Summary
The inherently error-prone nature of protein biosynthesis and turnover leads to a constant flux of destabilized proteins. Genetic mutations in conformational disease-associated proteins, as well as exposure to acute and chronic proteotoxic stresses, further increase the load of misfolded protein on the proteostasis network. During aging, this leads to enhanced instability of the proteome, failure to buffer destabilizing genetic mutations or polymorphisms, and cellular decline. The combination of cell-type-specific differences in the buffering capacity of the proteostasis network and destabilizing polymorphisms in the genetic background may account for some of the cell-type specificity observed in disease, even when the predominant disease-associated protein is widely expressed.
Introduction
Protein misfolding and aggregation underlies disease
Protein misfolding is recognized as the basis of numerous human diseases, known collectively as conformational diseases [1], including neurodegenerative diseases, certain cancers, type II diabetes and other metabolic pathologies, and various amyloidoses. In the cellular context, misfolding of a protein may lead to aberrant protein and membrane interactions, mislocalization, degradation, and aggregation, ultimately resulting in decreased availability of functional protein, as in Cystic Fibrosis, or gain-of-function toxicity, as in neurodegenerative diseases. Many neurodegenerative diseases, including Alzheimer’s Disease (AD), Parkinson’s Disease (PD), Amyotrophic Lateral Sclerosis (ALS), Huntington’s Disease (HD), and prion diseases, are characterized by the appearance of protein deposits, aggregates, or plaques in the postmortem tissues of affected individuals [2]. Although the identities of proteins involved are distinct, and corresponding diseases have different clinical manifestations, they have certain features suggestive of a common underlying mechanism of toxicity [3]. For example, all have delayed onset, with symptoms appearing usually in late adulthood. Most, with the exception of the polyglutamine diseases, such as HD, have both hereditary and sporadic forms, all cause cellular dysfunction and degeneration, and all are associated with protein misfolding and aggregation. Whether protein aggregation is causally linked to the pathogenesis of these conditions has been a focus of intense investigation, with much work done both from biophysical and biochemical perspective, and in cell culture and animal models.
Protein deposits associated with disease can be either extracellular or intracellular. Extracellular deposits usually consist of amyloid fibrils and are present in diseases such as light chain amyloidosis, senile systemic amyloidosis, type II diabetes, and familial cases of AD. Intracellular deposits - inclusion bodies or aggregates - can be ordered amyloid structures, non-amyloid fibrils, or amorphous or disordered aggregates. The currently favored hypothesis is that, during aggregation, certain molecular species resulting from conformational conversion of the native protein into non-native, often β-sheet-rich, forms initiate toxic events that ultimately lead to cellular dysfunction and degeneration [4]. While amyloid fibrils or larger, visible, aggregates were considered to be toxic at some point, the misfolded oligomeric forms, soluble oligomers, protofibrils, and even some monomeric species are currently thought of as toxic species [4–7]. This view is supported by the evidence that various non-disease-associated proteins become cytotoxic by conversion to β-sheet-rich oligomers [3]. Membrane disruption and depolarization, induction of oxidative stress, inappropriate activation of signaling pathways, and aberrant intracellular interactions, including partitioning of molecular chaperones and other cellular proteins into aggregates, have been proposed to explain the toxicity of these species [3,8,9]. However, despite much progress, certain aspects of conformational disease remain difficult to reconcile.
An intriguing aspect of neurodegenerative diseases is cell-type-selectivity with respect to the cells and tissues that undergo degeneration in different diseases. This selectivity occurs despite the fact that disease-associated proteins are expressed in a wide range of neuronal and non-neuronal tissues, often at much higher concentrations than in specific neuronal subtypes that are targeted in disease [10–12]. Perhaps consistent with this, in animal models, the disease-associated proteins are often expressed at high levels, and, in the case of polyQ-containing proteins, have longer polyQ lengths than are found in patients. Additionally, sensitized backgrounds have been used to expose their toxicity [13,14]. It appears that the aggregation and toxicity of these proteins depends on the cellular environment in which they are expressed.
Another characteristic aspect of disease is the delayed onset of aggregation and toxicity. Studies in animal models show that the age of onset of aggregation can be modulated genetically, by aging-signaling pathways, by enhancing the expression levels of molecular chaperones, and by activating stress-responsive pathways [15,16 Mojsilovic-Petrovic, 2009 #4711]. Moreover, in mouse models, genetic background has the capacity to override the toxic effects of the mutant protein [17–19]. There is also a large variability in the time of disease onset in patients, for example, in people harboring identical mutations in SOD1, or HD patients with identical or very similar near-threshold lengths of polyQ tract (the genetic determinants of disease) [20–23]. This implies that intrinsic misfolding, or aggregation propensity alone, is not sufficient to explain the initiation of toxicity by the disease-associated protein, and that the genetic background exerts a strong modulatory effect on aggregation and toxicity and therefore on disease severity and the age of onset.
Can some of the lessons learned from animal models of neurodegenerative diseases help refine our understanding of the underlying common mechanism(s) of toxicity? The questions that can be addressed by in vivo studies are those that look beyond the intrinsic structural properties of the disease-associated proteins, and consider their cellular context.
Proteostasis networks
One way in which disease-associated aggregation-prone proteins interact with their cellular environment is via the proteostasis networks. “Proteostasis” is, by our definition, the state of dynamic equilibrium in which protein synthesis and folding is balanced with degradation, while allowing for the conformational flexibility necessary for function, thus leading to a “healthy proteome” [24]. Genetic modifiers of aggregation and/or toxicity provide an insight into the nature of the interaction between the disease-associated, aggregation-prone proteins and the proteostasis networks. Molecular chaperones and the components of the degradation machinery are among the strongest and most consistent modifiers, suggesting that assistance in folding, prevention of aggregation and the disposal of the misfolded and aggregated species are crucial in vivo. Some of the modifiers identified in unbiased genetic screens in animal models point to a potential function of the affected proteins, for example genes involved in vesicular trafficking being identified as modifiers of α-syn toxicity [25]. Importantly, the hits obtained by screening for suppressors of polyQ or SOD1 aggregation in C. elegans showed a significant overlap with genes that are involved in the response to the osmotic stress-induced protein damage, while many suppressors of polyQ-expanded ataxin-3 in Drosophila also suppressed toxicity due to the generic increase in protein misfolding in a HSP70 hypomorphic strain [26–29]. These hits included proteins involved in RNA processing, protein synthesis, protein folding, and degradation, suggesting that a core set of factors function both in response to general stress-induced protein damage, and in response to the expression of disease-associated proteins. This core set of factors, representing the core of the proteostasis networks, may become compromised under conditions of disease and/or aging. The late onset and cell-type-restricted toxicity in conformational disease, despite often ubiquitous expression of disease-associated proteins, strongly support this view.
