Abstract
Over the past years, increasing numbers of distinct subsets have been discovered and identified for a T lymphocytes’ entity. Differentiation and function of each T cell subset are controlled by a specific master transcription factor. Importantly, Runt-related transcription factors, particularly Runx1 and Runx3, interplay with these master regulators in various aspects of T cells’ immunity. In this review article, we first explain roles of Th-Pok and Runx3 in differentiation of CD4 versus CD8 single positive cells, and later focus on cross-regulation of Th-Pok and Runx3 and their relationship with other factors such as TCR strength. Next, we provide evidences for the direct interplay of Runx1/3 with T-bet and GATA3 during Th1 versus Th2 commitment to activate or silence transcription of signature cytokine genes, IFNγ and IL4. Lastly, we explain feed-forward relationship between Runx1 and Foxp3 and discuss roles of Runx1 in regulatory T cells’ suppressive activity. This review highlights an essential importance of Runx molecules in controlling various T cell subsets’ differentiation and functions through molecular interplay with the master transcription factors in terms of protein-protein interaction as well as regulation of gene expression.
Keywords: cell differentiation, gene expression, gene targeting, T lymphocytes, transcription factor
Introduction
In this brief review article, we focus on the role of Runt-related transcription factors, Runx, in the differentiation and function of T lymphocytes. Runx has emerged as one of the most important regulatory factors in T-cell immunity. The Runx family is composed of three members, Runx1, Runx2 and Runx3, each of which forms a functional complex with a core binding factor β (Cbfβ) partner protein. Runx1 and Runx3 are known to be involved in T-cell immunity. T lymphocytes differentiate into subsets with distinct functions. In some T-cell subsets, Runx1 and Runx3 are equivalently expressed and exhibit redundant activities. However, in other T-cell subsets, Runx1 and Runx3 exert distinct functions. These differences depend mainly on the unique expression patterns of these proteins in a particular T-cell subset, but qualitative differences in Runx1 and Runx3 may also contribute to their differential activity in subsets of T cells. In a previously published review article, we described these common and distinct features of Runx factors that are relevant to each step of T-cell differentiation.1 In the present review, we emphasize the roles of Runx factors in three biologically and clinically important aspects of T-cell differentiation/function. These include lineage selection of helper versus killer cells, and T helper differentiation into various T helper cell and regulatory T-cell subsets.
Roles of Runx3 and Th-POK in killer versus helper lineage commitment
CD4+ CD8+ double-positive (DP) thymocytes ultimately differentiate into two lineages, CD4+ CD8− single-positive (CD4SP) helper T cells and CD4− CD8+ single-positive (CD8SP) killer T cells. Extensive studies of the molecular mechanisms and gene regulation involved in this lineage determination have led to the discovery of T helper-inducing POZ-Krüppel-like factor (Th-POK) as a key transcription factor in CD4SP commitment,2,3 and of Runx3 in CD8SP commitment.4–6 Furthermore, recent works have revealed that the interplay between Th-POK and Runx3 is pivotal in this thymocyte lineage determination process (Fig. 1).
Figure 1.
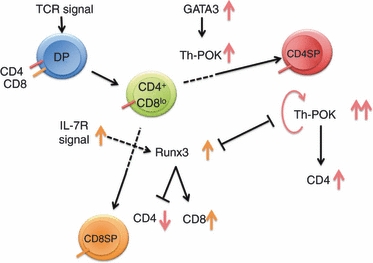
Roles of Runx3 and T helper-inducing POZ-Krüppel-like factor (Th-POK) in the differentiation of CD4 single-positive (SP) and CD8SP thymocytes. Double-positive (DP) cells first move to the CD4+ CD8lo stage, after which they differentiate into the CD4SP lineage if they receive a stronger/longer T-cell receptor (TCR) signal. Induction of Th-POK via GATA3 and maintenance of Th-POK through a positive auto-regulation mechanism are important in this step. If cells receive a weaker/shorter TCR signal, they differentiate into the CD8SP lineage. The interleukin-7 receptor (IL-7R) signal is somehow linked to the TCR signal for the induction of Runx3 expression. Runx3 then suppresses CD4 expression and simultaneously up-regulates CD8 expression. Runx3 and Th-POK mutually repress each other’s expression.
Th-POK in CD4SP differentiation
In 1998, a naturally occurring mutant mouse strain termed helper deficient (HD) was reported, and was so called because these mice lack CD4SP helper T cells.7 In these mice, MHC class II-restricted thymocytes that would ordinarily become CD4SP are re-directed into the CD8SP lineage. The HD mutation does not simply impair the differentiation of CD4SP cells but perturbs the choice between CD4SP and CD8SP lineages. In 2005, it was discovered that the HD phenotype could be attributed to a point mutation in the Zbtb7 gene, which encodes a zinc finger-containing transcription factor.2,3 This gene, also known as cKrox or Zfp67, is now most often referred to as Th-POK. Expression of Th-POK is limited to the CD4SP lineage, but is not detected in double negative (DN), DP and CD8SP cells. Over-expression of Th-POK can force class-I restricted cells that are destined to become CD8SP to re-differentiate into the CD4SP lineage. These observations, together with the phenotype of HD mice, strongly indicate that Th-POK is a master regulator of CD4SP differentiation.
