replaced by another trans factor following pilocarpine stimulation in vivo or BDNF treatment in vitro. Second, one also needs to take into account that the G196A SNP, which is located in the coding region, markedly reduces regulated secretion of BDNF (Egan et al., 2003; Chen et al., 2004). As it is well established that activation of acetylcholine muscarinic receptors by pilocarpine leads to BDNF mRNA and protein up-regulation and increased TrkB activation in the rat hippocampus in vivo (Knipper et al., 1994; Scharfman et al., 1999; Roberts et al., 2006; He et al., 2010), the G196A SNP may also block pilocarpine-induced dendritic trafficking of BDNF mRNA via its effect on BDNF secretion. Thus, the G196A SNP may produce a more severe impairment in dendritic localization of BDNF mRNA than translin deletion because it can affect both secretion of BDNF protein as well as the ability of trans factors, including translin, to bind to the transcript.
Recent studies performed in drosophila have provided a new perspective on the function of the translin/trax complex (Liu et al., 2009). In trying to reconstitute RNAi-mediated cleavage of target transcripts in vitro, they found that addition of recombinant translin/trax complex facilitates the ability of siRNA to activate this response. Furthermore, their results indicate that the translin/trax complex contains nuclease activity towards single-stranded RNA that mediates removal of the passenger strand of the siRNA duplex from the guide strand that is loaded onto the RISC complex. In pilot experiments, we have verified that the recombinant rat translin/trax complex displays a comparable nuclease activity (Z. Li, personal communication). Accordingly, it will be interesting to determine in future studies how the nuclease activity of the translin/trax complex is related to its role in mRNA transport into dendrites. One scenario which can explain both sets of observations is based on the ability of translin to form two types of complexes with RNA binding activity: homomeric translin complexes and heteromeric translin/trax complexes. Interestingly, Liu et al. (2009) found that only the latter possess nuclease activity, while homomeric translin complexes do not. Thus, it is possible that homomeric translin complexes mediate transport of dendritic mRNAs, as found for BDNF mRNA, and that heteromeric translin/trax complexes are involved in processing siRNA duplexes as found in drosophila. Alternatively, it is conceivable that heteromeric translin/trax complexes mediate dendritic trafficking of mRNAs, but that its nuclease activity is suppressed during mRNA transport. Accordingly, it will be of interest in future studies to test these models of translin’s dual role in mRNA transport and silencing.
Supplementary Material
Acknowledgments
This work was supported by PRIN (E.T.), National Alliance for Research on Schizophrenia and Depression (Z.L.) and National Institute on Drug Abuse (J.M.B.). We thank Dr. David Ginty for helpful discussions and Dr. Chung-Nan Sun for trax antibody purification and Dr. Junichiro Matsuda (NIBIO) for providing the translin ko mice.
References
- An JJ, Gharami K, Liao GY, Woo NH, Lau AG, Vanevski F, Torre ER, Jones KR, Feng Y, Lu B, Xu B. Distinct role of long 3’ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell. 2008;134:175–187. doi: 10.1016/j.cell.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K, Ishida R, Kasai M. Isolation and characterization of a cDNA encoding a Translin-like protein, TRAX. FEBS Lett. 1997;401:109–112. doi: 10.1016/s0014-5793(96)01444-5. [DOI] [PubMed] [Google Scholar]
