Abstract
All cytochrome P450s (CYPs) contain a cysteinate heme iron proximal ligand that plays a crucial role in their mechanism of action. Conversion of the proximal Cys436 to Ser in NH2-truncated microsomal CYP2B4 (ΔCYP2B4) transforms the enzyme into a two-electron NADPH oxidase producing H2O2 without monooxygenase activity [K.P. Vatsis, H.M. Peng, M.J. Coon, J. Inorg. Biochem. 91 (2002) 542–553]. To examine the effects of this ligation change on the heme iron spin-state and coordination structure of ΔC436S CYP2B4, the magnetic circular dichroism and electronic absorption spectra of several oxidation/ligation states of the variant have been measured and compared with those of structurally defined heme complexes. The spectra of the substrate-free ferric mutant are indicative of a high-spin five-coordinate structure ligated by anionic serinate. The spectroscopic properties of the dithionite-reduced (deoxyferrous) protein are those of a five-coordinate (high-spin) state, and it is concluded that the proximal ligand has been protonated to yield neutral serine (ROH-donor). Low-spin six-coordinate ferrous complexes of the mutant with neutral sixth ligands (NO, CO, and O2) examined are also likely ligated by neutral serine, as would be expected for ferric complexes with anionic sixth ligands such as the hydroperoxo-ferric catalytic intermediate. Ligation of the heme iron by neutral serine vs. deprotonated cysteine is likely the result of the large difference in their acidity. Thus, without the necessary proximal ligand push of the cysteinate, although the ΔC436S mutant can accept two electrons and two protons, it is unable to heterolytically cleave the O-O bond of the hydroperoxo-ferric species to generate Compound I and hydroxylate the substrate.
Keywords: CYP2B4, CYP2B4 heme axial mutant, CYP2B4(C436S) mutant, oxyferrous complex, electronic absorption spectroscopy, magnetic circular dichroism (MCD)
Introduction
The cytochrome P450s (P450s or CYPs) are heme-containing monooxygenases. All P450s share a common reaction cycle involving a cysteinate-ligated ferric heme iron active site. Initial substrate binding activates the catalytic cycle by formation of a high-spin complex that accepts an electron from NAD(P)H to form a deoxyferrous state, which binds dioxygen. Acceptance of a second electron followed by protonation leads to formation of reactive enzyme species for incorporation of a single oxygen atom into a wide range of organic substrates [1]. The P450s are unrivaled in the number of catalytic activities they can perform on substrates varying from retinoids, eicosanoids and steroids to xenobiotic compounds [1,2] (reviews by 15 leading P450 research groups compiled in [2]). As a result, this class of heme enzymes has been the target of a wide range of structure-function investigations for decades [2–4]. Nonetheless, the catalytic versatility and the ability of P450 to generate multiple activated oxygen species are not completely understood, although it is clear that the proximal cysteine plays a key mechanistic role via the thiolate “push” effect [5].
The importance of the cysteinate proximal heme ligand in P450 has been previously discussed [5–7]. There are two other important classes of proteins which also contain a cysteinate proximal ligand: C. fumago chloroperoxidase [8–10] and nitric oxide synthase [11–13], both of which show unique spectroscopic features like P450. The question of just how the cysteine residue facilitates this unparalleled catalytic activity has been a matter of great debate and has been tested through the preparation of several mutants. In P450cam (CYP101 from Pseudomonas putida), the axial cysteinate sulfur atom is surrounded by three amide protons of Leu358, Gly359 and Gln360 [7, 14]. These residues are within the H-bonding distance with the sulfur atom in the cysteine. Morishima and coworkers studied the role of the axial thiolate in cytochrome P450cam through the mutation of Gln360 to Pro and Leu358 to Pro amino acid residues [14]. In each of these mutants, one of the conserved amide protons proposed to neutralize the negative charge of the thiolate sulfur has been removed. This diminishes the hydrogen bond stabilization of the thiolate ligand and increases the electron donation (thiolate push) towards the iron center. As a consequence, the iron reduction potentials are shifted by −45 mV and −35 mV, respectively, for the Q360L and L358P P450cam mutants. The L358P mutant showed an increased push effect of the axial cysteine when compared with the wild-type enzyme, leading to increased protonation of the inner oxygen and decreased protonation of the outer oxygen atom in the putative iron-hydroperoxo intermediate of P450cam [15]. A related study that reinforces the above argument has also been done in which the role of the proximal thiolate iron ligand was examined by mutation of the Cys357 to His in cytochrome P450cam [16, 17]. The C357H mutant has little hydroxylation activity vs. camphor, but retains the catalytic properties of a proximal histidine-ligated protein as a peroxidase. For example, the C357H P450cam mutant was shown to be capable of performing hydrogen peroxide-dependent guaiacol peroxidation at a rate that was two orders of magnitude higher than wild-type P450cam. The loss of monooxygenase activity of C357H was attributed to the change in active site structure to six-coordinate (low-spin) as seen in the inactive form of the enzyme known as P420cam. A similar study has also been done by Coon and co-workers with CYP2B4 from rabbit liver microsomes, a member of the cytochrome P450 super family, by preparing a double mutant of A298E/C436H where the proximal Cys 436 was replaced with His [18]. As with the C357H P450cam mutant [16], there was a loss of the monooxygenase activity, but also a considerable enhancement of peroxidase activity (by ~100-fold with pyrogallol as substrate) vs. the wild-type CYP2B4 enzyme. These examples highlight the important role played by the proximal cysteinate ligand during oxygen activation by P450.