Runx3 in CD8SP differentiation
The Runx3 transcription factor, on the other hand, is a master regulator of CD8SP thymocyte differentiation. Runx3 deficiency causes a reduction in CD8SP thymocytes; however, this decrease is not as drastic as the reduction of CD4SP seen in Th-POK-deficient thymuses. In wild-type thymuses, expression of Runx3 protein is confined to the CD8SP subset, but Runx1 can be detected in all DN, DP, CD4SP and CD8SP cells.5 Therefore, lack of Runx3 in CD8SP cells probably causes a compensatory increase in Runx1 expression, thereby preventing a severe reduction in CD8SP in Runx3-deficient thymuses. When DP cells move to the CD8SP lineage, Runx3 suppresses CD4 expression by binding to the silencer region of the gene.4 It must be noted that along the DP to CD8SP pathway, expression of CD8 is first down-regulated (the CD4+ CD8lo stage), but is subsequently re-activated accompanying CD4 repression (the CD4− CD8+ stage). Runx3 mediates the re-activation of CD8 expression by binding to the enhancer region of the E8I gene.5 Hence, Runx3 regulates CD4 and CD8 expression negatively and positively, respectively, and provides a basis for mutually exclusive expression of CD4 and CD8 in the CD8SP subset.
Cross-regulation of Runx3 and Th-POK
In addition to regulating CD4 and CD8 expression, Runx3 can also indirectly regulate these genes by controlling Th-POK expression. Runx3 suppresses Th-POK expression by binding to its silencer region, which maps to a 3.1 kb upstream region of exon Ia (called RBS-1)8 containing a Runx consensus site, or to a distal responsive element.9 In cases of Runx3 deficiency, Th-POK expression is therefore de-repressed, which contributes to the re-direction of class I-restricted or CD8SP-oriented cells into the CD4SP lineage. Conversely, Th-POK can suppress Runx3 expression by binding to the Runx3 distal promoter.10,11 Therefore, both Th-POK and Runx3 are negative regulators of each other’s expression.10
This poses the question about what would happen to T-cell differentiation if neither Th-POK nor Runx3 were present? Unexpectedly, a substantial number of CD4SP and CD8SP cells are detected in both Th-POK- and Runx3-targeted thymuses.10 Therefore, Th-POK is probably not the sole master regulating the choice between the CD4 and CD8 lineage. In addition, the differentiation into the CD8SP lineage is probably not a default step that occurs automatically without the guidance of transcription factors. Runx3 appears to have functional significance other than its role in silencing Th-POK expression.
Various lines of evidence have indicated that the strength and duration of T-cell receptor (TCR) signals determines the lineage selection of DP thymocytes. Namely, a stronger and longer signal induces CD4SP differentiation, whereas a weaker and shorter signal results in CD8SP differentiation.12 Therefore, it is reasonable to link the stronger/longer TCR signal to Th-POK expression and the weaker/shorter signal to Runx3 expression. Expression of Th-POK protein inside cells is primarily determined at the transcriptional level. Th-POK transcription is initially induced by GATA3, whereas maintenance of Th-POK transcription relies on the Th-POK protein itself (positive auto-regulation).11,13 Therefore, the TCR signal together with GATA3 may induce Th-POK expression, while the strength/length of the TCR signal may stabilize Th-POK expression. In contrast, interleukin-7 receptor (IL-7R) and its downstream signalling molecule STAT5 are reported to induce Runx3 expression.14 It is not known at present how IL-7R signalling is linked to the weaker/shorter TCR signal.
In essence, Runx3 and Th-POK not only function antagonistically in the regulation of CD4/CD8 expression but also exert mutually suppressive activity on their own expression (Fig. 1). However, the activity attributed to Runx3 and Th-POK is necessary but not sufficient for lineage selection. For example, Runx3-binding to RBS-1 does not guarantee suppression of Th-POK expression.8 This suggests that some unknown factor other than Runx3/Th-POK might be involved in CD4SP/CD8SP lineage selection.
Roles of Runx, T-bet and GATA3 in T helper type 1 versus type 2 commitment
When encountering foreign antigens, peripheral CD4+ T cells initiate differentiation towards T helper type 1 (Th1), Th2, or other helper lineages, depending on the types of antigens encountered. The Th1 and Th2 phenotypes are characterized by the secretion of the representative cytokines interferon-γ (IFN-γ) and IL-4, which are controlled by their master regulators, the T-bet and GATA3 transcription factors, respectively. Recent advances have identified Runx factors as important regulators of T helper differentiation (Fig. 2).
Figure 2.
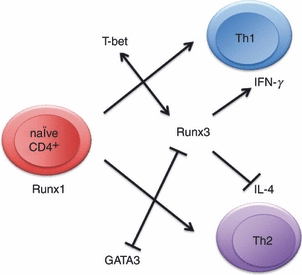
Roles of Runx factors in T helper differentiation. In naive CD4+ cells, Runx1 is detected, but its expression is down-regulated following T-cell receptor stimulation. Artificial abrogation of Runx1 skews the cells to a T helper type 2 (Th2) phenotype. However, Runx3 is induced in the Th1 differentiation pathway, and the Runx3–T-bet complex functions to enhance IFNγ and repress IL4 expression. Runx3 also interacts with and attenuates the activity of GATA3.