- Chen ZY, Patel PD, Sant G, Meng CX, Teng KK, Hempstead BL, Lee FS, et al. Variant brain-derived neurotrophic factor (BDNF) (Met66) alters the intracellular trafficking and activity-dependent secretion of wild-type BDNF in neurosecretory cells and cortical neurons. J Neurosci. 2004;24:4401–4411. doi: 10.1523/JNEUROSCI.0348-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chennathukuzhi V, Stein JM, Abel T, Donlon S, Yang S, Miller JP, Allman DM, Simmons RA, Hecht NB. Mice deficient for testis-brain RNA-binding protein exhibit a coordinate loss of TRAX, reduced fertility, altered gene expression in the brain, and behavioral changes. Mol Cell Biol. 2003;23:6419–6434. doi: 10.1128/MCB.23.18.6419-6434.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaruttini C, Sonego M, Baj G, Simonato M, Tongiorgi E. BDNF mRNA splice variants display activity-dependent targeting to distinct hippocampal laminae. Mol Cell Neurosci. 2008;37:11–19. doi: 10.1016/j.mcn.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Chiaruttini C, Vicario A, Li Z, Baj G, Braiuca P, Wu Y, Lee FS, Gardossi L, Baraban JM, Tongiorgi E. Dendritic trafficking of BDNF mRNA is mediated by translin and blocked by the G196A (Val66Met) mutation. Proc Natl Acad Sci U S A. 2009;106:16481–16486. doi: 10.1073/pnas.0902833106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberwine J, Belt B, Kacharmina JE, Miyashiro K. Analysis of subcellularly localized mRNAs using in situ hybridization, mRNA amplification, and expression profiling. Neurochem Res. 2002;10:1065–1077. doi: 10.1023/a:1020956805307. [DOI] [PubMed] [Google Scholar]
- Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, Bertolino A, Zaitsev E, Gold B, Goldman D, Dean M, Lu B, Weinberger DR. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112:257–269. doi: 10.1016/s0092-8674(03)00035-7. [DOI] [PubMed] [Google Scholar]
- Finkenstadt PM, Kang WS, Jeon M, Taira E, Tang W, Baraban JM. Somatodendritic localization of Translin, a component of the Translin/Trax RNA binding complex. J Neurochem. 2000;75:1754–1762. doi: 10.1046/j.1471-4159.2000.0751754.x. [DOI] [PubMed] [Google Scholar]
- Finkenstadt PM, Jeon M, Baraban JM. Masking of the Translin/Trax complex by endogenous RNA. FEBS Lett. 2001;498:6–10. doi: 10.1016/s0014-5793(01)02470-x. [DOI] [PubMed] [Google Scholar]
- Finkenstadt PM, Jeon M, Baraban JM. Trax is a component of the Translin-containing RNA binding complex. J Neurochem. 2002;83:202–210. doi: 10.1046/j.1471-4159.2002.01158.x. [DOI] [PubMed] [Google Scholar]
- Fukuda Y, Ishida R, Aoki K, Nakahara K, Takashi T, Mochida K, Suzuki O, Matsuda J, Kasai M. Contribution of Translin to hemapoietic regeneration after sublethal ionizing irradiation. Biol Pharm Bull. 2008;31:207–211. doi: 10.1248/bpb.31.207. [DOI] [PubMed] [Google Scholar]
- He XP, Pan E, Sciarretta C, Minichello L, McNamara JO. Disruption of TrkB-mediated phospholipase Cγ signaling inhibits limbic epileptogenesis. J Neurosci. 2010;30:6188–6196. doi: 10.1523/JNEUROSCI.5821-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang EJ, Reichardt LF. Neurotrophins: roles in neuronal development and function. Annu Rev Neurosci. 2001;24:677–736. doi: 10.1146/annurev.neuro.24.1.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindler S, Wang H, Richter D, Tiedge H. RNA transport and local control of translation. Annu Rev Cell Dev Biol. 2005;21:223–245. doi: 10.1146/annurev.cellbio.21.122303.120653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipper M, da Penha Berzaghi M, Blöchl A, Breer H, Thoenen H, Lindholm DP. Positive feedback between acetylcholine and the neurotrophins nerve growth factor and brain-derived neurotrophic factor in the rat hippocampus. Eur J Neurosci. 1994;6:668–71. doi: 10.1111/j.1460-9568.1994.tb00312.x. [DOI] [PubMed] [Google Scholar]