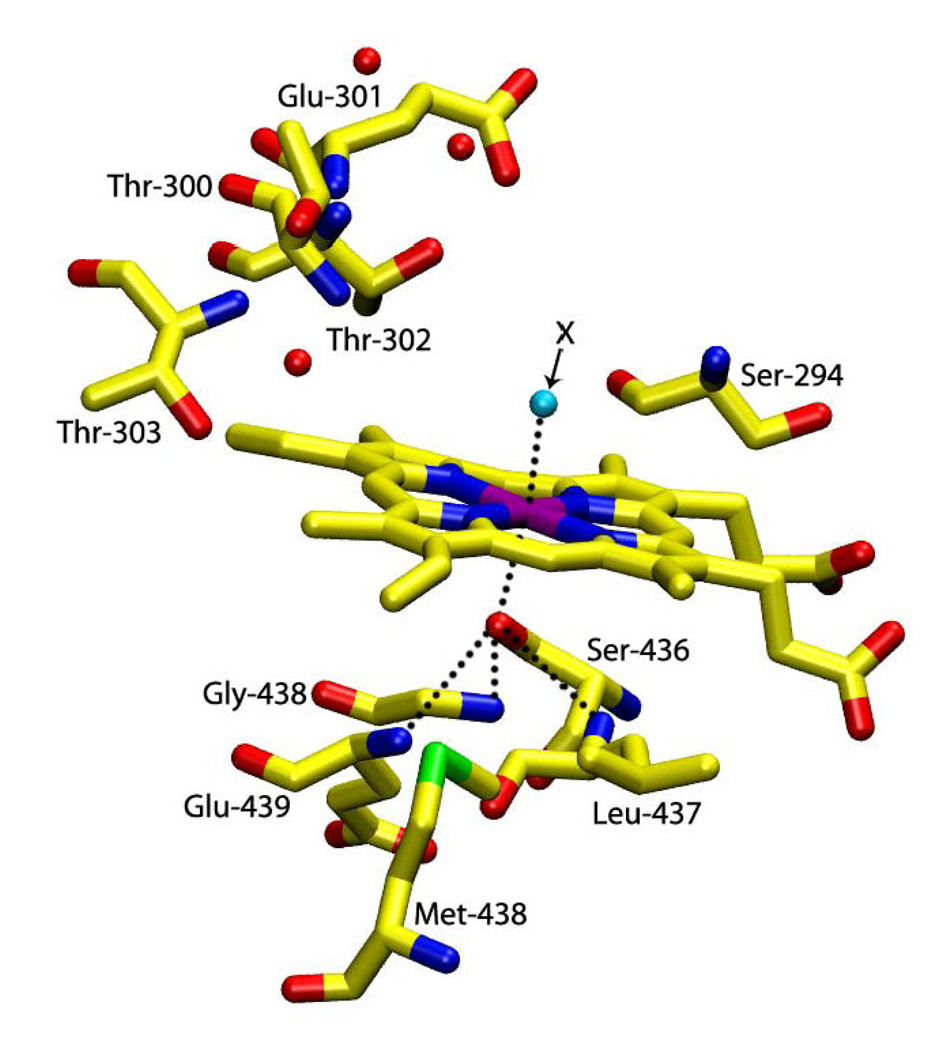
CYP2B4 from rabbit liver microsomes is responsible for the oxidative metabolism of a wide variety of physiologically important as well as foreign (xenobiotic) substrates. It is membrane-bound and accepts electrons from a microsomal NADPH-cytochrome P450 reductase containing FAD and FMN [3,19]. To achieve a better understanding of the role of the axial thiolate ligand in P450 catalysis, the cysteine residue, Cys 436 (Fig. 1) [19], was mutated to serine (C436S) by Vatsis, Peng and Coon [5]. The C436S CYP2B4 mutant was unable to oxidatively metabolize compounds such as d-benzphetamine, 1-phenylethanol and 4-fluorophenol [5]. Nonetheless, the mutant was still able to function as a two-electron oxidase, accepting two electrons from NADPH and reducing dioxygen to hydrogen peroxide. Furthermore, the C436S mutant did not show any catalase (H2O2 disproportionation) activity. Consequently, since the C436S mutant is enzymatically active as a two-electron oxidase (i.e., it is not an enzymatically inactive P420 form), it should be a useful system to separate the contribution of the proximal ligand to different aspects of the P450 mechanism such as substrate binding, electron transfer, changes in spin and coordination properties and the generation of catalytically active intermediates. To do so, it is first necessary to better understand the coordination structure of the C436S mutant.
Figure 1.
Simulated schematic representation of the active site structure of C436S mutant of CYP2B4 prepared using the Visual Molecular Dynamic (VMD) program (pdb file 1SUO [19]). In this study, trans-axial X stands for H2O, NO, CO and O2 ligands.
In the present work, we have examined the ferric and ferrous states of the truncated C436S mutant of CYP2B4 (ΔC436S CYP2B4) and several of its ligand adducts (Fig. 1) with electronic absorption (EA) and magnetic circular dichroism (MCD) spectroscopy. MCD spectroscopy has been repeatedly shown to be a very sensitive probe of the coordination structure of heme iron centers in proteins by comparison of the MCD spectra of new heme proteins or mutants with those of structurally defined heme complexes [20]. The H93G myoglobin (Mb) “cavity mutant” is an especially useful system for modeling heme proteins [20,21,22]. We have compared the MCD and EA spectra of ΔC436S CYP2B4 to those of H93G Mb with different O-donor ligands, such as phenolate (PhO−), benzoate (BzO−) and acetate (AcO−) [23,24], as well as to relevant states of H93Y Mb, wild-type Mb and horseradish peroxidase (HRP), in order to determine the ligation state and establish the heme iron coordination structure of this interesting P450 mutant.
2. Experimental Procedures
2.1 Materials
Nitric oxide gas was obtained from Matheson Co. All other chemicals were purchased from Sigma Aldrich and used as received.
2.2 Wild-Type and Mutant Heme Proteins
Truncated wild-type CYP2B4 (ΔCYP2B4) and its C436S mutant [ΔC436S CYP2B4 (ΔC436S)], expressed in E. coli and purified as described [16,25,26], were gifts of Drs. K. P. Vatsis and M. J. Coon. H93Y human myoglobin, prepared as reported [27], was a gift from Dr. Masao Ikeda-Saito. Imidazole-free sperm whale H93G Mb was prepared as reported [28,29].
2.3 Preparations of Samples for EA and MCD Spectroscopy
Unless otherwise noted, spectral measurements were carried out at 4 °C (~50 µM) in 100 mM potassium phosphate buffer (pH 7.0) for H93G Mb and H93Y Mb and in 0.1 M potassium phosphate buffer, pH 7.4, containing 5% glycerol for ΔC436S CYP2B4. Preparation of the H93G Mb phenolate (PhO−) adduct was achieved by addition of phenol in dimethyl sulfoxide (DMSO) to exogenous-ligand-free ferric protein (~50 µM) [23]. After examination of the ferric state of each protein (~1.5 mL in a 1 cm septum-sealed cuvette), the samples were extensively purged over 30 min with a continuous flow of nitrogen while on ice. Reduction to the deoxyferrous state was achieved by addition of 1 µL of 100 mM sodium dithionite solution. The ferrous-NO, ferrous-O2 and ferrous-CO complexes were generated by bubbling NO, O2 and CO gases, respectively, using a needle into the deoxyferrous sample in a cuvette fitted with a rubber septum.
2.4 Spectroscopic Techniques
EA spectra were recorded with a Cary 400 spectrometer (at ~4 °C) or a JASCO J600A spectropolarimeter (at ~4 °C). MCD spectra were measured in a 1 cm or 0.2 cm cuvette at a magnetic field of 1.41T with the JASCO J600A spectropolarimeter as described [30]. Data acquisition and manipulation were done as reported [30], with JASCO software.
3. Results and discussion
Previous work on the C436S mutant of ΔCYP2B4 focused on the importance of the axial cysteine residue and what happens to the activity of the protein when that residue was altered [5]. The data supported the idea that change of the thiolate ligand of Cys to a serinate ligand in ΔC436S led to the loss of hydroxylation activity in the oxidative metabolism of several compounds. However, the mutant was still enzymatically active as a two-electron oxidase in which the reduction of oxygen to hydrogen peroxide was coupled to the oxidation of NADPH [5]. The present study has centered on the investigation of the coordination structure and spin state of the mutant protein. Hence, we have prepared exogenous ligand (X)-bound (X = NO, O2 and CO) forms of the mutant (Fig. 1) in the ferric and ferrous states to generate complexes for spectroscopic examination.