Expression profiles of Runx proteins during T helper differentiation
Runx1, but not Runx3, is found in naive CD4+ T cells whose TCR stimulation immediately down-regulates Runx1 protein expression.15 Interestingly, when cells are cultivated under conditions that skew their T helper differentiation, Runx1 is re-activated or the expression of Runx3 is newly induced. Hence, Th1-committed cells express only Runx3, whereas Th2 cells express both Runx1 and Runx3.15–18
Runx1 represses GATA3 and IL4 expression in naive CD4+ cells
Is modulation of the expression pattern of Runx an obligatory step for T helper differentiation? A potential answer to this question has been furnished by a report about Runt-transgenic cells.15 The Runt domain is the DNA-binding domain of Runx, and functions in a dominant negative fashion against the intact Runx protein. Upon TCR activation, the differentiation of Runt-transgenic CD4+ cells is more skewed towards the Th2 phenotype than that of non-transgenic cells. This effect has been attributed to the up-regulation of GATA3 expression. However, transduction of Runx1 into naive wild-type cells attenuates Th2 skewing, and is accompanied by cessation of GATA3 expression. Runx1 also affects Th differentiation by binding to a DNaseI hypersensitive site IV in the IL4 silencer, thereby repressing IL4 expression.16 Therefore, a drop in the level of Runx1 expression after TCR stimulation might be a prerequisite for naive CD4+ cells to commence Th2 differentiation.
Runx3 and T-bet co-operatively augment Th1 differentiation
We next focus on the induction of Runx3 expression during T helper differentiation. When CD4+ cells are cultured under Th1-skewing conditions, T-bet is initially induced, and Runx3 is subsequently up-regulated in a T-bet-dependent manner.17 T-bet and Runx3 proteins have been shown to physically associate to form theT-bet/Runx3 complex which then binds to the IL4 silencer (hypersensitive site IV) to repress IL4 expression, and to the IFNγ promoter to enhance IFNγ expression. It is notable that Runx3 is up-regulated at a rather late stage of TCR activation under Th1-skewing conditions. A positive, feed-forward interplay of Runx3 and T-bet is therefore likely to be involved in enforcement of commitment to the Th1 lineage, but not in the determination of the lineage itself. Runx3 protein also appears to contribute to the strength of the Th1 phenotype by physically interacting with and attenuating the activity of GATA3 protein.18 In other words, GATA3 can block Runx3-mediated IFNγ expression by interacting with Runx3.19 In contrast to Th1 cells, the significance of Runx1 and Runx3 expression in Th2-skewed cells is not clear. One possibility is that Runx functions to down-modulate excessive Th2 reactivity.
Disturbance of the Th1/Th2 balance in Runx-modulated mice
Collectively, both Runx1 and Runx3 attenuate Th2 and favour Th1 phenotypes so Runx-abrogated mice with the Cbfbf/f;CD4-Cre-tg mutation are prone to Th2-type diseases such as bronchial asthma,16 in which inflammation along the airway and elevated serum levels of IgA, IgG1 and IgE (Th2-associated immunoglobulin subclasses) are observed. In contrast, when Runx3 is transduced into mice as a transgene, expression of Th1-type cytokines (IFN-γ, tumour necrosis factor-α, IL-12 and IL-18) is uniformly increased even in unstimulated CD4+ cells.18 Antigen injection of Runx3-tg mice results in higher serum levels of IgG2a and IgG2b (Th1-associated immunoglobulin subclasses).
Runx1 and Th17
Th17 is another distinct T helper subset into which CD4+ cells differentiate, and RORγt is a master regulator of this Th17 lineage.20 As for the involvement of Runx1 in Th17 differentiation, retroviral transduction of Runx1 has been shown to induce the Th17 phenotype, whereas Runx1 siRNA antagonizes Th17 differentiation.21 Runx1 appears to favour the Th17 phenotype by forming a complex with RORγt, and up-regulates IL17 expression by binding to its enhancer and promoter. Runx1 promotes RORγt expression as well. It remains to be seen if these observed effects of Runx1 on Th17 differentiation also hold in vivo, using Runx1-engineered mice.
Differentiation and function of regulatory T cells
Excessive immune activity against self or foreign antigens is suppressed by a subset of regulatory CD4+ T (Treg) cells whose activity prevents autoimmune disease by increasing tolerance towards the antigen. Treg cells, which are characterized by high expression of the Foxp3 transcription factor, arise naturally as Treg cells (nTreg) in the thymus, or as induced Treg cells (iTreg) in peripheral lymphoid tissues.22–24 A sustained high level of Foxp3 expression is vital for expression of various Treg-cell-associated genes as well as for differentiation and maintenance of Treg cells.25 Mutations in the Foxp3 gene lead to a catastrophic autoimmune disease termed IPEX (immune dysregulation, polyendocrinopathy, enteropathy, X-linked) syndrome in humans or to the scurfy phenotype in mice.26,27
Pathologies observed in mice with Runx1-deficient Treg cells
The function of the Runx1/Cbfβ complex in Treg cells has been intensively investigated. An in vitro study demonstrated that the physical interaction of Runx1 with FoxP3 is necessary for Treg cells to exert their suppressive activity on immunological reactions.28 Subsequently, two in vivo studies confirmed that the Runx1/Cbfβ complex is indispensable for Treg-cell function. In one report,29Runx1F/F:FIC and CbfβF/F:FIC mice were generated in which Runx1 or Cbfβ was targeted specifically in Treg cells. These mice develop gastritis with elevated serum IgE, autoantibodies, splenomegaly and lymphadenopathy. In another report,30Cbfβfl/fl:Foxp3YFP-cre mice harbouring Cbfβ-deleted Treg cells were generated. These mice showed lymphohistocyte and plasmacyte infiltration into multiple organs, and developed inflammatory diseases, such as pneumonitis and arteritis in the lungs. It is noteworthy that the pathologies observed in Treg-specific Runx1- and Cbfβ-deficient mice are similar to, but relatively milder than those seen in FoxP3-deficient mice.22
Runx1 is essential for the suppressive action of Treg cells
Is the occurrence of autoimmune disease in the aforementioned Treg-cell-specific, Runx1/Cbfβ-deleted mice attributable to compromised, suppressive Treg-cell activity? Adoptive transfer of Cbfβ-deficient Treg cells into severe combined immunodeficiency disorder (SCID) mice failed to prevent the development of CD4+ CD25− CD45RBhi cell-induced colitis.29 When cultured in vitro, Cbfβ-deficient Treg cells failed to suppress the proliferation of responder CD4+ T cells.29–31 Furthermore, knockdown of Runx1 with small interfering (si) RNA impaired the in vitro suppressive activity of human primary or induced Treg cells.28,32 These observations indicate that the abrogation of Runx1/Cbfβ impairs Treg-cell suppressive activity. Hence, in mice with Runx1- or Cbfβ-deleted Treg cells, the number of active CD4+ Foxp3− conventional T cells is increased.29,30 Such activated T cells tend to secrete pro-inflammatory cytokines such as IFN-γ and IL-4, which presumably drive vicious cycles of inflammation, eventually causing destructive hyper-reactivity.