- Li Z, Baraban JM. High affinity binding of the Translin/Trax complex to RNA does not require the presence of Y or H elements. Mol Brain Res. 2004;120:123–129. doi: 10.1016/j.molbrainres.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Li Z, Wu Y, Baraban JM. The Translin/Trax RNA binding complex: clues to function in the nervous system. Biochim Biophys Acta. 2008;1779:479–485. doi: 10.1016/j.bbagrm.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Ye X, Jiang F, Liang C, Chen D, Peng J, Kinch LN, Grishin NV, Liu Q. C3PO, an endoribonuclease that promotes RNAi by facilitating RISC activation. Science. 2009;325:750–753. doi: 10.1126/science.1176325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Christian K, Lu B. BDNF: a key regulator for protein synthesis-dependent LTP and long-term memory? Neurobiol Learning and Mem. 2008;89:312–323. doi: 10.1016/j.nlm.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattabiraman PP, Tropea D, Chiaruttini C, Tongiorgi E, Cattaneo A, Domenici L. Neuronal activity regulates the developmental expression and subcellular localization of cortical BDNF mRNA isoforms in vivo. Mol Cell Neurosci. 2005;28:556–570. doi: 10.1016/j.mcn.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Poo MM. Neurotrophins as synaptic modulators. Nat Rev Neurosci. 2001;2:24–32. doi: 10.1038/35049004. [DOI] [PubMed] [Google Scholar]
- Righi M, Tongiorgi E, Cattaneo A. Brain-derived neurotrophic factor (BDNF) induces dendritic targeting of BDNF and tyrosine kinase B mRNAs in hippocampal neurons through a phosphatidylinositol-3 kinase-dependent pathway. J Neurosci. 2000;20:3165–3174. doi: 10.1523/JNEUROSCI.20-09-03165.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DS, Hu Y, Lund IV, Brooks-Kayla AR, Russek SJ. Brain-derived neurotrophic factor (BDNF)-induced synthesis of early growth response factor 3 (Egr3) controls the levels of type A GABA receptor a4 subunits in hippocampal neurons. J Biol Chem. 2006;281:29431–29435. doi: 10.1074/jbc.C600167200. [DOI] [PubMed] [Google Scholar]
- Scharfman HE, Goodman JH, Sollas AL. Actions of brain-derived neurotrophic factor in slices from rats with spontaneous seizures and mossy fiber sprouting in the dentate gyrus. J Neurosci. 1999;19:5619–5631. doi: 10.1523/JNEUROSCI.19-13-05619.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuman EM, Dynes JL, Steward O. Synaptic regulation of translation of dendritic mRNAs. J Neurosci. 2006;26:7143–7146. doi: 10.1523/JNEUROSCI.1796-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun CN, Cheng HC, Chou JL, Lee SY, Lin YW, Lai HL, Chen HM, Chern Y. Rescue of p53 blockage by the A(2A) adenosine receptor via a novel interacting protein, translin-associated protein X. Mol Pharmacol. 2006;70:454–466. doi: 10.1124/mol.105.021261. [DOI] [PubMed] [Google Scholar]
- Sutton MA, Schuman EM. Dendritic protein synthesis, synaptic plasticity, and memory. Cell. 2006;127:49–58. doi: 10.1016/j.cell.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Taira E, Finkenstadt PM, Baraban JM. Identification of Translin and Trax as components of the GS1 strand-specific DNA binding complex enriched in brain. J Neurochem. 1998;71:471–477. doi: 10.1046/j.1471-4159.1998.71020471.x. [DOI] [PubMed] [Google Scholar]
- Thoenen H. Neurotrophins and activity-dependent plasticity. Prog Brain Res. 2000;128:183–191. doi: 10.1016/S0079-6123(00)28016-3. [DOI] [PubMed] [Google Scholar]
- Tongiorgi E, Righi M, Cattaneo A. Activity-dependent dendritic targeting of BDNF and TrkB mRNAs in hippocampal neurons. J Neurosci. 1997;17:9492–9505. doi: 10.1523/JNEUROSCI.17-24-09492.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tongiorgi E, Armellin M, Giulianini PG, Bregola G, Zucchini S, Paradiso B, Steward O, Cattaneo A, Simonato M. Brain-derived neurotrophic factor mRNA and protein are targeted to discrete dendritic laminas by events that trigger epileptogenesis. J Neurosci. 2004;24:6842–6852. doi: 10.1523/JNEUROSCI.5471-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tongiorgi E. Activity-dependent expression of brain-derived neurotrophic factor in dendrites: facts and open questions. Neurosci Res. 2008;61:335–346. doi: 10.1016/j.neures.2008.04.013. [DOI] [PubMed] [Google Scholar]
- Turski L, Ikonomidou C, Turski WA, Bortolotto ZA, Cavalheiro EA. Review: cholinergic mechanisms and epileptogenesis. The seizures induced by pilocarpine: a novel experimental model of intractable epilepsy. Synapse. 1989;3:154–171. doi: 10.1002/syn.890030207. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