In previous studies, the H93G Mb cavity mutant has been used successfully to understand the heme coordination structures in heme proteins [20,32]. The cavity generated by replacing the proximal His 93 with Gly can be filled with exogenous ligands, such as imidazole, amine, phenolate, thiolate etc. to obtain a wide variety of models for heme proteins [20]. Therefore, EA and MCD spectra of the ferric, ferric-NO, deoxyferrous, ferrous-NO, oxyferrous, and ferrous-CO states of the mutant C436S CYP2B4 have been measured and the resulting spectra have been compared with those of parallel derivatives of H93G Mb O-donor adducts (phenolate or acetate), as well as in some cases with H93Y Mb, wild-type Mb, and wild-type HRP, in order to establish the spin state and coordination structure of C436S CYP2B4.
3.1 Ferric form of the C436S CYP2B4 mutant
First of all, it is important to note the differences seen between each of the states of wild-type ΔCYP2B4 and the ΔC436S CYP2B4 mutant. The EA spectra for the ferric species had a shift in the Soret peak position from 417 nm for the wild-type (data not shown) to 404 nm for the mutant [5]. The EA and MCD spectra of the ΔC436S CYP2B4 mutant in the ferric state are most similar to those of the five-coordinate tyrosinate-ligated ferric H93Y Mb [33] as well as the phenolate adduct of ferric H93G Mb [23](Fig. 2, A and C). In Fig. 2, the EA spectra show sharp Soret absorption maxima with the most intense feature at ~404 nm and a broad pπ-dπ charge transfer band at 597 nm for both ferric H93Y Mb and the ΔC436S CYP2B4 mutant. At the same time, the MCD spectra of ferric ΔC436S CYP2B4 and H93Y Mb show derivative-shaped features centered at ~410 nm and another derivative-shaped feature centered at ~610 nm in the visible region along with small peaks at ~475 and ~510 nm. These spectral features of ΔC436S CYP2B4 and H93Y Mb are also similar to those of H93G(PhO−) Mb adduct and, to some extent, to those of H93G(AcO−) Mb (Fig. 2, B and C). However, the high-spin charge transfer band for ferric H93(AcO−) Mb adducts is red-shifted to 617 nm (vs. 604 nm for the phenolate complex) and the MCD spectrum of acetate-bound H93G Mb in this region is non-symmetric (with more intense trough at ~630 nm than a peak at ~605 nm). It has recently been established by crystallography that the acetate adduct of ferric H93G Mb is six-coordinate with acetate and water as ligands [23]. Based mainly on the close spectral similarity to ferric H93Y Mb (Fig. 2A and C), we conclude that ferric ΔC436S CYP2B4 contains a five-coordinate serinate-ligated heme center.
Figure 2.
MCD (A and B) and electronic (UV-Vis) absorption (C) spectra of the ferric state of the ΔC436S CYP2B4 mutant (solid line), human H93Y Mb (dashed line), phenolate (PhO−)-(dotted line) and acetate (AcO−)-ligated (dot-dashed line) ferric H93G Mb. The phenolate (PhO−)- and acetate (AcO−)-ligated ferric H93G Mb were prepared by adding 36 and 850 mM phenol and acetate, respectively, to exogenous-ligand-free ferric H93G Mb. The spectra were recorded in 0.1 M potassium phosphate at pH 7.0 at 4°C.
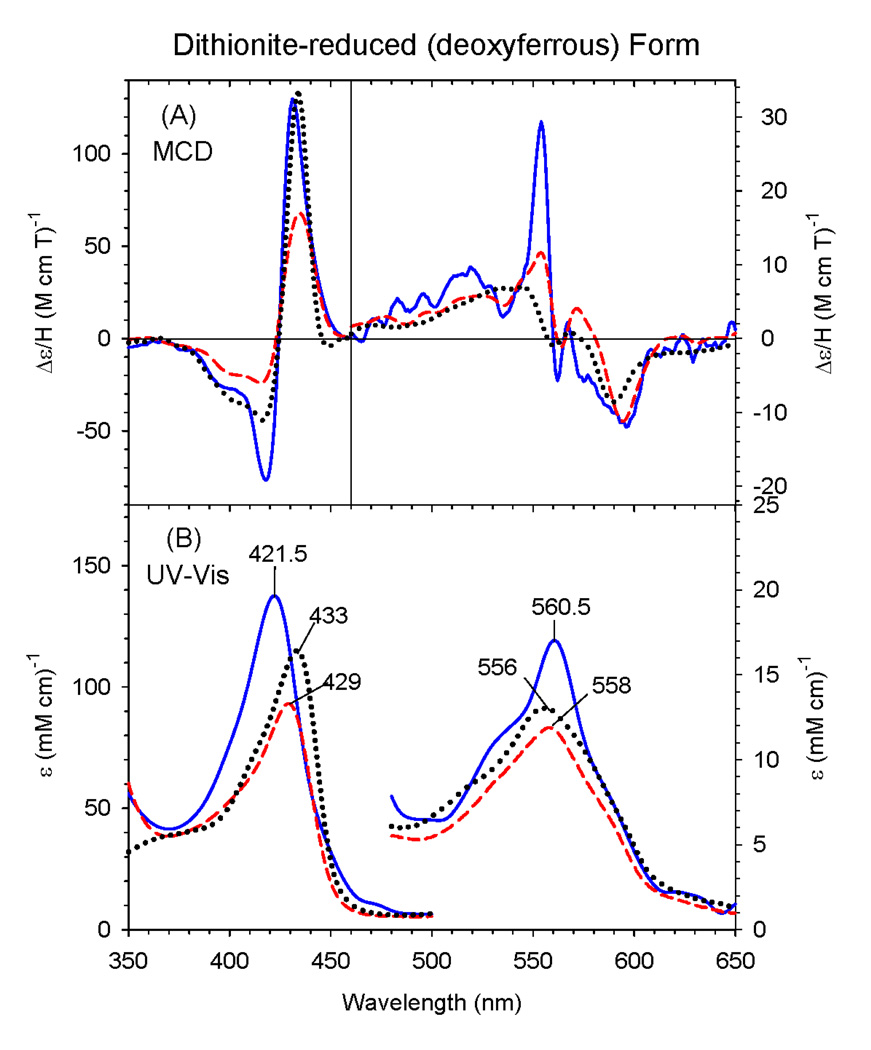
3.2 Deoxyferrous Form of the C436S mutant
Fig. 3 shows the EA and MCD spectra of deoxyferrous (dithionite-reduced) ΔCYP2B4 C436S overlaid with those of human deoxyferrous H93Y Mb and wild-type sperm whale Mb. The Soret peak for the ΔC436S mutant (421.5 nm) is considerably blue-shifted compared with those of human H93Y Mb (429 nm) and wild-type Mb (433 nm), while the visible region EA spectral peak positions (556 – 561 nm) for the three proteins are somewhat similar. In their MCD spectra, the Soret region band shapes for these three heme proteins are similar except for some intensity differences. In the visible region, the MCD spectra of the three proteins are overall similar except that the ΔC436S mutant has an additional sharp derivative-shaped band centered around 560 nm. Therefore, although deoxyferrous ΔC436S (Fig. 3) shows spectral features that are mainly indicative of a five-coordinate complex as does deoxyferrous wild-type Mb, it is apparent that it has some six-coordination as well that would exhibit the sharp derivative-shaped feature as mentioned above [34]. The sixth ligand would likely be a water molecule, as is seen in the ferric state. This is similar to the previously reported data for ferrous human H93Y Mb which were interpreted as being due to mainly five-coordination with a small amount of six-coordination, again from the presence of the small derivative-shaped MCD feature centered around 560 nm [24,35]. Further, the relatively intense Soret MCD peak (~130 M−1cm−1T−1) and the trough near 580 nm in the MCD spectrum of the ΔC436S mutant are typical of a five-coordinate ferrous complex. Some additional features (e.g., a prominent shoulder at ~530 nm) appear in visible region of the EA spectrum due to slight six-coordination in ΔC436S in contrast to the broadly single peak in the visible region at 558 nm for H93Y Mb.