Regulation of Foxp3 expression by Runx1
How does the Runx1/Cbfβ complex regulate Treg cells? Accumulated evidence supports a feed-forward relationship between Runx1/Cbfβ and Foxp3 (Fig. 3). Namely, Runx1/Cbfβ induces Foxp3 gene expression, and subsequently, Runx1/Cbfβ interacts with Foxp3 protein to regulate expression of Foxp3 itself as well as other Treg-cell-associated genes.32 We will first discuss the regulation of Foxp3 by Runx1/Cbfβ.
Figure 3.
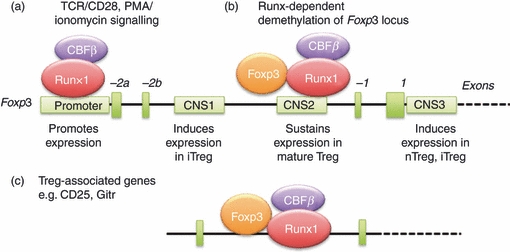
The Runx1/Cbfβ complex regulates Foxp3 expression in regulatory T (Treg) cells through a complicated feed-forward mechanisms. Foxp3 regulatory elements are schematically illustrated, including a promoter and three conserved non-coding sequences (CNS). CNS1 is important for induction of Foxp3 expression in induced Treg cells, CNS2 maintains sustained Foxp3 expression in mature Treg cells, and CNS3 induces Foxp3 expression in both natural Treg and induced Treg cells. Multiple Runx consensus sites have been identified in the promoter and CNS2 regions. (a) Runx1/Cbfβ binds to a Foxp3 promoter in response to T-cell receptor signalling. Other transcription factors such as NFAT and AP1 (not shown) are also involved in driving Foxp3 expression. (b) Runx1/Cbfβ participates in demethylation of the Foxp3 locus. Upon demethylation, Foxp3 protein forms a complex with Runx1/Cbfβ that then binds to the demethylated CNS2 region in mature Treg cells and maintains sustained Foxp3 expression. (c) Foxp3 together with Runx1/Cbfβ binds to and regulates the expression of Treg-associated target genes, such as CD25 and GITR. Dark green boxes: exons, light green boxes: regulatory DNA elements.
In peripheral iTreg cells derived from Cbfβfl/fl:Foxp3YFP-cre or CbfβF/F:FIC mice, expression of Foxp3 is substantially reduced.29,30 Similarly, treatment of human CD4+ CD25hi T cells with Runx1-siRNA significantly attenuated expression of Foxp3, and this effect was even stronger when cells were treated with both Runx1- and Runx3-siRNA.29,31 However, retroviral transduction of Runx3 into naive CD4+ T cells can elevate Foxp3 protein levels.32 Above, we discussed the indispensable role of Runx/Cbfβ in Treg-cell suppressive activity. Interestingly, transduction of Foxp3 into Cbfβ-deficient Treg cells restores their suppressive activity30, suggesting that Runx/Cbfβ contributes to Treg-cell function indirectly through the regulation of Foxp3 gene expression.
Foxp3 gene transcription is controlled by a promoter region (Fig. 3a) and by three conserved non-coding sequences (CNS) in introns (Fig. 3b).33 The Foxp3 promoter contains three putative Runx sites at 333, 287 and 53 bp upstream of the transcriptional start site. Runx1 binds to these sites as demonstrated by promoter enzyme immunoassay, promoter tiling and chromatin immunoprecipitation assays.29–32 In a luciferase reporter assay using PMA/ionomycin-stimulated human CD4+ T cells, transduction of Runx1 greatly enhanced Foxp3 promoter activity, whereas mutation of the Runx consensus sites abrogated that activity.29,31
Runx1/Cbfβ also binds to the CNS2 element,29,30 which contains CpG islands and is a differentially methylated region. In footprint analysis using Treg cells, consensus Runx sites in Foxp3 were accessible to DNaseI, suggesting an open structure for CNS2.32 The CNS2-bound Runx1 appears to regulate Foxp3 expression by modifying its chromatin structure. When H3K4me3 or H3K4me9 modifications of histone H3 were evaluated, CNS2 was more methylated in Cbfβ-deficient Treg cells than in wild-type Treg cells.30 In other words, CNS2 is demethylated in a Cbfβ-dependent manner in wild-type Treg cells. This demethylation appears to allow Foxp3 to bind to CNS2, which then probably contributes to the maintenance of Foxp3 expression.32,33 As expected, Foxp3 binding to CNS2 is impaired in Cbfβ-deficient cells.33 The above observations collectively suggest a crucial role for Runx1/Cbfβ in the maintenance of demethylation at the Foxp3 locus, and by inference, in increased Foxp3 expression in Treg cells.