Proteostasis networks and aging
The aging-related decline in the functionality of proteostasis networks is well documented, and the levels of functional chaperones and the capacity of the clearance mechanisms are affected during aging. Increased protein damage, transcriptional and translational dysregulation, aberrant signaling, and other changes also accompany this decline. These aging-related changes could result in local shifts in the equilibrium between properly folded and misfolded or unfolded states. In this scenario, the susceptibility to conformational disease could be imparted by the cell-type-specific differences in the composition of proteostasis networks and in the robustness of adaptive stress responses.
The molecular interactions between the genetic pathways regulating lifespan and proteostasis are mediated, in part, by factors that detect and respond to misfolded proteins. These factors include molecular chaperones, the heat-shock transcription factor HSF1, which is activated when rapid increase in protein misfolding is induced, the FOXO transcription factor DAF-16, which is regulated by the ILS pathway, and other stress-inducible transcription factors. Downregulation of HSF1 activity in C. elegans suppresses the ILS-mediated lifespan extension and protection against proteotoxicity. It also leads to decreased normal lifespan and an accelerated aging phenotype, while overexpression of HSF1 extends lifespan [15,30]. Moreover, both HSF1 and DAF-16 are regulated by the NAD-dependent sirtuin, SIRT1, providing further evidence linking these stress-inducible transcription factors to aging and to the metabolic state of the cell [31–33]. The functional relationship between ILS and protein folding homeostasis can be demonstrated by the induction of both thermotolerance and life span extension either by mutations in the ILS pathway, or by a sublethal heat stress [34]. Furthermore, cells from naturally long-lived [35] or lifespan mutant [36] rodents appear to be resistant to multiple proteotoxic stresses. At the cellular level, the transcriptional output of aging signaling pathways and stress responsive pathways converge on common components of proteostasis networks involved in folding, clearance and detoxification processes, as well as metabolic components.
The integration of proteostasis and aging networks in conformational diseases is underscored by a decrease in aggregation/toxicity of disease-associated proteins upon overexpression of individual molecular chaperones [37], or upon activation of HSF1 or DAF-16. Likewise, proteasomal adaptation (by modulation of substrate accessibility to the proteasome core) to environmental stress in C. elegans ensures both resistance to proteotoxic conditions, including polyQ aggregation, and maintenance of life span under normal conditions, arguably through regulating degradation of misfolded proteins [38]. We are only beginning to understand how organismal regulation of growth, metabolism and aging is integrated with the maintenance of proteostasis at the cellular level. For example, recent evidence from C. elegans shows that the ability of individual cells in an organism to respond to the proteotoxic conditions is controlled by the activity of a subset of neurons [39].
Proteostasis is not robust
Heat shock proteins (HSPs) function as molecular chaperones in the absence of stress by guiding conformational transitions during synthesis, folding, translocation, assembly, and degradation of proteins [40–42]. Under conditions of stress, such as heat shock, HSPs are upregulated to counteract the deleterious consequences of protein misfolding. The ability of the cell to manage widespread protein damage during these proteotoxic stress conditions is often taken to indicate that the abundant expression of chaperones under basal conditions [43,44], together with the adaptive stress responses, provides sufficient ‘folding capacity’ to buffer unexpected folding requirements. However, the accumulation of misfolded and aggregated proteins in aging-related conformational diseases, and the associated toxicity, challenge this view and indicate a failure of these pathways.
Several possible, not mutually exclusive, explanations may account for such failure. First, HSPs levels or activity are often not enhanced in symptomatic cells, despite the accumulation of misfolded and aggregated proteins [45]. For example, in C. elegans, intracellular aggregation of polyQ proteins only sporadically activated HSP expression [46]. This is consistent either with the idea of an organismal override of the cell-autonomous stress responses [39], or with the accumulation of misfolded proteins in conformational diseases being too gradual, or not reaching the threshold necessary for the activation of the heat shock response. Second, molecular chaperones, components of degradative machinery and other proteins are often found trapped in aggregates, potentially mimicking hypomorphic phenotypes [46–49]. Third, there is evidence that expression of disease-associated misfolded proteins can interfere with key components of the proteostasis network such as the proteasome [50,51], and inhibits the heat shock response. This could potentially indicate that cells (or organisms) can adapt to the chronic expression of misfolded proteins by actively preventing stress induction. Finally, stress responses may simply fail to adequately address chronic misfolding. For example, a mouse sti mutation in the tyrosyl-tRNA synthetase, a model for a subtype of Charcot-Marie-Tooth neuropathy [52], leads to the production of heterogeneous misfolded proteins, accompanied by increased expression of chaperones in the cytoplasm and the endoplasmic reticulum (ER) [53]. Although an observed increase in chaperone expression suggests that adaptive transcriptional responses are indeed activated, the cellular dysfunction and neurodegeneration in this mouse model indicate that chronic protein misfolding may overwhelm the proteostasis networks. An interesting question, therefore, is how much misfolding is too much misfolding. We must further consider whether all misfolded and damaged proteins are recognized, refolded or cleared with equal efficiency, or, alternatively, whether certain proteins, such as those implicated in neurodegenerative diseases, are particularly challenging for the quality control machineries.