Figure 3.
MCD (A) and electronic (UV-Vis) absorption (B) spectra of deoxyferrous ΔC436S CYP2B4 mutant (solid line), human H93Y Mb (dashed line) and SW Mb (dotted line). The spectra for the H93Y Mb and SW Mb were obtained in 0.1 M potassium phosphate (pH 7.0) at 4 °C and for the ΔC436S CYP2B4 mutant in 70% (v/v) glycerol and 30% 0.3 M potassium phosphate (pH 8.0) at −50 °C.
Since P450 normally has an anionic proximal ligand (cysteinate form) in the ferrous state, proximal side of the P450 is setup to stabilize this additional charge which accumulates during reduction from ferric to ferrous. [9]. This is due to the fact that the proximal donor atom is adjacent to the amide protons of Gly438 (3.30 Å) and Leu437 (3.54 Å) in CYP2B4 that can form one or two hydrogen bonds [19]. The system to which it is being compared, deoxyferrous Mb, is not setup to stabilize an anionic ligand in the ferrous state. In particular, our efforts to form anionic adducts of ferrous H93G Mb have consistently failed [20]. For example, thiol addition to ferrous H93G Mb leads to thiol (i.e., not thiolate) ligation regardless of thiol acidity and/or pH [31]. Since alcohols are less acidic than thiols, it is even less likely that an alcohol would deprotonate upon binding to ferrous H93G Mb. Egeberg et al. have examined the identity of a heme iron ligand(s) of proximal or distal His mutants of sperm whale Mb and determined that ferrous (dithionite-reduced) sperm whale H93Y Mb is five-coordinate high spin, but does not have distal His ligated [36]. Instead, they concluded that either water or another amino acid might be the ligand to the ferrous H93Y Mb heme iron. On the other hand, an analogous human H93Y Mb mutant appears to form the distal His (H64)-ligated five-coordinate high spin ferrous species [27]. Based on our results, we propose that a neutral tyrosine is an even more likely axial ligand in ferrous sperm whale H93Y Mb. Hence, it is reasonable to assume that neutral serine is the proximal axial ligand in the ferrous state of ΔC436S CYP2B4 mutant as well.
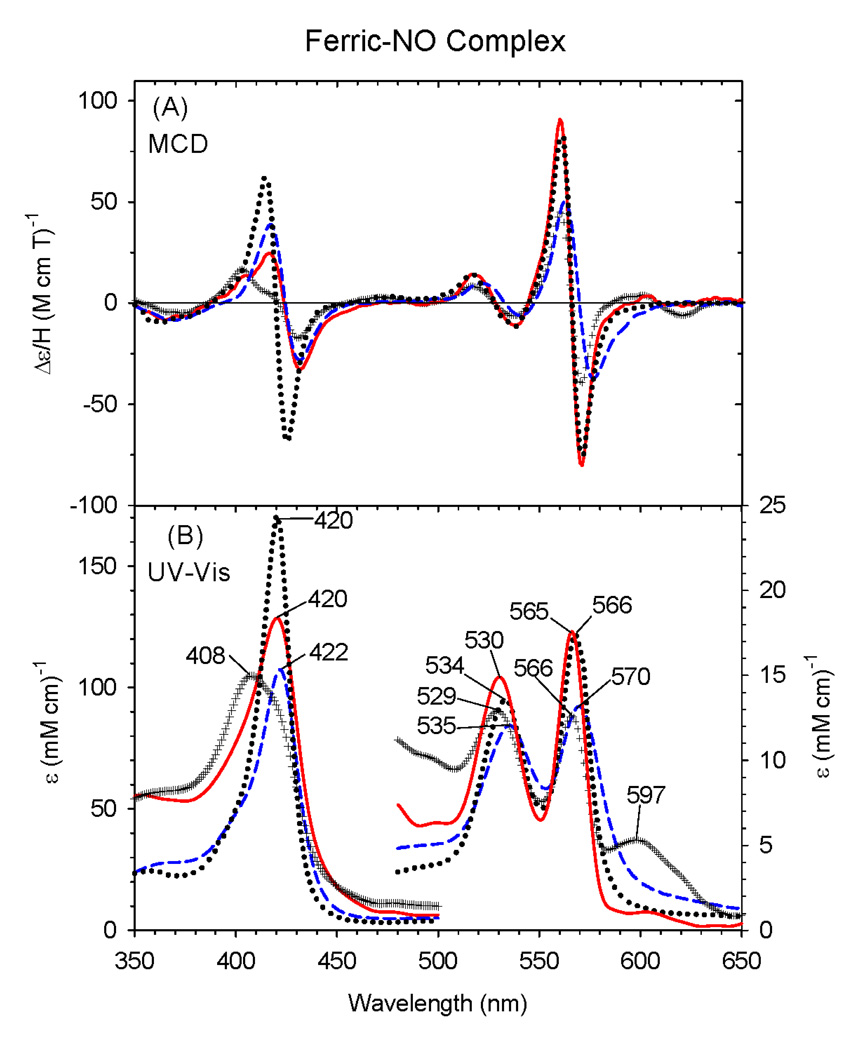
3.3 Ferric- and Ferrous-NO complexes
Comparison of the MCD and EA spectra of ferric-NO ΔC436S CYP2B4 and ferric-NO H93Y human Mb [27] in Fig. 4 (black cross vs. blue dashed lines) shows that the former does not fully bind NO to form a complete six-coordinate complex as judged by the EA peak at 597 nm (ε597 = ~ 4.9 mM−1 cm−1) indicative of remaining NO-free native ferric ΔC436S CYP2B4 (ε597 = ~ 9.6 mM−1 cm−1) (Fig. 2). Furthermore, formation of the ferric-NO H93Y complex was not complete due to some NO-free protein (~10%) (data not shown). To calculate by extrapolation the spectra of the 100% ferric-NO for ΔC436S CYP2B4 complex, 50% (= ~4.9/9.6) of NO-free ferric ΔC436S is subtracted from the observed spectrum based on an assumption that NO-saturated ferric ΔC436S CYP2B4 should not have an absorption peak at ~600 nm. The resulting MCD spectrum (Fig. 4A, red solid line) compares favorably with those of ferric-NO H93Y Mb (calculated for 100% NO saturation, Fig. 4, blue dashed line) and are typical of low-spin six-coordination with an intense derivative-shaped feature centered at ~570 nm. This can also be confirmed by comparing it to the ferric-NO complex of HRP as shown in Fig. 4. However, the main difference is the relatively high intensity of the derivative-shaped Soret MCD band centered at ~420 nm for ferric-NO HRP. Since ferric hemes prefer to have an anionic rather than neutral ligands to achieve charge neutrality at the heme center (the overall charge of which is a sum of the individual charges of ferric iron (3+), porphyrin dianion (2−) and the axial ligand), the serine is likely to be deprotonated (1−) in the ferric-NO complex of the ΔC436S mutant.