Runx1-Foxp3 proteins interact to regulate Treg cells
Runx1 and Foxp3 proteins also exert their activities by forming a complex together (Fig. 3c). The Runx1–Foxp3 interaction was demonstrated by co-immunoprecipitation experiments and by in vitro glutathione S-transferase pulldown assays.28 Studies using deletion mutants demonstrated that the regions responsible for the interaction were located between amino acids 362–402 (C-terminal) of Runx1 and 278–336 (between the forkhead domain and the leucine zipper motif) of Foxp3. Transduction of full-length Foxp3 (but not Foxp3 lacking a Runx1 interaction domain) can induce expression of Treg-associated cell surface molecules [CD25, cyctotoxic T-lymphocyte antigen 4 (CTLA-4) and glucocorticoid induced tumour necrosis factor receptor (GITR)]. This is because both Runx1 and Foxp3 bind to intron 1 of CD25 and GITR, and work co-operatively in gene induction.28,34 The above data collectively suggest that the Runx1–Foxp3 complex induces expression of Treg-cell-associated genes (and exerts Treg-cell-suppressive activity). It is noteworthy that, in Th17 cells, Foxp3 forms a complex with Runx1 and RORγt to suppress IL-17 production.21
In short, the indispensability of Runx1/Cbfβ for Treg-cell differentiation and stability has been gradually unveiled. Runx1/Cbfβ exerts its effects through a unique feed-forward relationship with a Treg-cell signature transcription factor, Foxp3.
Involvement of Runx transcription factors in other aspects of T-cell differentiation/function
Finally, we summarize the roles of Runx factors in T-cell biology that have been published so far with references that are classified according to the steps of T-cell differentiation/function involved (Table 1). The involvement of Runx in non-canonical γδT, natural killer T, natural killer and dendritic cell subsets is also summarized. This information provides an up-to-date snapshot of the interplay between T-cell differentiation and function and Runx that should be useful for researchers working in and around this field.
Table 1.
Summary of the roles of Runx in the differentiation and function of T lymphocytes
| T-cell subsets | Roles of Runx | Refs mentioned in the text | Refs not mentioned in the text |
|---|---|---|---|
| Reviews | On Runx and T, in general | 1 | |
| Reviews | On CD4+ versus CD8+ | 35–38 | |
| DN thymocytes | Runx1 is necessary at multiple steps in the differentiation of DN thymocytes. Runx3 is not a main factor | 39–44 | |
| DP thymocytes | Runx1 influences apoptotic sensitivity of DP thymocytes | 45 | |
| CD4+ cells | Runx1 is important for the differentiation of CD4+ thymocytes and homeostasis of peripheral CD4+ cells | 42,46 | |
| CD8+ cells | Both Runx3 and Runx1 are necessary for the differentiation of CD8+ cells | 4–6,14 | 40,42,47–50 |
| CD4+ versus CD8+ selection, Runx and ThPOK | A transcription factor network is involved in the selection of either the CD4+ or CD8+ lineage | 8–11,13 | 51 |
| Th1/Th2 | Runx1 and Runx3 function in favour of Th1 differentiation and antagonize Th2 differentiation | 15–19 | |
| Th17 | Runx1 and RORγt interact with each other | 21 | |
| Treg | Foxp3 segregates the Treg lineage from the conventional CD4+ lineage by interacting with Runx1 | 28–33 | |
| γδT and natural killer T | Runx3 is necessary for the emergence of skin-residing γδT cells. Runx1 is necessary for the development of natural killer T cells in thymus | 52,53 | |
| Natural killer and dendritic cells | Runx3 is important for the full maturation of the natural killer cell lineage. Runx3 negatively regulates dendritic cell maturation | 54–56 | |
| Domain analyses of Runx | Intra-Runx domains necessary for CD4 repression | 57–60 | |
| Runx expression | Runx expression in T cells is regulated transcriptionally and post-transcriptionally | 61–63 |
DN, double-negative; DP, double-positive; Th1, T helper type 1; Th-POK, T helper-inducing POZ-Krüppel-like factor; Treg, regulatory T.
Acknowledgments
W.F.W. is supported by Japan Society for the Promotion of Science (JSPS) postdoctoral fellowship for foreign researchers, Japan. M.S. is a member of GCOE, Network Medicine founded at Tohoku University, Japan.
Disclosures
None.