Specificity in proteostasis networks
Our understanding of the underlying mechanisms responsible for the failure of protein folding homeostasis in conformational disease partly depends on the knowledge of the cell- and tissue-specific composition, regulation and limitations of proteostasis networks. While there are clear indications of cell-type specific expression of some molecular chaperones and components of degradation machinery [54], a comprehensive definition of tissue- and cell-specific expression patterns during organismal development and aging is lacking. Beyond expression, the substrate specificities of different chaperones, even within the same family, their co-chaperone interactions, and the regulation of their activity within the cell are also not well understood. Defining specific chaperone requirements for the folding, maturation and maintenance of the native state for different disease-associated proteins should begin to address these gaps. For example, the recognition and retention of misfolded forms of mutant CFTR specifically depends on a HSP90 co-chaperone AHA1, and downregulation of AHA1 improves CFTR trafficking [55]. This sets the important paradigm that differences in expression or regulation of AHA1 itself, other components of the HSP90 complex, or even other substrates of AHA1, could modulate the fate of CFTR. Co-chaperones, and chaperones with a more restricted expression pattern or substrate repertoire may be particularly important to account for specificity. For example, HSP70 chaperones have been demonstrated in many studies to be important in the regulation of folding, misfolding and aggregation of both normal cellular proteins and of disease-associated proteins. However, apart from compartment-specific HSP70s, we have a rather limited understanding of the specific contributions of each of the ~12 members of HSP70 family to this process, and of their specificity toward protein substrates. A study of J domain-containing proteins in yeast, undertaken for the purpose of molecularly dissecting the HSP70 – J protein network [56], revealed that while many J proteins performed “generalized” functions, a subset of them had non-redundant functions, perhaps by targeting HSP70s to specific substrates [56]. In agreement with this, dHDJ1, but not dHDJ2, synergized with HSP70 in suppression of polyglutamine toxicity in Drosophila [57].
Chaperones regulate both folding and aggregation
It is currently thought that the toxicity of disease-associated aggregation-prone proteins in vivo reflects a combination of their intrinsic aggregation propensity and their interaction with individual components of the proteostasis network, such as molecular chaperones. It is well documented that molecular chaperones can influence both the folding trajectories and aggregation pathways of their client proteins [57,58]. A mechanistic insight into this process is provided by a study of the folding and aggregation of the variable domain of immunoglobulin light chain (VL), mutations in which cause light chain amyloidosis [59]. The HSP70 chaperone, BiP, is necessary for the efficient folding of the VL in vivo [60], and also regulates its amyloid aggregation in vitro. Strikingly, one of the two dominant BiP-binding sites on the VL was directly involved in the formation of amyloid fibrils, as the synthetic peptide containing this site specifically inhibited amyloidogenesis by the VL [61]. Thus, this chaperone’s normal function in promoting efficient folding of its substrate in the cell can also serve to prevent amyloid formation by the mutant forms of this protein in disease, with the same binding site potentially mediating both functions. Another study that echoes this conclusion showed that the anti-oligomer antibody, A11, recognizes a subset of molecular chaperones, and that A11 binding to these chaperones can interfere with the suppression of aggregation or the refolding of substrates [62]. An increased competition for such chaperone from other substrates, particularly if uncompensated for by the stress response, may thus favor the shift toward amyloidogenesis and the formation of toxic species by the disease-associated protein.
Cellular implications of competition for folding resources
The apparent lack of robustness of the proteostasis networks to chronic misfolding is unexpected and has significant implications for late-onset conformational diseases. The aging of an organism is accompanied by an increase in the accumulation of damaged proteins and a decrease in fidelity of biosynthetic processes. If compensatory adaptive responses fail, accumulated misfolded proteins could deplete essential components of the proteostasis machinery [48,51,63]. With as much as 70% of rare missense alleles in human population predicted to be mildly deleterious [64], and approximately half of the genetic changes in inherited disease (in OMIM and HGMD databases) being due to nonsynonymous substitutions [65], such compromise should manifest in a gradual increase in cellular dysfunction and onset of disease. In this scenario, the specific complement of mutations and polymorphisms in expressed genes, in addition to the strength and composition of the proteostatis networks, will establish a dynamic threshold for the onset of dysfunction, both in a cell-specific, and an individual organism-specific manner.
Endogenous metastable proteins suffer when disease proteins misfold
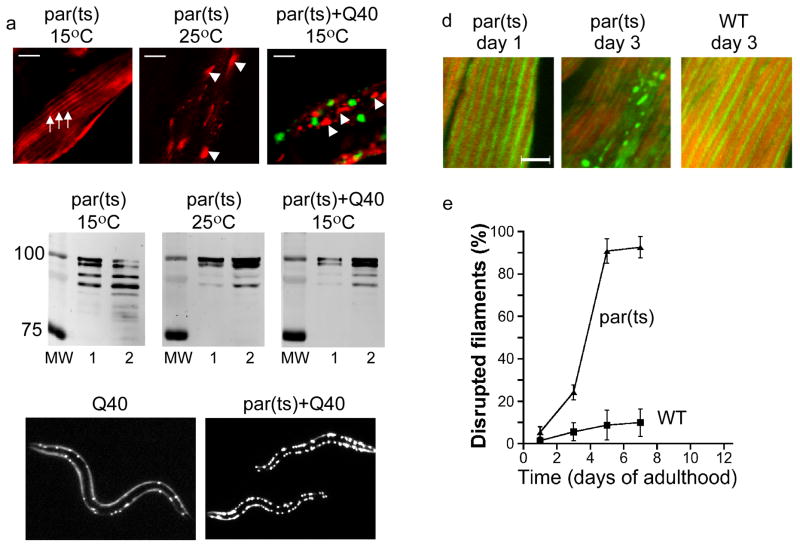
Recent studies in C. elegans have shown that expression of expanded polyQ proteins leads to the global disruption of cellular protein folding [66]. This global effect manifests itself in destabilization or misfolding of other expressed metastable proteins, modeled by endogenous temperature-sensitive (ts) folding mutants, with a feedback to polyQ aggregation (Figure 1a–c). As there was no detectable co-aggregation, this mutual destabilization was likely mediated by the competition for folding resources, necessary for maintaining metastablets proteins in their folded and functional conformations [66]. In the absence of polyQs, these ts mutants are responsive to the modulation of the folding environment by aging signaling pathways and HSF1 at permissive temperatures, and also show delayed onset of ts phenotypes (Figure 1d,e) [67]. This delayed onset of dysfunction appears to correlate to the onset of misfolding or the mislocalization of the corresponding metastable protein. Likewise, the phenotypic exposure of ts phenotypes in polyQ-containing strains also stemmed from misfolding and aggregation of the ts proteins. Thus, at least in this model, mild folding variants in the genetic background appear to function as a ‘trigger’ for the expression of the toxicity of the aggregation-prone protein, while also channeling the toxicity to tissue-specific phenotypes. Similar destabilization of susceptible proteins, that are either naturally highly dependent on molecular chaperones for their stability and activity, or encoded by destabilizing polymorphisms, could contribute significantly to conformational disease.