Figure 4.
MCD (A) and electronic (UV-Vis) absorption (B) spectra of the ferric-NO complex of the ΔC436S CYP2B4 mutant (solid line), human H93Y Mb (dashed line) and HRP (dotted line). The spectra for the ΔC436S CYP2B4 mutant and H93Y Mb were obtained by calculating the experimental spectra (+ + + line for the ΔC436S CYP2B4 mutant and not shown for H93Y Mb) to 100% NO saturation assuming that the experimentally obtained spectra were 50% and 90% NO-saturated, respectively. The spectra were recorded in 0.1 M potassium phosphate at pH 7.0 at 4°C.
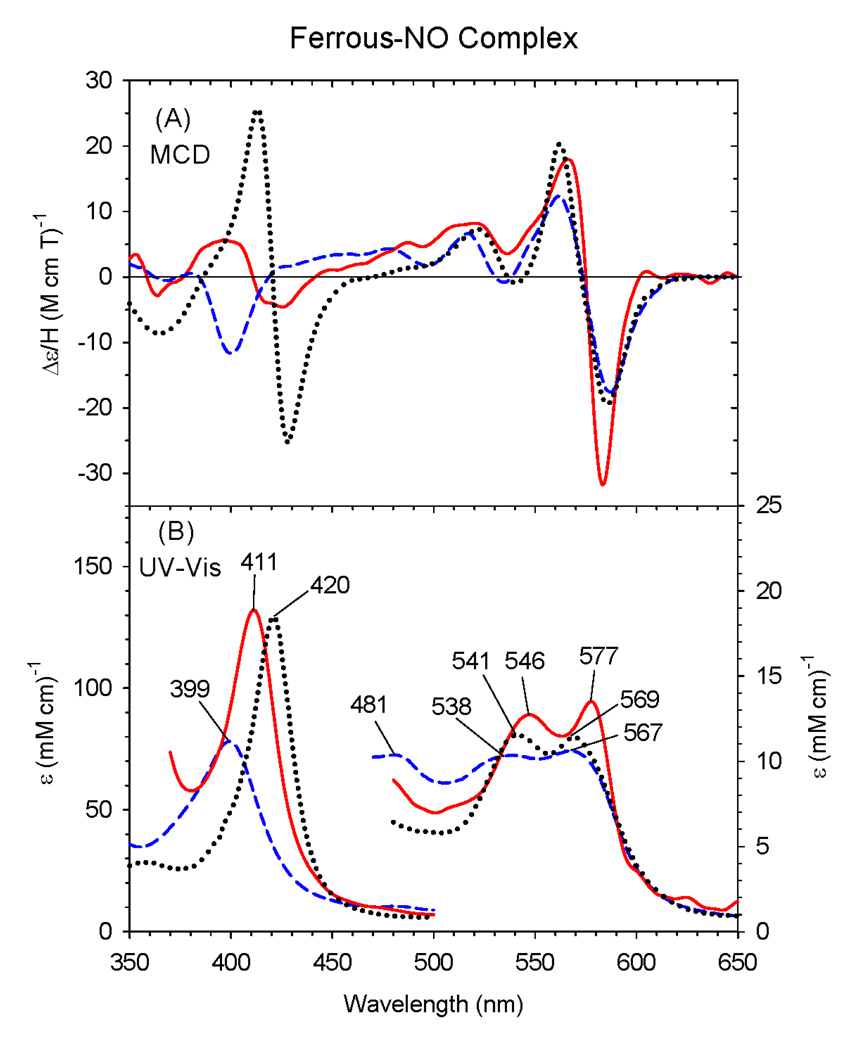
The EA and MCD spectra of ferrous-NO ΔC436S CYP2B4, H93Y Mb and wild-type Mb are overlaid in Fig. 5. We have previously reported that ferrous H93Y Mb forms a five-coordinate NO complex [33] (i.e., the tyrosine ligand present in the ferric state is no longer bound to the iron). This conclusion was reached based on the presence of a prominent MCD Soret trough at about 400 nm for ferrous-NO H93Y Mb together with the Soret EA peak around 400 nm. In contrast, the EA and MCD spectra of ferrous-NO ΔC436S are typical of six-coordinate ferrous-NO complexes based on the presence of a distinct (although not sharp) derivative-shaped MCD Soret feature, centered at ~411 nm, and the Soret EA peak at 411 nm [22]. The EA and MCD spectral features for ferrous-NO ΔC436S CYP2B4 are also reasonably similar to those of the six-coordinate ferrous-NO HRP adduct. Previously we have reported that the ferrous-NO complex of P450epoK in the presence of substrate likely has a neutral cysteine (thiol) ligated with the Soret absorption peak at 413 nm in the presence of substrate [37]. Therefore, from the presence of the Soret absorption peak at 411 nm, we can presume that ferrous-NO C436S CYP2B4 has a neutral serine ligand trans to NO. To the best of our knowledge, this is the first report of a six-coordinate, ferrous-NO complex with a neutral oxygen donor proximal ligand in active site of heme protein.
Figure 5.
MCD (A) and electronic (UV-Vis) absorption (B) spectra of the ferrous-NO complex of the ΔC436S CYP2B4 mutant (solid line), human H93Y Mb (dashed line) and HRP (dotted line). The spectra were recorded in 0.1 M potassium phosphate at pH 7.0 at 4°C.