References
- 1.Kohu K, Kubo M, Ichikawa H, Ohno S, Habu S, Sato T, Satake M. Pleiotropic roles of Runx transcription factors in the differentiation and function of T lymphocytes. Current Immunol Rev. 2008;4:101–15. [Google Scholar]
- 2.He X, He X, Dave VP, et al. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 2005;433:826–33. doi: 10.1038/nature03338. [DOI] [PubMed] [Google Scholar]
- 3.Sun G, Liu X, Mercado P, Jenkinson SR, Kypriotou M, Feigenbaum L, Galéra P, Bosselut R. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat Immunol. 2005;6:373–81. doi: 10.1038/ni1183. [DOI] [PubMed] [Google Scholar]
- 4.Taniuchi I, Osato M, Egawa T, Sunshine MJ, Bae SC, Komori T, Ito Y, Littman DR. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 2002;111:621–33. doi: 10.1016/s0092-8674(02)01111-x. [DOI] [PubMed] [Google Scholar]
- 5.Sato T, Ohno S, Hayashi T, Sato C, Kohu K, Satake M, Habu S. Dual functions of Runx proteins for re-activating CD8 and silencing CD4 at the commitment process into CD8 thymocytes. Immunity. 2005;22:317–28. doi: 10.1016/j.immuni.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 6.Kohu K, Sato T, Ohno S, et al. Overexpression of the Runx3 transcription factor increases the proportion of mature thymocytes of the CD8 single positive lineage. J Immunol. 2005;174:2627–36. doi: 10.4049/jimmunol.174.5.2627. [DOI] [PubMed] [Google Scholar]
- 7.Dave VP, Allman D, Keefe R, Hardy RR, Kappes DJ. HD mice: a novel mouse mutant with a specific defect in the generation of CD4+ T cells. Proc Natl Acad Sci USA. 1998;95:8187–92. doi: 10.1073/pnas.95.14.8187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Setoguchi R, Tachibana M, Naoe Y, Muroi S, Akiyama K, Tezuka C, Okuda T, Taniuchi I. Repression of the transcription factor Th-POK by Runx complexes in cytotoxic T cell development. Science. 2008;319:822–5. doi: 10.1126/science.1151844. [DOI] [PubMed] [Google Scholar]
- 9.He X, Park K, Wang H, He X, Zhang Y, Hua X, Li Y, Kappes DJ. CD4–CD8 lineage commitment is regulated by a silencer element at the ThPOK transcription-factor locus. Immunity. 2008;28:346–58. doi: 10.1016/j.immuni.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 10.Egawa T, Littman DR. ThPOK acts late in specification of the helper T cell lineage and suppresses Runx-mediated commitment to the cytotoxic T cell lineage. Nat Immunol. 2008;9:1131–9. doi: 10.1038/ni.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muroi S, Naoe Y, Miyamoto C, Akiyama K, Ikawa T, Masuda K, Kawamoto H, Taniuchi I. Cascading suppression of transcriptional silencers by ThPOK seals helper T cell fate. Nat Immunol. 2008;9:1113–21. doi: 10.1038/ni.1650. [DOI] [PubMed] [Google Scholar]
- 12.Singer A, Adoro S, Park JH. Lineage fate and intense debate: myths, models and mechanisms of CD4- versus CD8-lineage choice. Nat Rev Immunol. 2008;8:788–801. doi: 10.1038/nri2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L, Wildt KF, Zhu J, et al. Distinct functions for the transcription factors GATA-3 and ThPOK during intrathymic differentiation of CD4+ T cells. Nat Immunol. 2008;9:1122–30. doi: 10.1038/ni.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park J-H, Adoro S, Guinter T, et al. Signaling by intrathymic cytokines, not T cell antigen receptors, specifies CD8 lineage choice and promotes the differentiation of cytotoxic-lineage T cells. Nat Immunol. 2010;11:257–64. doi: 10.1038/ni.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Komine O, Hayashi K, Natsume W, et al. The Runx1 transcription factor inhibits the differentiation of naive CD4+ T cells into the Th2 lineage by repressing GATA3 expression. J Exp Med. 2003;198:51–61. doi: 10.1084/jem.20021200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naoe Y, Setoguchi R, Akiyama K, Muroi S, Kuroda M, Hatam F, Littman DR, Taniuchi I. Repression of interleukin-4 in T helper type 1 cells by Runx/Cbf β binding to the Il4 silencer. J Exp Med. 2007;204:1749–55. doi: 10.1084/jem.20062456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Djuretic IM, Levanon D, Negreanu V, Groner Y, Rao A, Ansel KM. Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nat Immunol. 2007;8:145–53. doi: 10.1038/ni1424. [DOI] [PubMed] [Google Scholar]
- 18.Kohu K, Ohmori H, Wong W-F, et al. The Runx3 transcription factor augments TH1 and down-modulates TH2 phenotypes by interacting with and attenuating GATA3. J Immunol. 2009;183:7817–24. doi: 10.4049/jimmunol.0802527. [DOI] [PubMed] [Google Scholar]
- 19.Yagi R, Junttila IS, Wei G, Urban JF, Jr, Zhao K, Paul WE, Zhu J. The transcription factor GATA3 actively represses RUNX3 protein-regulated production of interferon-γ. Immunity. 2010;32:507–17. doi: 10.1016/j.immuni.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Littman DR, Rudensky AY. Th17 and regulatory T cells in mediating and restraining inflammation. Cell. 2010;140:845–58. doi: 10.1016/j.cell.2010.02.021. [DOI] [PubMed] [Google Scholar]