Figure 1.
a–b. Expression of Q40-YFP (Q40, green) in nematodes carrying a destabilizing mutation in paramyosin (par(ts), red) causes mislocalization (a) and misfolding (b) of paramyosin. a, immunostaining and epifluorescence; b, limited proteolysis. c. Q40-YFP itself displays early onset of aggregation when expressed in a ts background. a–c are adapted from Gidalevitz et. al. Science 2006; 311(5766):1471–4. d. Mislocalized par(ts) (green) in an otherwise WT background is seen as early as day 3 of adulthood, and is similar in appearance to the par(ts) at the restrictive temperature (a). Actin filaments counterstained in red. e. Quantification of the number of cells with disrupted myofilaments in par(ts) or WT worms. d–e are adapted from Ben-Zvi et. al., Proc Natl Acad Sci U S A. 2009;106(35):14914–9.
The failure to support metastable proteins in their functional state appears to be a common consequence of expression of disease-associated aggregation-prone proteins. It was also observed in a model of SOD1 misfolding, where expression of three different SOD1 mutant proteins – G85R, G93A and a truncated 127X – in strains harboring ts mutations in unrelated genes also resulted in exposure of ts phenotypes at permissive conditions [68]. These mutants of SOD1 exhibit different structural properties and folding trajectories, from G93A that partitions between the native state, characterized by dismutase activity, and tightly aggregated, SDS-sensitive material, to 127X, present only in an unstructured aggregated state, probably due to the large truncation [68]. Considering the likely structured, SDS-resistant aggregation of polyQ expanded proteins, these findings support the view that intracellular toxicity does not need to depend on specific oligomeric structures or folding conformers. In agreement with this view, many of the modifiers of toxicity of polyQ-expanded ataxin-3 in Drosophila also rescue the generic toxicity of protein misfolding due to the reduced function of HSP70 [29], and many of the modifiers of polyQ in C. elegans also regulate the protection from osmotic-stress-induced protein damage [26,28].
What is the nature of proteins and pathways affected by aggregation-prone disease-associated proteins?
The buffering of ts phenotypes by proteostasis networks is reminiscent of the ability of molecular chaperone HSP90 to buffer phenotypic variation due to cryptic mutations [69]. These cryptic mutations were proposed to be exposed only under (proteotoxic) stress conditions, when the functional availability of HSP90 is limited. Indeed, the phenotypes exposed by the limitation of HSP90 in Drosophila correlated to specific genetic backgrounds, and were also affected by temperature. A study in zebra fish showed that developmental phenotypes commonly observed upon HSP90 limitation reflected underlying polymorphisms, whose frequency was strain-specific, while phenotypes that were rarely seen were unique to specific mutant carrier strains [70]. The authors suggested that a similar buffering of underlying polymorphisms may explain an incomplete penetrance observed in human disease. Given that HSP90 is thought to recognize near-native or metastable proteins, these studies imply that, even in the genetically restricted context of laboratory strains of model organisms, there is a surprising amount of normally buffered protein variants that are capable of allowing exposure of phenotypic traits.
Genetic mutations and polymorphisms, genomic instability, mistranslation, or incorporation of amino-acid analogues (such as certain antibiotics or plant metabolites), all have the potential to affect folding pathways and the stability of the native state [52,53,59,71,72]. Coding polymorphisms are not rare and are estimated to occur at an average of two per coding sequence [73], while misincorporation during translation might cause up to 18% of expressed proteins to contain an amino acid substitution [74]. Recent work on the evolution of protein sequences suggests that selection against the toxicity of misfolding due to mistranslation may represent an important evolutionary pressure, when considering highly expressed proteins [74]. From a physiological perspective, this may indicate that the flux of misfolded or destabilized proteins in a cell bears a significant fitness cost. This cost is not only due to the loss-of-function of the misfolded proteins, but also due to the toxic effects of induced aggregation, the consequences of inappropriate intermolecular interactions [9,24,59], and the abnormal engagement of molecular chaperones and degradative machineries. Thus, the cell-type specific complement of expressed protein polymorphisms in functional pathways and complexes could contribute to both the threshold for the onset of proteotoxicity and the ensuing phenotypes in disease.
A model for the cell-specific toxicity in neurodegenerative diseases
We propose a model where the disease-associated aggregation-prone protein is buffered, at least initially, by the cellular folding environment (Figure 2b). Misfolding and aggregation of the disease-associated protein may then reflect the decreased availability of or increased competition for certain limiting components of the proteostasis networks. Such limiting components are likely to be specific to the particular disease-related protein (i.e. polyQ vs. SOD1), and may or may not be cell-type specific. Aging-related changes in proteostasis networks and stress responses may alone be sufficient to initiate the cascade of misfolding and aggregation of the disease-associated protein. The initiation of the misfolding and aggregation cascade could be accompanied by the general toxicity due to the presence of misfolded species and aggregation intermediates (oligomers), and the altered or inappropriate interactions of these species with other cellular components, in a manner dependent on the identity of the disease-associated protein. On the other hand, if other proteins in the cell (target proteins) compete for the same limiting component(s), the misfolding and aggregation of the disease-associated protein is greatly favored, even before the general compromise of the folding environment. This competition could be either due to the intrinsic instability and chaperone dependence of the target protein, its low translational robustness, or due to destabilizing polymorphisms. The mutual destabilization of the disease-associated protein and the target protein will additionally contribute to disease as the function of the cellular pathways and protein complexes containing the target protein become compromised, thus imparting the cell- and tissue-specific characteristics to the disease phenotypes (Figure 2d).
Figure 2.
a–b. Proteostasis networks buffer metastable and misfolded species, ensuring normal cellular function. c–d. Failure of proteostasis networks to buffer misfolding results in onset of dysfunction. Accumulated protein damage, proteotoxic stresses and aging all contribute to the increase in misfolding and thus to the decline in the proteostasis network capacity. Either a single dominant mutation (d), or a cumulative effect of milder destabilizing mutations and polymorphisms (c) may lead to the cell-specific dysfunction of sensitized pathways and protein complexes, by competing for a shared limiting component(s) of the proteostasis network.