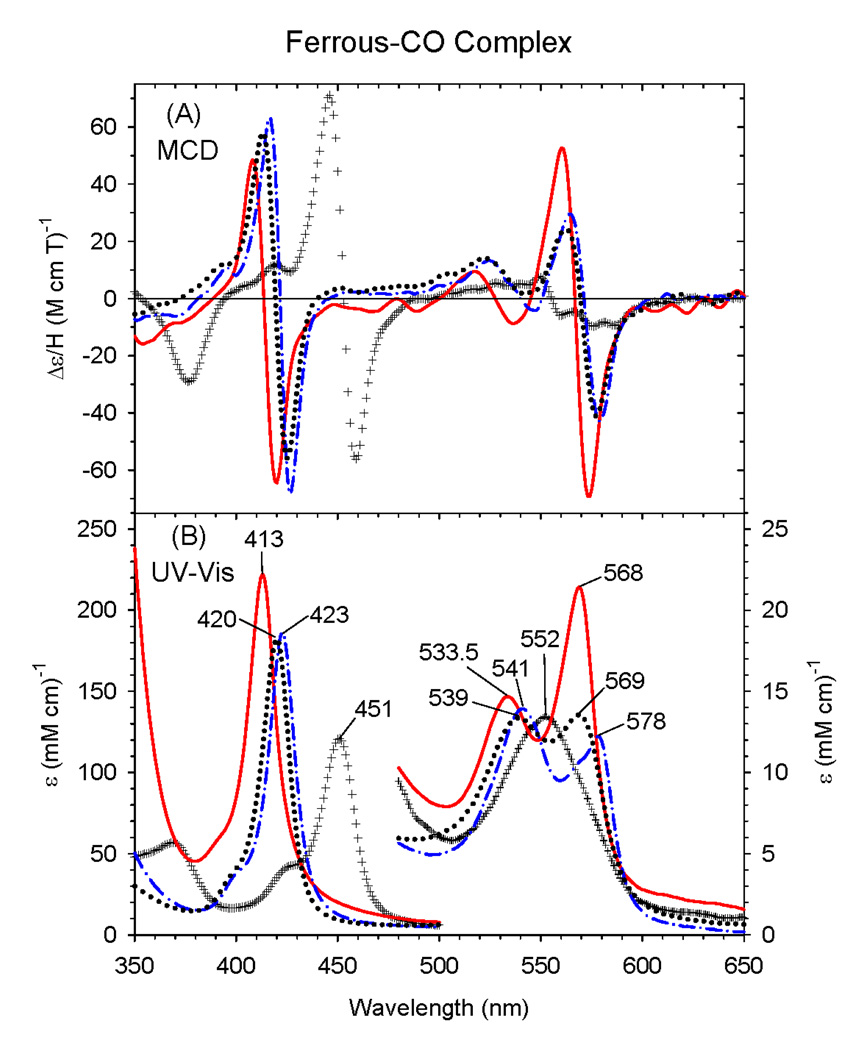
3.4 Ferrous-CO complex
There are also significant changes in the spectral properties of the ferrous-CO complex of ΔC436S CYP2B4 protein compared to the wild-type protein (Fig. 6). The Soret absorption peak for the wild-type enzyme is at 451 nm with a single visible peak at 552 nm [6]. In contrast, the ΔC436S CYP2B4 mutant shows a sharp Soret EA peak at 413 nm with peaks in the visible region at 533.5 nm and 568 nm [6]. This significant blue-shift in the Soret peak provides the clearest indication that the mutant has a dramatically changed coordination structure due to the neutral serine proximal ligand in ferrous state. The MCD spectra of ferrous-CO ΔC436S CYP2B4, H93Y Mb, wild-type ΔCYP2B4 and wild-type Mb are overlaid in Fig. 6. Despite the difference in the position of the Soret EA peak for ferrous-CO ΔC436S, the overall MCD spectral features of ΔC436S CYP2B4 are similar to those of the ferrous-CO complex of H93Y Mb mutant (with the peaks at 420, 539 and 569 nm). The Soret EA peak for ferrous-CO bovine liver catalase and coral allene oxide synthase, presumably ligated by anionic tyrosinate, is found at 425 nm [38,39]. Given that protonation of the thiolate sulfur donor (thiolate → thiol) leads to a 30 nm blue-shift in the position of the ferrous-CO Soret maximum (~450 to 420 nm), the 12 nm blue-shift (425 to 413 nm) seen when comparing ferrous-CO bovine liver catalase and coral allene oxide synthase vs. ferrous-CO ΔC436S is likely the result of ligation of a neutral rather than anionic serine oxygen donor to the heme trans to bound CO in the latter.
Figure 6.
MCD (A) and electronic (UV-Vis) absorption (B) spectra of the ferrous-CO complex of the ΔC436S CYP2B4 mutant (solid line), wild-type ΔCYP2B4 (+ + + line), human H93Y Mb (dotted line), and sperm whale Mb (dot-dashed line). The spectra were recorded in 100 mM potassium phosphate at pH 7.0 at 4°C.
Interestingly, the proximal His25Ala cavity mutant of human liver heme oxygenase (HO) has also been shown to exhibit an unusually blue-shifted Soret absorption peak at 411 nm (with 533 and 566 nm visible region peaks) in the ferrous-CO state [39]. Examinations of ferrous-CO HO with resonance Raman spectroscopy [υ(CO) and υ (Fe-CO), which are 1960 and 529 cm^-1, respectively] relative to empirical correlations between these two parameters for various Fe(II)-CO heme systems, led the authors to conclude that the proximal ligand for ferrous-CO H25A HO is a much weaker ligand than a histidine and that the carbon monoxide binding site is hydrophobic [40]. In addition, the close similarities between the resonance Raman parameters of the ferrous-CO complex of H25A HO to those of the ferrous-CO complexes of Fe(deuteroheme IX dimethyl ester)-THF and Fe(Tpiv)-THF [41] complexes (where THF and Tpiv stand for tetrahydrofuran and α,α,α,α-meso-tetrakis(o-pivalamidophenyl)porphyrin, respectively), where the axial ligand trans to bound CO is most likely to be the oxygen atom of THF, further supported this conclusion.
Another case of such an unusually blue-shifted Soret absorption peak for a ferrous-CO protoheme complex has been found for the heme binding NEAr iron Transporter (NEAT) domains of IsdC (NEAT-C), a protein involved in heme scavenging by Staphylococcus aureus, which exhibits MCD and EA (Soret peak at 412 nm) spectral features that are similar to those of ferrous-CO ΔC436S. Even though the authors interpreted the unique spectra of the ferrous-CO NEAT-C protein as attributable to formation of a protein-free bis-carbonmonoxy-Fe(II)-heme [42], the same explanation is unlikely to apply to ferrous-CO ΔC436S CYP2B4 because this derivative as well as the ferric, ferrous (dithionite-reduced) and ferrous-O2 forms of the ΔC436S protein all have clearly detectable CD signals between 300 and 700 nm (data not shown). Since the free heme exhibits no optical activity due to its highly symmetric structure, the presence of CD signals indicates that the heme is associated with the protein [43].
