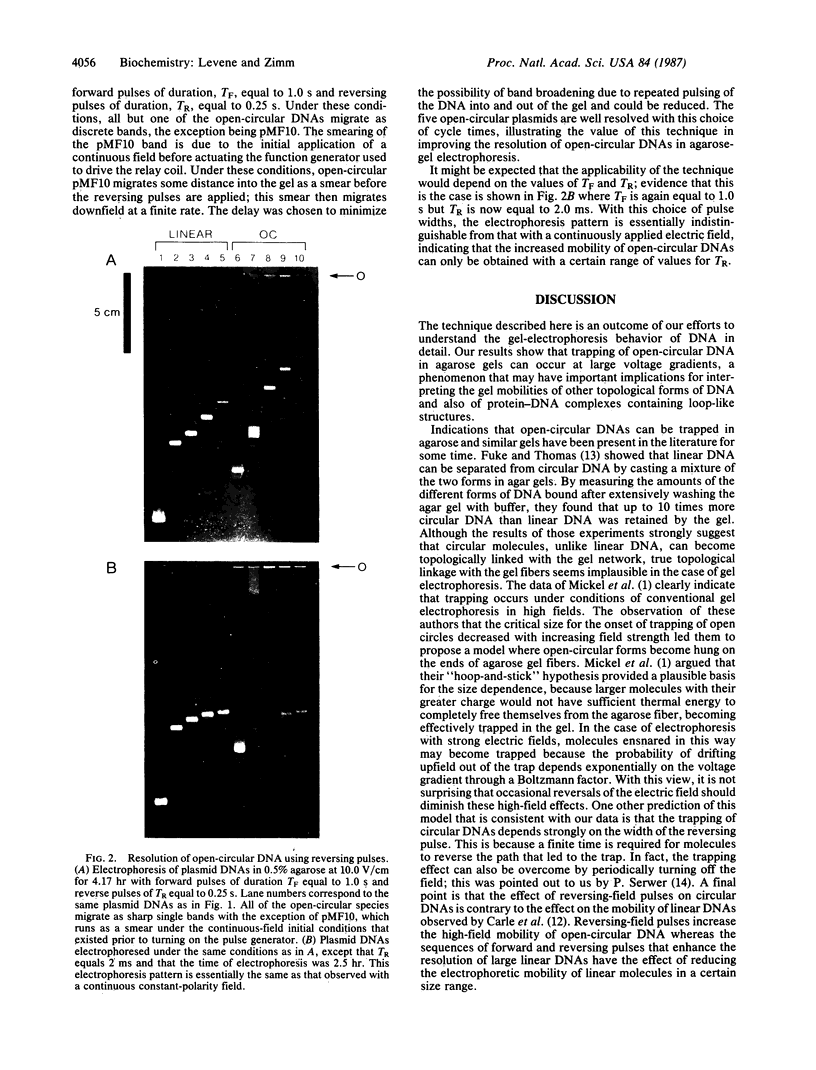
Abstract
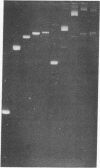
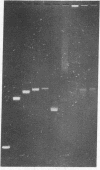
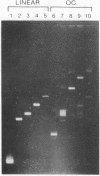
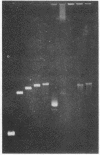
The effect of high electric fields on the gel-electrophoretic mobility of open-circular DNA in agarose differs dramatically from that on linear molecules of the same molecular weight. At high fields, sufficiently large circular forms are prevented from migrating into the gel whereas linear molecules and smaller circular DNAs migrate normally. This effect is strongly field dependent, affecting circular molecules of decreasing size with increasing field strength. We have studied this effect with a series of plasmid DNAs ranging from 2.9 to 56 kilobase pairs using continuous and reversing-pulse electric fields. Application of reversing pulses abolishes the effect under certain conditions and supports the model for the gel electrophoresis of open-circular DNA where circular forms are trapped by engaging the free end of an agarose gel fiber.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Carle G. F., Frank M., Olson M. V. Electrophoretic separations of large DNA molecules by periodic inversion of the electric field. Science. 1986 Apr 4;232(4746):65–68. doi: 10.1126/science.3952500. [DOI] [PubMed] [Google Scholar]
- Fuke M., Thomas C. A., Jr Isolation of open-circular DNA molecules by retention in agar gels. J Mol Biol. 1970 Sep 14;52(2):395–397. doi: 10.1016/0022-2836(70)90040-9. [DOI] [PubMed] [Google Scholar]
- Johnson P. H., Grossman L. I. Electrophoresis of DNA in agarose gels. Optimizing separations of conformational isomers of double- and single-stranded DNAs. Biochemistry. 1977 Sep 20;16(19):4217–4225. doi: 10.1021/bi00638a014. [DOI] [PubMed] [Google Scholar]
- Lumpkin O. J., Déjardin P., Zimm B. H. Theory of gel electrophoresis of DNA. Biopolymers. 1985 Aug;24(8):1573–1593. doi: 10.1002/bip.360240812. [DOI] [PubMed] [Google Scholar]
- McDonell M. W., Simon M. N., Studier F. W. Analysis of restriction fragments of T7 DNA and determination of molecular weights by electrophoresis in neutral and alkaline gels. J Mol Biol. 1977 Feb 15;110(1):119–146. doi: 10.1016/s0022-2836(77)80102-2. [DOI] [PubMed] [Google Scholar]
- Mickel S., Arena V., Jr, Bauer W. Physical properties and gel electrophoresis behavior of R12-derived plasmid DNAs. Nucleic Acids Res. 1977;4(5):1465–1482. doi: 10.1093/nar/4.5.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortle D., Nathans D. Local mutagenesis: a method for generating viral mutants with base substitutions in preselected regions of the viral genome. Proc Natl Acad Sci U S A. 1978 May;75(5):2170–2174. doi: 10.1073/pnas.75.5.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Measurement of DNA length by gel electrophoresis. Anal Biochem. 1979 Dec;100(2):319–323. doi: 10.1016/0003-2697(79)90235-5. [DOI] [PubMed] [Google Scholar]
- Stellwagen N. C. Effect of the electric field on the apparent mobility of large DNA fragments in agarose gels. Biopolymers. 1985 Dec;24(12):2243–2255. doi: 10.1002/bip.360241207. [DOI] [PubMed] [Google Scholar]