Many neurodegenerative and other conformational diseases exist in both familial and sporadic forms. The variants share clinical manifestations but appear to have different underlying causes of disease. The familial variants are associated with mutations that encode aggregation-prone proteins, while the factors contributing to the sporadic disease are largely unknown. Our model suggests that sporadic disease may reflect the dysfunction of the same sensitized pathways and protein complexes that are targeted by the expression of the disease-associated proteins in familial disease (Figure 2c). Several different mechanisms may contribute to this dysfunction. First, there could be a general compromise of the proteostasis network. Such a general effect could be caused by aging-related changes, or by environmental factors, such as proteotoxic stresses, inflammatory and febrile conditions, etc. A general effect could also be caused by genetic mutations that impair the proteostasis network by altering its capacity or robustness. Second, mutations in factors that are not usually associated with familial forms of disease could lead to an increased competition for the limiting components of the proteostasis network. In this case, either a single substrate destabilized by sporadic mutation, or several mildly destabilized substrates (for example due to coding polymorphisms or mistranslation), could mimic the competition present in the familial disease. Finally, the target pathways or complexes themselves may be affected directly, by damage or by mutations/polymorphisms in their different protein components, thereby causing disease.
Identification of TDP43 [75] as a protein commonly affected both in familial and sporadic cases of FTLD-U [76] and ALS [77] could provide an example for this paradigm: the dysfunction of TDP43 itself, or of the cellular pathway in which it functions, brought about by different mechanisms, could underlie the cellular dysfunction in these diseases. Interestingly, TDP43 pathology is not present in the familial ALS cases associated with SOD1 mutations, potentially indicating that ALS with and without SOD1 mutations are distinct disorders. Alternatively, the cellular pathway in which TDP43 functions may itself be affected by the expression of mutant SOD1. This underscores the importance of identifying the target pathway that is directly responsible for the cellular toxicity in conformational diseases for both diagnosis and disease intervention.
Acknowledgments
E.A.K was supported by an individual postdoctoral fellowship from the National Institutes of Health (NINDS); research in the laboratory of R.I.M. was supported by grants from the National Institutes of Health (NIGMS and NIA), the HDSA Coalition for the Cure, and the ALS Association.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1**.Carrell RW, Lomas DA. Conformational disease. Lancet. 1997;350:134–138. doi: 10.1016/S0140-6736(97)02073-4. This review represents the first recognition that certain diseases, termed conformational diseases, are caused by abnormal unfolding and aggregation of the underlying protein(s) [DOI] [PubMed] [Google Scholar]
- 2.Soto C, Estrada LD. Protein misfolding and neurodegeneration. Arch Neurol. 2008;65:184–189. doi: 10.1001/archneurol.2007.56. [DOI] [PubMed] [Google Scholar]
- 3*.Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo J, Taddei N, Ramponi G, Dobson CM, Stefani M. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 2002;416:507–511. doi: 10.1038/416507a. This paper shows that proteins not normally associated with disease can be induced to form highly cytotoxic aggregated species, thereby providing evidence that protein aggregation is a common underlying feature in diseases such as AD, HD, etc. [DOI] [PubMed] [Google Scholar]
- 4.Meredith SC. Protein denaturation and aggregation: Cellular responses to denatured and aggregated proteins. Ann N Y Acad Sci. 2005;1066:181–221. doi: 10.1196/annals.1363.030. [DOI] [PubMed] [Google Scholar]
- 5.Sanchez I, Mahlke C, Yuan J. Pivotal role of oligomerization in expanded polyglutamine neurodegenerative disorders. Nature. 2003;421:373–379. doi: 10.1038/nature01301. [DOI] [PubMed] [Google Scholar]
- 6**.Walsh DM, Klyubin I, Fadeeva JV, Cullen WK, Anwyl R, Wolfe MS, Rowan MJ, Selkoe DJ. Naturally secreted oligomers of amyloid beta protein potently inhibit hippocampal long-term potentiation in vivo. Nature. 2002;416:535–539. doi: 10.1038/416535a. Demonstrates that A oligomers, in the absence of monomers or amyloid fibrils, are toxic to rats, thereby leading to the toxic oligomer hypothesis for disease. [DOI] [PubMed] [Google Scholar]
- 7.Nagai Y, Inui T, Popiel HA, Fujikake N, Hasegawa K, Urade Y, Goto Y, Naiki H, Toda T. A toxic monomeric conformer of the polyglutamine protein. Nat Struct Mol Biol. 2007;14:332–340. doi: 10.1038/nsmb1215. [DOI] [PubMed] [Google Scholar]
- 8.Lashuel HA, Hartley D, Petre BM, Walz T, Lansbury PT., Jr Neurodegenerative disease: amyloid pores from pathogenic mutations. Nature. 2002;418:291. doi: 10.1038/418291a. [DOI] [PubMed] [Google Scholar]
- 9.Stefani M, Dobson CM. Protein aggregation and aggregate toxicity: new insights into protein folding, misfolding diseases and biological evolution. J Mol Med. 2003;81:678–699. doi: 10.1007/s00109-003-0464-5. [DOI] [PubMed] [Google Scholar]
- 10.Strong TV, Tagle DA, Valdes JM, Elmer LW, Boehm K, Swaroop M, Kaatz KW, Collins FS, Albin RL. Widespread expression of the human and rat Huntington’s disease gene in brain and nonneural tissues. Nat Genet. 1993;5:259–265. doi: 10.1038/ng1193-259. [DOI] [PubMed] [Google Scholar]
- 11.Trottier Y, Devys D, Imbert G, Saudou F, An I, Lutz Y, Weber C, Agid Y, Hirsch EC, Mandel JL. Cellular localization of the Huntington’s disease protein and discrimination of the normal and mutated form. Nat Genet. 1995;10:104–110. doi: 10.1038/ng0595-104. [DOI] [PubMed] [Google Scholar]
- 12.Li SH, Schilling G, Young WS, 3rd, Li XJ, Margolis RL, Stine OC, Wagster MV, Abbott MH, Franz ML, Ranen NG, et al. Huntington’s disease gene (IT15) is widely expressed in human and rat tissues. Neuron. 1993;11:985–993. doi: 10.1016/0896-6273(93)90127-d. [DOI] [PubMed] [Google Scholar]