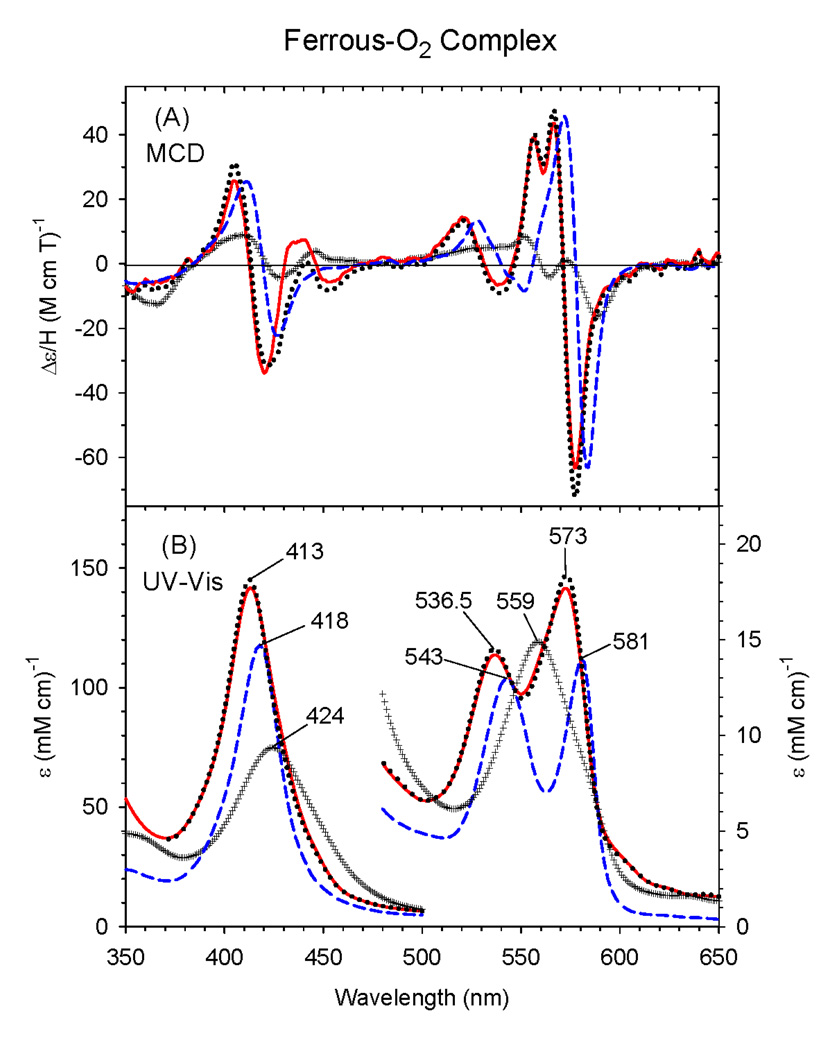
3.5 Ferrous-O2 complex
The EA and MCD spectra of oxyferrous C436S CYP2B4 prepared at ~-50 °C in 100 mM potassium phosphate buffer (at pH 7.0) in the presence of 70% glycerol are displayed in Fig. 7. The corresponding spectra of wild-type ΔCYP2B4 and wild-type Mb are included for comparison. Interestingly, the spectra of this proximal side mutant exhibit a blue-shifted Soret absorption peak at 413 nm in its oxyferrous state compared to 424 nm for WT Δ2B4 or 418 nm for wild-type Mb. In the visible region of the EA spectrum of the oxy-ΔC436S CYP2B4 mutant, there are two peaks at 536.5 nm and 573 nm. In comparison, wild-type ΔCYP2B4 has only one peak at 559 nm. The MCD spectrum of the oxyferrous ΔC436S CYP2B4 mutant thus generated is quite different from that of oxy-wild-type Δ2B4, but there are some similarities with the wild-type Mb spectrum, especially the overall enhanced intensity of the features and the derivative-shaped bands in the Soret and visible regions. Furthermore, there is a unique split in the visible region peak (around 562nm) of the MCD spectrum of ferrous-O2 ΔC436S CYP2B4. In addition, an apparently extra MCD spectral feature between 430 – 470 nm (a broad peak and trough at ~435 and ~455 nm, respectively) suggests the presence of a minor, second species in the O2 complex of the ferrous ΔC436S mutant. The slight broadening on the low-energy side of the Soret EA peak suggests that there could be some contamination by deoxyferrous protein due to incomplete O2 binding; subtraction of 10% of the spectrum of that derivative from the recorded spectrum (red solid line) removes some, but not all of this minor contribution (black dotted line). Nonetheless, the overall spectral properties of the oxyferrous ΔC436S mutant are clearly distinct from those of either the wild-type protein or native Mb, consistent with the conclusion that the new species has a different coordination structure(s), presumably the first example of a neutral alcohol-ligated ferrous-O2 adduct in a protein.
Figure 7.
MCD (A) and electronic (UV-Vis) absorption (B) spectra of the ferrous-O2 complex of the ΔC436S CYP2B4 mutant (solid line) [as well as 10% deoxyferrous subtracted spectrum (dotted line)], wild-type ΔCYP2B4 (+ + + line) and sperm whale Mb (dashed line). The spectra were measured at ~−50 °C in a 30/70 (v/v) mixture of 100 mM potassium phosphate buffer (at pH 7.0) and glycerol.
4. Conclusions
In this study, we have sought to establish a better understanding of the ligation and coordination states of ΔC436S CYP2B4 mutant by preparing and spectroscopically characterizing the ferric, ferrous and ferrous ligand (X) adducts (X = CO, NO and O2) of the protein. The EA and MCD spectral features in ferric-ΔC436S mutant are indicative of a five-coordinate species, which is different from those of wild-type substrate-free ΔCYP2B4 even though both proteins have an anionic proximal axial ligand, deprotonated serine (serinate) and cysteine (cysteinate), respectively. The EA and MCD spectra of the several ferrous ΔC436S CYP2B4 derivatives examined most closely resemble those of the parallel ferrous H93Y Mb forms and are most likely the result of ligation by a neutral alcohol ligand. In particular, the spectra of deoxyferrous ΔC436S CYP2B4 have been compared to those of several deoxyferrous heme centers and are most similar to the spectra of ferrous H93Y human Mb. The additional information we see with NO in ferric and deoxyferrous states suggest that the Ser coordination to iron is mostly intact during the sixth ligand binding, even though we did not generate homogeneous complexes. The ferrous-NO derivative of ΔC436S CYP2B4 mutant is six-coordinate and most likely has a neutral serine ligand. The MCD spectra of ferrous-CO and –O2 complexes of the mutant exhibit distinct features that are consistent with the formation of six-coordinate low-spin complexes, again likely with neutral alcohol ligation.
Previously we have reported that proximal neutral cysteine coordination in the ferrous state in monooxygenases can lead to loss of catalytic activities [31]. Those results demonstrated that retention of thiolate ligation throughout the P450 reaction cycle is only possible due to the careful design of the proximal heme environment to stabilize the anionic thiolate ligand in the ferrous states of the enzyme. The present results reveal that in the ferrous state, the proximal heme environment of P450 is not able to stabilize the deprotonated, alcoholate, form of the significantly less acidic serine alcohol ligand (pKa = ~13 for serine vs. ~8.3 for cysteine). The inability of ΔC436S CYP2B4 to retain the anionic serinate proximal ligand in the ferrous state with neutral sixth ligands is likely to extend to anionic ligand-bound ferric states (which also have an overall neutral heme unit) including the hydroperoxo-ferric intermediate, the key precursor of the Compound I species. This is the most probable cause for its negligible monooxygenase activity. Nonetheless, despite the switch of the axial ligand donor atom from sulfur to oxygen, the ΔC436S mutant maintains the ability to accept two electrons and two protons, i.e., NADPH oxidase activity. However, without the necessary proximal ligand push [4,9], the ΔC436S mutant is unable to heterolytically cleave the O-O bond of the hydroperoxo-ferric intermediate to generate Compound I and hydroxylate the substrate. Thus, the present results provide new information about the coordination structure of the ΔC436S mutant that sheds light on our basic understanding of how the proximal ligand “push effect” influences the catalytic activity of cytochrome P450.
Acknowledgments
We thank Professor Steven G. Boxer for the H93G expression system, Dr. Kostas P. Vatsis and Professor Minor J. Coon for the truncated wild-type CYP2B4 and its ΔC436S mutant, and Professor M. Ikeda-Saito for the human H93Y Mb protein. Support from the NIH (GM 26730) and Research Corp. (to J.H.D.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
We dedicate this article to Professor Minor J. (Jud) Coon in recognition of his brilliant contributions and outstanding leadership to the field of cytochrome P450 over the past five decades. We wish to especially draw attention to his exceptional qualities as a role model, not just for how to do world-class science, but also for how to be a wonderfully supportive colleague and friend.
