Abstract
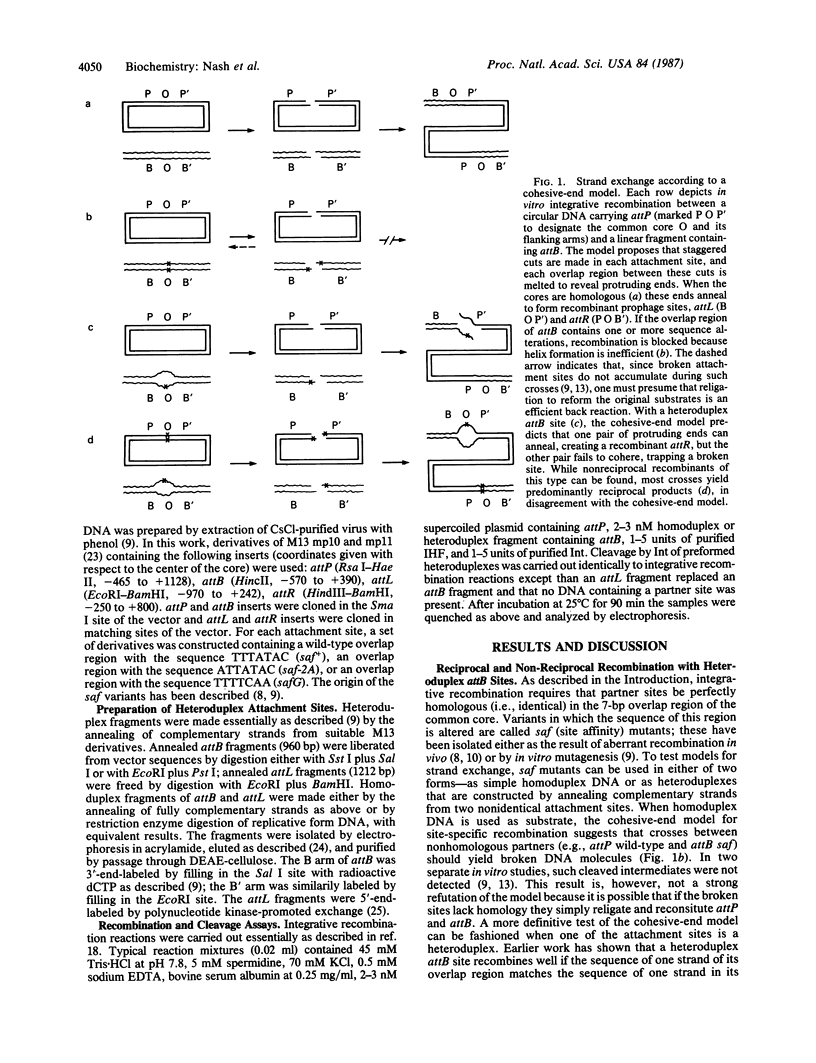
Bacteriophage lambda integration and excision take place at specific loci called attachment sites. Earlier work has shown that efficient recombination requires the identical sequence to be present in both attachment sites throughout the seven-base-pair region between the points of strand exchange. A plausible model for the role of homology postulates that Int, the site-specific recombinase, makes double-strand breaks at attachment sites such that each broken end has a short single-strand protrusion. Recombination would then depend upon the capacity of these protrusions to form Watson-Crick helices--i.e., to anneal--a process that might require perfect complementarity between the cohesive ends. To test this model, we have studied Int-promoted crosses in which one attachment site is a heteroduplex. Specifically, we constructed sites in which the seven-base-pair region between the points of strand exchange contains one or more noncomplementary pairs. The double-strand break and annealing mechanism predicts that crosses with these heteroduplex sites should yield one completed recombinant and one broken site. We find that such nonreciprocal recombination is uncommon and that the typical outcome of crosses involving a heteroduplex site is a reciprocal recombinant in which both products are resealed. Moreover, the occasional appearance of nonreciprocal products can be explained by our finding that Int can cleave heteroduplex attachment sites after recombination is completed. Taken together, our data strongly indicate that bacteriophage lambda recombination does not proceed by the homology-dependent annealing of cohesive ends; acceptable alternatives for the role of homology are discussed.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bauer C. E., Gardner J. F., Gumport R. I. Extent of sequence homology required for bacteriophage lambda site-specific recombination. J Mol Biol. 1985 Jan 20;181(2):187–197. doi: 10.1016/0022-2836(85)90084-1. [DOI] [PubMed] [Google Scholar]
- Bauer C. E., Hesse S. D., Gardner J. F., Gumport R. I. DNA interactions during bacteriophage lambda site-specific recombination. Cold Spring Harb Symp Quant Biol. 1984;49:699–705. doi: 10.1101/sqb.1984.049.01.079. [DOI] [PubMed] [Google Scholar]
- Craig N. L., Nash H. A. The mechanism of phage lambda site-specific recombination: site-specific breakage of DNA by Int topoisomerase. Cell. 1983 Dec;35(3 Pt 2):795–803. doi: 10.1016/0092-8674(83)90112-5. [DOI] [PubMed] [Google Scholar]
- Echols H., Green L. Some properties of site-specific and general recombination inferred from int-initiated exchanges by bacteriophage lambda. Genetics. 1979 Oct;93(2):297–307. doi: 10.1093/genetics/93.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enquist L. W., Nash H., Weisberg R. A. Strand exchange in site-specific recombination. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1363–1367. doi: 10.1073/pnas.76.3.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner J. F., Nash H. A. Role of Escherichia coli IHF protein in lambda site-specific recombination. A mutational analysis of binding sites. J Mol Biol. 1986 Sep 20;191(2):181–189. doi: 10.1016/0022-2836(86)90255-x. [DOI] [PubMed] [Google Scholar]
- Honigman A., Hu S. L., Szybalski W. Regulation of integration by coliphage lambda: activation of int transcription by the cII and cIII proteins. Virology. 1979 Jan 30;92(2):542–556. doi: 10.1016/0042-6822(79)90156-9. [DOI] [PubMed] [Google Scholar]