- 13.Faber PW, Voisine C, King DC, Bates EA, Hart AC. Glutamine/proline-rich PQE-1 proteins protect Caenorhabditis elegans neurons from huntingtin polyglutamine neurotoxicity. Proc Natl Acad Sci U S A. 2002;99:17131–17136. doi: 10.1073/pnas.262544899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Voisine C, Varma H, Walker N, Bates EA, Stockwell BR, Hart AC. Identification of potential therapeutic drugs for huntington’s disease using Caenorhabditis elegans. PLoS ONE. 2007;2:e504. doi: 10.1371/journal.pone.0000504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- 16**.Cohen E, Bieschke J, Perciavalle RM, Kelly JW, Dillin A. Opposing activities protect against age-onset proteotoxicity. Science. 2006;313:1604–1610. doi: 10.1126/science.1124646. The authors showed that HSF1 promotes disaggregation of disease-associated proteins, while DAF-16 enhances the formation of inert aggregates from toxic oligomers. These findings provide a link between the ILS pathway and protection against proteotoxicity. [DOI] [PubMed] [Google Scholar]
- 17*.Kunst CB, Messer L, Gordon J, Haines J, Patterson D. Genetic mapping of a mouse modifier gene that can prevent ALS onset. Genomics. 2000;70:181–189. doi: 10.1006/geno.2000.6379. Here, the authors demonstrate that the expression of the “ALS phenotype” in mice carrying a mutation SOD1 is highly dependent on genetic background. This is consistent with genetic polymorphisms having a profound modulatory effect on mutant SOD1 toxicity. [DOI] [PubMed] [Google Scholar]
- 18.Heiman-Patterson TD, Deitch JS, Blankenhorn EP, Erwin KL, Perreault MJ, Alexander BK, Byers N, Toman I, Alexander GM. Background and gender effects on survival in the TgN(SOD1-G93A)1Gur mouse model of ALS. J Neurol Sci. 2005;236:1–7. doi: 10.1016/j.jns.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 19.Lloret A, Dragileva E, Teed A, Espinola J, Fossale E, Gillis T, Lopez E, Myers RH, MacDonald ME, Wheeler VC. Genetic background modifies nuclear mutant huntingtin accumulation and HD CAG repeat instability in Huntington’s disease knock-in mice. Hum Mol Genet. 2006;15:2015–2024. doi: 10.1093/hmg/ddl125. [DOI] [PubMed] [Google Scholar]
- 20.Cajavec B, Herzel H, Bernard S. Death of neuronal clusters contributes to variance of age at onset in Huntington’s disease. Neurogenetics. 2006;7:21–25. doi: 10.1007/s10048-005-0025-x. [DOI] [PubMed] [Google Scholar]
- 21.Wexler NS, Lorimer J, Porter J, Gomez F, Moskowitz C, Shackell E, Marder K, Penchaszadeh G, Roberts SA, Gayan J, et al. Venezuelan kindreds reveal that genetic and environmental factors modulate Huntington’s disease age of onset. Proc Natl Acad Sci U S A. 2004;101:3498–3503. doi: 10.1073/pnas.0308679101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orrell RW, King AW, Hilton DA, Campbell MJ, Lane RJ, de Belleroche JS. Familial amyotrophic lateral sclerosis with a point mutation of SOD-1: intrafamilial heterogeneity of disease duration associated with neurofibrillary tangles. J Neurol Neurosurg Psychiatry. 1995;59:266–270. doi: 10.1136/jnnp.59.3.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ikeda M, Abe K, Aoki M, Sahara M, Watanabe M, Shoji M, St George-Hyslop PH, Hirai S, Itoyama Y. Variable clinical symptoms in familial amyotrophic lateral sclerosis with a novel point mutation in the Cu/Zn superoxide dismutase gene. Neurology. 1995;45:2038–2042. doi: 10.1212/wnl.45.11.2038. [DOI] [PubMed] [Google Scholar]
- 24*.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. This review describes how the field has converged on an understanding of the role of protein folding homeostasis in stress aging, and disease, and serves to define the term “proteostasis.”. [DOI] [PubMed] [Google Scholar]
- 25.van Ham TJ, Thijssen KL, Breitling R, Hofstra RM, Plasterk RH, Nollen EA. C. elegans model identifies genetic modifiers of alpha-synuclein inclusion formation during aging. PLoS Genet. 2008;4:e1000027. doi: 10.1371/journal.pgen.1000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26*.Nollen EA, Garcia SM, van Haaften G, Kim S, Chavez A, Morimoto RI, Plasterk RH. Genome-wide RNA interference screen identifies previously undescribed regulators of polyglutamine aggregation. Proc Natl Acad Sci U S A. 2004;101:6403–6408. doi: 10.1073/pnas.0307697101. This paper marks the first genome-wide screen to identify modifiers of polyQ aggregation. The results highlight the important fact that numerous cellular processes involved in protein folding homeostasis impact polyQ protein aggregation, and thereby begin to elucidate the factors that comprise the “proteostasis network”. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J, Farr GW, Hall DH, Li F, Furtak K, Dreier L, Horwich AL. An ALS-linked mutant SOD1 produces a locomotor defect associated with aggregation and synaptic dysfunction when expressed in neurons of Caenorhabditis elegans. PLoS Genet. 2009;5:e1000350. doi: 10.1371/journal.pgen.1000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamitina T, Huang CG, Strange K. Genome-wide RNAi screening identifies protein damage as a regulator of osmoprotective gene expression. Proc Natl Acad Sci U S A. 2006;103:12173–12178. doi: 10.1073/pnas.0602987103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bilen J, Bonini NM. Genome-wide screen for modifiers of ataxin-3 neurodegeneration in Drosophila. PLoS Genet. 2007;3:1950–1964. doi: 10.1371/journal.pgen.0030177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morley JF, Morimoto RI. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol Biol Cell. 2004;15:657–664. doi: 10.1091/mbc.E03-07-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31*.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. Demonstrates that SIRT1 regulates lifespan and the cellular stress response by modulating, via deacetylation, the FOXO transcription factors. [DOI] [PubMed] [Google Scholar]
- 32.Lee SJ. alpha-synuclein aggregation: a link between mitochondrial defects and Parkinson’s disease? Antioxid Redox Signal. 2003;5:337–348. doi: 10.1089/152308603322110904. [DOI] [PubMed] [Google Scholar]