- 21.Zhang F, Meng G, Strober W. Interactions among the transcription factors Runx1, RORγt and Foxp3 regulate the differentiation of interleukin 17-producing T cells. Nat Immunol. 2008;9:1297–307. doi: 10.1038/ni.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fontenot JD, Gavin MA, Rudensky AY. FoxP3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4:330–6. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 23.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor FoxP3. Science. 2003;299:1057–61. doi: 10.1126/science.1079490. [DOI] [PubMed] [Google Scholar]
- 24.Khattri R, Cox T, Yasayko SA, Ramsdell F. An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nat Immunol. 2003;4:337–42. doi: 10.1038/ni909. [DOI] [PubMed] [Google Scholar]
- 25.Williams LM, Rudensky AY. Maintenance of the FoxP3-dependent developmental program in mature regulatory T cells requires continued expression of FoxP3. Nat Immunol. 2007;8:277–84. doi: 10.1038/ni1437. [DOI] [PubMed] [Google Scholar]
- 26.Bennett CL, Christie J, Ramsdell F, et al. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutation of FOXP3. Nat Genet. 2001;27:20–1. doi: 10.1038/83713. [DOI] [PubMed] [Google Scholar]
- 27.Brunkow ME, Jeffery EW, Hjerrild KA, et al. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet. 2001;27:68–73. doi: 10.1038/83784. [DOI] [PubMed] [Google Scholar]
- 28.Ono M, Yaguchi H, Ohkura N, et al. Foxp3 controls regulatory T-cell function by interacting with AML1/Runx1. Nature. 2007;446:685–9. doi: 10.1038/nature05673. [DOI] [PubMed] [Google Scholar]
- 29.Kitoh A, Ono M, Naoe Y, et al. Indispensable role of the Runx1-Cbfβ transcription complex for in vivo-suppressive function of FoxP3+ regulatory T cells. Immunity. 2009;31:609–20. doi: 10.1016/j.immuni.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 30.Rudra D, Egawa T, Chong MMW, Treuting P, Littman DR, Rudensky AY. Runx-CBFβ complexes control expression of the transcription factor Foxp3 in regulatory T cells. Nat Immunol. 2009;10:1170–8. doi: 10.1038/ni.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klunker S, Chong MMW, Mantel P-Y, et al. Transcription factors RUNX1 and RUNX3 in the induction and suppressive function of Foxp3+ inducible regulatory T cells. J Exp Med. 2009;206:2701–15. doi: 10.1084/jem.20090596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bruno L, Mazzarella L, Hoogenkamp M, et al. Runx proteins regulate Foxp3 expression. J Exp Med. 2009;206:2329–37. doi: 10.1084/jem.20090226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng Y, Josefowicz S, Chaudhry A, Peng XP, Forbush K, Rudensky AY. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 2010;463:808–13. doi: 10.1038/nature08750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu Y, Borde M, Heissmeyer V, et al. FoxP3 controls regulatory T cell function through cooperation with NFAT. Cell. 2006;126:375–87. doi: 10.1016/j.cell.2006.05.042. [DOI] [PubMed] [Google Scholar]
- 35.Egawa T, Taniuchi I. Antagonistic interplay between ThPOK and Runx in lineage choice of thymocytes. Blood Cells Mol Dis. 2009;43:27–9. doi: 10.1016/j.bcmd.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 36.Egawa T. Runx and ThPOK: a balancing act to regulate thymocyte lineage commitment. J Cell Biochem. 2009;107:1037–45. doi: 10.1002/jcb.22212. [DOI] [PubMed] [Google Scholar]
- 37.Wang L, Bosselut R. CD4-CD8 lineage differentiation: Thpok-ing into the nucleus. J Immunol. 2009;183:2903–9. doi: 10.4049/jimmunol.0901041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Collins A, Littman DR, Taniuchi I. RUNX proteins in transcription factor networks that regulate T-cell lineage choice. Nat Rev Immunol. 2009;9:106–15. doi: 10.1038/nri2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Talebian L, Li Z, Guo Y, et al. T-lymphoid, megakaryocyte, and granulocyte development are sensitive to decreases in CBFβ dosage. Blood. 2007;109:11–21. doi: 10.1182/blood-2006-05-021188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woolf E, Xiao C, Fainaru O, et al. Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc Natl Acad Sci USA. 2003;100:7731–6. doi: 10.1073/pnas.1232420100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ichikawa M, Asai T, Saito T, et al. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat Med. 2004;10:299–304. doi: 10.1038/nm997. [DOI] [PubMed] [Google Scholar]
- 42.Egawa T, Tillman RE, Naoe Y, Taniuchi I, Littman DR. The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J Exp Med. 2007;204:1945–57. doi: 10.1084/jem.20070133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sato T, Ito R, Nunomura S, Ohno S, Hayashi K, Satake M, Habu S. Requirement of transcription factor AML1 in proliferation of developing thymocytes. Immunol Lett. 2003;89:39–46. doi: 10.1016/s0165-2478(03)00103-2. [DOI] [PubMed] [Google Scholar]
- 44.Wong W-F, Nakazato M, Watanabe T, et al. Over-expression of Runx1 transcription factor impairs the development of thymocytes from the double negative to double positive stages. Immunology. 2010;130:243–53. doi: 10.1111/j.1365-2567.2009.03230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abe N, Kohu K, Ohmori H, et al. Reduction of Runx1 transcription factor activity up-regulates Fas and Bim expression and enhances the apoptotic sensitivity of double positive thymocytes. J Immunol. 2005;175:4475–82. doi: 10.4049/jimmunol.175.7.4475. [DOI] [PubMed] [Google Scholar]