- Hsu P. L., Landy A. Resolution of synthetic att-site Holliday structures by the integrase protein of bacteriophage lambda. Nature. 1984 Oct 25;311(5988):721–726. doi: 10.1038/311721a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda H., Moriya K., Matsumoto T. In vitro study of illegitimate recombination: involvement of DNA gyrase. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):399–408. doi: 10.1101/sqb.1981.045.01.054. [DOI] [PubMed] [Google Scholar]
- Kaiser A. D., Wu R. Structure and function of DNA cohesive ends. Cold Spring Harb Symp Quant Biol. 1968;33:729–734. doi: 10.1101/sqb.1968.033.01.083. [DOI] [PubMed] [Google Scholar]
- Landy A., Ross W. Viral integration and excision: structure of the lambda att sites. Science. 1977 Sep 16;197(4309):1147–1160. doi: 10.1126/science.331474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Miller H. I. Primary structure of the himA gene of Escherichia coli: homology with DNA-binding protein HU and association with the phenylalanyl-tRNA synthetase operon. Cold Spring Harb Symp Quant Biol. 1984;49:691–698. doi: 10.1101/sqb.1984.049.01.078. [DOI] [PubMed] [Google Scholar]
- Nash H. A., Mizuuchi K., Enquist L. W., Weisberg R. A. Strand exchange in lambda integrative recombination: genetics, biochemistry, and models. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):417–428. doi: 10.1101/sqb.1981.045.01.056. [DOI] [PubMed] [Google Scholar]
- Nash H. A., Pollock T. J. Site-specific recombination of bacteriophage lambda. The change in topological linking number associated with exchange of DNA strands. J Mol Biol. 1983 Oct 15;170(1):19–38. doi: 10.1016/s0022-2836(83)80225-3. [DOI] [PubMed] [Google Scholar]
- Nash H. A. Purification and properties of the bacteriophage lambda Int protein. Methods Enzymol. 1983;100:210–216. doi: 10.1016/0076-6879(83)00057-9. [DOI] [PubMed] [Google Scholar]
- Nash H. A., Robertson C. A. Purification and properties of the Escherichia coli protein factor required for lambda integrative recombination. J Biol Chem. 1981 Sep 10;256(17):9246–9253. [PubMed] [Google Scholar]
- Ross W., Landy A. Patterns of lambda Int recognition in the regions of strand exchange. Cell. 1983 May;33(1):261–272. doi: 10.1016/0092-8674(83)90355-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski P. Site-specific recombinases: changing partners and doing the twist. J Bacteriol. 1986 Feb;165(2):341–347. doi: 10.1128/jb.165.2.341-347.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman M., Gottesman M. Attachment site mutants of bacteriophage lambda. J Mol Biol. 1973 Dec 25;81(4):461–482. doi: 10.1016/0022-2836(73)90517-2. [DOI] [PubMed] [Google Scholar]
- Signer E. R., Weil J. Site-specific recombination in bacteriophage lambda. Cold Spring Harb Symp Quant Biol. 1968;33:715–719. doi: 10.1101/sqb.1968.033.01.081. [DOI] [PubMed] [Google Scholar]
- Weisberg R. A., Enquist L. W., Foeller C., Landy A. Role for DNA homology in site-specific recombination. The isolation and characterization of a site affinity mutant of coliphage lambda. J Mol Biol. 1983 Oct 25;170(2):319–342. doi: 10.1016/s0022-2836(83)80151-x. [DOI] [PubMed] [Google Scholar]
- de Massy B., Studier F. W., Dorgai L., Appelbaum E., Weisberg R. A. Enzymes and sites of genetic recombination: studies with gene-3 endonuclease of phage T7 and with site-affinity mutants of phage lambda. Cold Spring Harb Symp Quant Biol. 1984;49:715–726. doi: 10.1101/sqb.1984.049.01.081. [DOI] [PubMed] [Google Scholar]