- 33*.Westerheide SD, Anckar J, Stevens SM, Jr, Sistonen L, Morimoto RI. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science. 2009;323:1063–1066. doi: 10.1126/science.1165946. The authors show that HSF1 activity is attenuated by acetylation and enhanced by the deacatylation activity of SIRT1. These data thereby establish a role for SIRT1 in proteostasis and the heat shock response. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34*.Lithgow GJ, White TM, Melov S, Johnson TE. Thermotolerance and extended life-span conferred by single-gene mutations and induced by thermal stress. Proc Natl Acad Sci U S A. 1995;92:7540–7544. doi: 10.1073/pnas.92.16.7540. This work provides an important connection between an animal’s resistance to stress and life expectancy, suggesting that the ability to respond to stress limits the life expectancy of metaozoans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salmon AB, Sadighi Akha AA, Buffenstein R, Miller RA. Fibroblasts from naked mole-rats are resistant to multiple forms of cell injury, but sensitive to peroxide, ultraviolet light, and endoplasmic reticulum stress. J Gerontol A Biol Sci Med Sci. 2008;63:232–241. doi: 10.1093/gerona/63.3.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salmon AB, Murakami S, Bartke A, Kopchick J, Yasumura K, Miller RA. Fibroblast cell lines from young adult mice of long-lived mutant strains are resistant to multiple forms of stress. Am J Physiol Endocrinol Metab. 2005;289:E23–29. doi: 10.1152/ajpendo.00575.2004. [DOI] [PubMed] [Google Scholar]
- 37.Muchowski PJ, Wacker JL. Modulation of neurodegeneration by molecular chaperones. Nat Rev Neurosci. 2005;6:11–22. doi: 10.1038/nrn1587. [DOI] [PubMed] [Google Scholar]
- 38.Yun C, Stanhill A, Yang Y, Zhang Y, Haynes CM, Xu CF, Neubert TA, Mor A, Philips MR, Ron D. Proteasomal adaptation to environmental stress links resistance to proteotoxicity with longevity in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2008;105:7094–7099. doi: 10.1073/pnas.0707025105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39*.Prahlad V, Cornelius T, Morimoto RI. Regulation of the cellular heat shock response in Caenorhabditis elegans by thermosensory neurons. Science. 2008;320:811–814. doi: 10.1126/science.1156093. This paper demonstrates that the heat shock response is non-cell-autonomous in a multicellular organism, unlike previously assumed, but is instead controlled by the thermosensory neuronal network. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hartl FU. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–579. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 41.Morimoto RI, Kline MP, Bimston DN, Cotto JJ. The heat-shock response: regulation and function of heat-shock proteins and molecular chaperones. Essays Biochem. 1997;32:17–29. [PubMed] [Google Scholar]
- 42.Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 43.Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 44.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 45.Zourlidou A, Gidalevitz T, Kristiansen M, Landles C, Woodman B, Wells DJ, Latchman DS, de Belleroche J, Tabrizi SJ, Morimoto RI, et al. Hsp27 overexpression in the R6/2 mouse model of Huntington’s disease: chronic neurodegeneration does not induce Hsp27 activation. Hum Mol Genet. 2007;16:1078–1090. doi: 10.1093/hmg/ddm057. [DOI] [PubMed] [Google Scholar]
- 46.Satyal SH, Schmidt E, Kitagawa K, Sondheimer N, Lindquist S, Kramer JM, Morimoto RI. Polyglutamine aggregates alter protein folding homeostasis in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2000;97:5750–5755. doi: 10.1073/pnas.100107297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stenoien DL, Cummings CJ, Adams HP, Mancini MG, Patel K, DeMartino GN, Marcelli M, Weigel NL, Mancini MA. Polyglutamine-expanded androgen receptors form aggregates that sequester heat shock proteins, proteasome components and SRC-1, and are suppressed by the HDJ-2 chaperone. Hum Mol Genet. 1999;8:731–741. doi: 10.1093/hmg/8.5.731. [DOI] [PubMed] [Google Scholar]
- 48.Kim S, Nollen EA, Kitagawa K, Bindokas VP, Morimoto RI. Polyglutamine protein aggregates are dynamic. Nat Cell Biol. 2002;4:826–831. doi: 10.1038/ncb863. [DOI] [PubMed] [Google Scholar]
- 49.Matsumoto G, Stojanovic A, Holmberg CI, Kim S, Morimoto RI. Structural properties and neuronal toxicity of amyotrophic lateral sclerosis-associated Cu/Zn superoxide dismutase 1 aggregates. J Cell Biol. 2005;171:75–85. doi: 10.1083/jcb.200504050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 51.Holmberg CI, Staniszewski KE, Mensah KN, Matouschek A, Morimoto RI. Inefficient degradation of truncated polyglutamine proteins by the proteasome. Embo J. 2004;23:4307–4318. doi: 10.1038/sj.emboj.7600426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jordanova A, Irobi J, Thomas FP, Van Dijck P, Meerschaert K, Dewil M, Dierick I, Jacobs A, De Vriendt E, Guergueltcheva V, et al. Disrupted function and axonal distribution of mutant tyrosyl-tRNA synthetase in dominant intermediate Charcot-Marie-Tooth neuropathy. Nat Genet. 2006;38:197–202. doi: 10.1038/ng1727. [DOI] [PubMed] [Google Scholar]
- 53**.Lee JW, Beebe K, Nangle LA, Jang J, Longo-Guess CM, Cook SA, Davisson MT, Sundberg JP, Schimmel P, Ackerman SL. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006;443:50–55. doi: 10.1038/nature05096. The authors demonstrate that perturbing the fidelity of translation in neurons leads to the accumulation of misfolded proteins and, eventually, to neuronal death. [DOI] [PubMed] [Google Scholar]
- 54*.Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem. 2009;78:959–991. doi: 10.1146/annurev.biochem.052308.114844. This recent review proposes that proteostasis is influenced both by the proteostasis network capacity and by protein folding energetics. It further proposes that small molecules can enhance proteostasis by stabilizing specific proteins or by enhancing the proteostasis network capacity. [DOI] [PubMed] [Google Scholar]
- 55*.Wang X, Venable J, LaPointe P, Hutt DM, Koulov AV, Coppinger J, Gurkan C, Kellner W, Matteson J, Plutner H, et al. Hsp90 cochaperone Aha1 downregulation rescues misfolding of CFTR in cystic fibrosis. Cell. 2006;127:803–815. doi: 10.1016/j.cell.2006.09.043. HSP90 cochaperone AHA1 is shown to modulate the HSP90-dependent folding of CFTR. The authors discuss how the chaperone environment can differentiate between deleterious mutations and mild polymorphisms. [DOI] [PubMed] [Google Scholar]