- 46.Hayashi K, Natsume W, Watanabe T, et al. Diminution of the AML1 transcription factor function causes differential effects on the fates of CD4 and CD8 single positive T cells. J Immunol. 2000;165:6816–24. doi: 10.4049/jimmunol.165.12.6816. [DOI] [PubMed] [Google Scholar]
- 47.Ehlers M, Laule-Kilian K, Petter M, et al. Morpholino antisense oligonucleotide-mediated gene knockdown during thymocyte development reveals role for Runx3 transcription factor in CD4 silencing during development of CD4−/CD8+ thymocytes. J Immunol. 2003;171:3594–604. doi: 10.4049/jimmunol.171.7.3594. [DOI] [PubMed] [Google Scholar]
- 48.Grueter B, Petter M, Egawa T, et al. Runx3 regulates integrin αE/CD103 and CD4 expression during development of CD4−/CD8+ T cells. J Immunol. 2005;175:1694–705. doi: 10.4049/jimmunol.175.3.1694. [DOI] [PubMed] [Google Scholar]
- 49.Hayashi K, Abe N, Watanabe T, Obinata M, Ito M, Sato T, Habu S, Satake M. Overexpression of AML1 transcription factor drives thymocytes into the CD8 single positive lineage. J Immunol. 2001;167:4957–65. doi: 10.4049/jimmunol.167.9.4957. [DOI] [PubMed] [Google Scholar]
- 50.Zamisch M, Tian L, Grenningloh R, Xiong Y, Wildt KF, Ehlers M, Ho I-C, Bosselut R. The transcription factor Ets1 is important for CD4 repression and Runx3 up-regulation during CD8 T cell differentiation in the thymus. J Exp Med. 2009;206:2685–99. doi: 10.1084/jem.20092024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wildt KF, Sun G, Grueter B, Fischer M, Zamisch M, Ehlers M, Bosselut R. The transcription factor Zbtb7b promotes CD4 expression by antagonizing Runx-mediated activation of the CD4 silencer. J Immunol. 2007;179:4405–14. doi: 10.4049/jimmunol.179.7.4405. [DOI] [PubMed] [Google Scholar]
- 52.Woolf E, Brenner O, Goldenberg D, Levanon D, Groner Y. Runx3 regulates dendritic epidermal T cell development. Dev Biol. 2007;303:703–14. doi: 10.1016/j.ydbio.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 53.Egawa T, Eberl G, Taniuchi I, Benlagha K, Geissmann F, Hennighausen L, Bendelac A, Littman DR. Genetic evidence supporting selection of the Vα14i NKT cell lineage from double-positive thymocyte precursors. Immunity. 2005;22:705–16. doi: 10.1016/j.immuni.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 54.Ohno S, Sato T, Kohu K, Takeda K, Okumura K, Satake M, Habu S. Runx proteins are involved in regulation of CD122, Ly49 family and IFNγ expression during NK cell differentiation. Int Immunol. 2007;20:71–9. doi: 10.1093/intimm/dxm120. [DOI] [PubMed] [Google Scholar]
- 55.Fainaru O, Woolf E, Lotem J, et al. Runx3 regulates mouse TGF-β-mediated dendritic cell function and its absence results in airway inflammation. EMBO J. 2004;23:969–79. doi: 10.1038/sj.emboj.7600085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fainaru O, Shseyov D, Hantisteanu S, Groner Y. Accelerated chemokine receptor7-mediated dendritic cell migration in Runx3 knockout mice and the spontaneous development of asthma-like disease. Proc Natl Acad Sci USA. 2005;102:10598–603. doi: 10.1073/pnas.0504787102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yarmus M, Woolf E, Bernstein Y, Fainaru O, Negreanu V, Levanon D, Groner Y. Groucho/transducin-like Enhancer-of-split (TLE)-dependent and -independent transcriptional regulation by Runx3. Proc Natl Acad Sci USA. 2006;103:7384–9. doi: 10.1073/pnas.0602470103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nishimura M, Fukushima-Nakase Y, Fujita Y, Nakao M, Toda S, Kitamura N, Abe T, Okuda T. VWRPY motif-dependent and -independent roles of AML1/Runx1 transcription factor in murine hematopoietic development. Blood. 2004;103:562–70. doi: 10.1182/blood-2003-06-2109. [DOI] [PubMed] [Google Scholar]
- 59.Kawazu M, Asai T, Ichikawa M, et al. Functional domains of Runx1 are differentially required for CD4 repression, TCRβ expression, and CD4/8 double-negative to CD4/8 double-positive transition in thymocyte development. J Immunol. 2005;174:3526–33. doi: 10.4049/jimmunol.174.6.3526. [DOI] [PubMed] [Google Scholar]
- 60.Telfer JC, Hedblom EE, Anderson MK, Laurent MN, Rothenberg EV. Localization of the domains in Runx transcription factors required for the repression of CD4 in thymocytes. J Immunol. 2004;172:4359–70. doi: 10.4049/jimmunol.172.7.4359. [DOI] [PubMed] [Google Scholar]
- 61.Telfer JC, Rothenberg EV. Expression and function of a stem cell promoter for the murine CBFα2 gene: distinct roles and regulation in natural killer and T cell development. Dev Biol. 2001;229:363–82. doi: 10.1006/dbio.2000.9991. [DOI] [PubMed] [Google Scholar]
- 62.Fujii M, Hayashi K, Niki M, et al. Overexpression of AML1 renders a T hybridoma resistant to T cell receptor-mediated apoptosis. Oncogene. 1998;17:1813–20. doi: 10.1038/sj.onc.1202087. [DOI] [PubMed] [Google Scholar]
- 63.Chung DD, Honda K, Cafuir L, McDuffie M, Wotton D. The Runx3 distal transcript encodes an additional transcriptional activation domain. FEBS J. 2007;274:3429–39. doi: 10.1111/j.1742-4658.2007.05875.x. [DOI] [PubMed] [Google Scholar]