- 56*.Sahi C, Craig EA. Network of general and specialty J protein chaperones of the yeast cytosol. Proc Natl Acad Sci U S A. 2007;104:7163–7168. doi: 10.1073/pnas.0702357104. The authors performed a comprehensive analysis of 13 S. cerevisiae J-domain proteins. They found that some J proteins were genetically non-redundant, thereby providing evidence that certain Hsp70 functions require specific J proteins, possibly for interaction with specific substrates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chan HY, Warrick JM, Gray-Board GL, Paulson HL, Bonini NM. Mechanisms of chaperone suppression of polyglutamine disease: selectivity, synergy and modulation of protein solubility in Drosophila. Hum Mol Genet. 2000;9:2811–2820. doi: 10.1093/hmg/9.19.2811. [DOI] [PubMed] [Google Scholar]
- 58*.Warrick JM, Chan HY, Gray-Board GL, Chai Y, Paulson HL, Bonini NM. Suppression of polyglutamine-mediated neurodegeneration in Drosophila by the molecular chaperone HSP70. Nat Genet. 1999;23:425–428. doi: 10.1038/70532. The authors demonstrate that HSP70 overexpression suppresses the toxic effects of a polyQ-containing protein in an invertebrate model. The effect of HSP70 on aggregation of polyQ protein is shown to be through modifying the aggregation pathway, rather than changing the extent of aggregation. [DOI] [PubMed] [Google Scholar]
- 59.Stevens FJ, Argon Y. Pathogenic light chains and the B-cell repertoire. Immunol Today. 1999;20:451–457. doi: 10.1016/s0167-5699(99)01502-9. [DOI] [PubMed] [Google Scholar]
- 60.Melnick J, Dul JL, Argon Y. Sequential interaction of the chaperones BiP and GRP94 with immunoglobulin chains in the endoplasmic reticulum. Nature. 1994;370:373–375. doi: 10.1038/370373a0. [DOI] [PubMed] [Google Scholar]
- 61.Davis PD, Raffen R, Dul LJ, Vogen MS, Williamson KE, Stevens JF, Argon Y. Inhibition of amyloid fiber assembly by both BiP and its target peptide. Immunity. 2000;13:433–442. doi: 10.1016/s1074-7613(00)00043-1. [DOI] [PubMed] [Google Scholar]
- 62.Yoshiike Y, Minai R, Matsuo Y, Chen YR, Kimura T, Takashima A. Amyloid oligomer conformation in a group of natively folded proteins. PLoS One. 2008;3:e3235. doi: 10.1371/journal.pone.0003235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Suhr ST, Senut MC, Whitelegge JP, Faull KF, Cuizon DB, Gage FH. Identities of sequestered proteins in aggregates from cells with induced polyglutamine expression. J Cell Biol. 2001;153:283–294. doi: 10.1083/jcb.153.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kryukov GV, Pennacchio LA, Sunyaev SR. Most rare missense alleles are deleterious in humans: implications for complex disease and association studies. Am J Hum Genet. 2007;80:727–739. doi: 10.1086/513473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ng PC, Henikoff S. Predicting the effects of amino acid substitutions on protein function. Annu Rev Genomics Hum Genet. 2006;7:61–80. doi: 10.1146/annurev.genom.7.080505.115630. [DOI] [PubMed] [Google Scholar]
- 66**.Gidalevitz T, Ben-Zvi A, Ho KH, Brignull HR, Morimoto RI. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science. 2006;311:1471–1474. doi: 10.1126/science.1124514. Taking advantage of thermodynamically unstable proteins as folding sensors, the authors show that aggregation-prone disease-associated proteins disrupt the cellular folding environment, and, importantly, are themselves more aggregation-prone in the background of metastable proteins. [DOI] [PubMed] [Google Scholar]
- 67*.Ben-Zvi A, Miller EA, Morimoto RI. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0902882106. The authors employ the same metastable proteins as in Gidalevitz, et. al., 2006, as folding sensors in an otherwise wild-type genetic background, and find that the cellular folding environment becomes detectably compromised very early during aging. This provides evidence that the proteostasis network is not robust, likely having consequences on conformational disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gidalevitz T, Krupinski T, Garcia S, Morimoto RI. Destabilizing protein polymorphisms in the genetic background direct phenotypic expression of mutant SOD1 toxicity. PLoS Genet. 2009;5:e1000399. doi: 10.1371/journal.pgen.1000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- 70.Yeyati PL, Bancewicz RM, Maule J, van Heyningen V. Hsp90 selectively modulates phenotype in vertebrate development. PLoS Genet. 2007;3:e43. doi: 10.1371/journal.pgen.0030043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ng AP, Howe Fong J, Sijin Nin D, Hirpara JL, Asou N, Chen CS, Pervaiz S, Khan M. Cleavage of misfolded nuclear receptor corepressor confers resistance to unfolded protein response-induced apoptosis. Cancer Res. 2006;66:9903–9912. doi: 10.1158/0008-5472.CAN-06-0002. [DOI] [PubMed] [Google Scholar]
- 72.Suckow J, Markiewicz P, Kleina LG, Miller J, Kisters-Woike B, Muller-Hill B. Genetic studies of the Lac repressor. XV. 4000 single amino acid substitutions and analysis of the resulting phenotypes on the basis of the protein structure. J Mol Biol. 1996;261:509–523. doi: 10.1006/jmbi.1996.0479. [DOI] [PubMed] [Google Scholar]
- 73.Sachidanandam R, Weissman D, Schmidt SC, Kakol JM, Stein LD, Marth G, Sherry S, Mullikin JC, Mortimore BJ, Willey DL, et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- 74**.Drummond DA, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008;134:341–352. doi: 10.1016/j.cell.2008.05.042. This paper describes a computational evolutionary simulation that takes into account selection against cytotoxic misfolded proteins that are generated by errors in translation. A model is proposed by which thermodynamically unstable proteins caused by translational errors may contribute to neurodegenerative diseases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 76.Cairns NJ, Neumann M, Bigio EH, Holm IE, Troost D, Hatanpaa KJ, Foong C, White CL, 3rd, Schneider JA, Kretzschmar HA, et al. TDP-43 in familial and sporadic frontotemporal lobar degeneration with ubiquitin inclusions. Am J Pathol. 2007;171:227–240. doi: 10.2353/ajpath.2007.070182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, Ackerley S, Durnall JC, Williams KL, Buratti E, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]