References
- 1.Sono M, Roach MP, Coulter ED, Dawson JH. Chem. Rev. 1996;96:2841–2888. doi: 10.1021/cr9500500. [DOI] [PubMed] [Google Scholar]
- 2.Ortiz de Montellano PR, editor. Cytochrome P450: Structure, Mechanism, and Biochemistry. 2nd. New York: Plenum; 1995. [Google Scholar]
- 3.Bridges A, Gruenke L, Chang YT, Vakser IA, Loew G, Waskell L. J. Biol. Chem. 1998;273:17036–17049. doi: 10.1074/jbc.273.27.17036. [DOI] [PubMed] [Google Scholar]
- 4.Meunier B, de Visser SP, Shaik S. Chem. Rev. 2004;104:3947–3980. doi: 10.1021/cr020443g. [DOI] [PubMed] [Google Scholar]
- 5.Dawson JH. Science. 1988;240:433–439. doi: 10.1126/science.3358128. [DOI] [PubMed] [Google Scholar]
- 6.Vatsis KP, Peng HM, Coon MJ. J. Inorg. Biochem. 2002;91:542–553. doi: 10.1016/s0162-0134(02)00438-5. [DOI] [PubMed] [Google Scholar]
- 7.Poulos TL, Finzel BC, Gunsalus IC, Wagner GC, Kraut J. J. Biol. Chem. 1985;260:16122–16130. [PubMed] [Google Scholar]
- 8.Hollenberg PF, Hager LP. J. Biol. Chem. 1973;248:2630–2633. [PubMed] [Google Scholar]
- 9.Sundaramoorthy M, Terner J, Poulos TL. Structure. 1995;3:1367–1377. doi: 10.1016/s0969-2126(01)00274-x. [DOI] [PubMed] [Google Scholar]
- 10.Dawson JH, Sono M. Chem. Rev. 1987;87:1255–1276. [Google Scholar]
- 11.Crane BR, Arvai AS, Ghosh DK, Wu C, Getzoff ED, Stuehr DJ, Tainer JA. Science. 1998;279:2121–2126. doi: 10.1126/science.279.5359.2121. [DOI] [PubMed] [Google Scholar]
- 12.Marletta MA, Hurshman AR, Rusche KM. Curr. Opin. Chem. Biol. 1998;2:656–663. doi: 10.1016/s1367-5931(98)80098-7. [DOI] [PubMed] [Google Scholar]
- 13.Poulos TL. Drug Metab. Dispos. 2005;33:10–18. doi: 10.1124/dmd.104.002071. [DOI] [PubMed] [Google Scholar]
- 14.Yoshioka S, Tosha T, Takahashi S, Ishimori K, Hori H, Morishima I. J. Am. Chem. Soc. 2002;124:14571–14579. doi: 10.1021/ja0265409. [DOI] [PubMed] [Google Scholar]
- 15.Yoshioka S, Takahashi S, Ishimori K, Morishima I. J. Inorg. Biochem. 2000;81:141–151. doi: 10.1016/s0162-0134(00)00097-0. [DOI] [PubMed] [Google Scholar]
- 16.Auclair K, Moenne-Loccoz P, Ortiz de Montellano PR. J. Am. Chem. Soc. 2001;123:4877–4885. doi: 10.1021/ja0040262. [DOI] [PubMed] [Google Scholar]
- 17.Yoshioka S, Takahashi S, Hori H, Ishimori K, Morishima I. Eur. J. Biochem. 2001;268:252–259. doi: 10.1046/j.1432-1033.2001.01872.x. [DOI] [PubMed] [Google Scholar]
- 18.Vatsis KP, Peng HM, Coon MJ. Arch. Biochem. Biophys. 2005;434:128–138. doi: 10.1016/j.abb.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 19.Scott EE, White MA, He YA, Johnson EF, Stout CD, Halpert JR. J. Biol. Chem. 2004;279:27294–27301. doi: 10.1074/jbc.M403349200. [DOI] [PubMed] [Google Scholar]
- 20.Perera R, Dawson JH. J. Porphyrins Phthalocyanines. 2004;8:246–254. [Google Scholar]
- 21.Pond AE, Roach MP, Sono M, Rux AH, Franzen S, Hu R, Thomas MR, Wilks A, Dou Y, Ikeda-Saito M, Ortiz de Montellano PR, Woodruff WH, Boxer SG, Dawson JH. Biochemistry. 1999;38:7601–7608. doi: 10.1021/bi9825448. [DOI] [PubMed] [Google Scholar]
- 22.Pond AE, Roach MP, Thomas MR, Boxer SG, Dawson JH. Inorg. Chem. 2000;39:6061–6066. doi: 10.1021/ic0007198. [DOI] [PubMed] [Google Scholar]
- 23.Qin J, Perera R, Lovelace LL, Dawson JH, Lebioda L. Biochemistry. 2006;45:3170–3177. doi: 10.1021/bi052171s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roach MP, Puspita WJ, Watanabe Y. J. Inorg. Biochem. 2000;81:173–182. doi: 10.1016/s0162-0134(00)00101-x. [DOI] [PubMed] [Google Scholar]
- 25.Vaz AD, Pernecky SJ, Raner GM, Coon MJ. Proc. Natl. Acad. Sci. U. S. A. 1996;93:4644–4648. doi: 10.1073/pnas.93.10.4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pernecky SJ, Olken NM, Bestervelt LL, Coon MJ. Arch. Biochem. Biophys. 1995;318:446–456. doi: 10.1006/abbi.1995.1253. [DOI] [PubMed] [Google Scholar]
- 27.Adachi S, Nagano S, Ishimori K, Watanabe Y, Morishima I, Egawa T, Kitagawa T, Makino R. Biochemistry. 1993;32:241–252. doi: 10.1021/bi00052a031. [DOI] [PubMed] [Google Scholar]
- 28.Sigman JA, Pond AE, Dawson JH, Lu Y. Biochemistry. 1999;38:11122–11129. doi: 10.1021/bi990815o. [DOI] [PubMed] [Google Scholar]
- 29.Barrick D. Biochemistry. 1994;33:6546–6554. doi: 10.1021/bi00187a023. [DOI] [PubMed] [Google Scholar]
- 30.Huff AM, Chang CK, Cooper DK, Smith KM, Dawson JH. Inorg. Chem. 1993;32:1460–1466. [Google Scholar]
- 31.Perera R, Sono M, Sigman JA, Pfister TD, Lu Y, Dawson JH. Proc. Natl. Acad. Sci. U. S. A. 2003;100:3641–3646. doi: 10.1073/pnas.0737142100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dawson JH, Pond AE, Roach MP. Biopolymers. 2002;67:200–206. doi: 10.1002/bip.10087. [DOI] [PubMed] [Google Scholar]
- 33.Voegtle HL, Sono M, Adak S, Pond AE, Tomita T, Perera R, Goodin DB, Ikeda-Saito M, Stuehr DJ, Dawson JH. Biochemistry. 2003;42:2475–2484. doi: 10.1021/bi0271502. [DOI] [PubMed] [Google Scholar]
- 34.Perera R, Sono M, Kinloch R, Zhang H, Tarasev M, Im SC, Waskell L, Dawson JH. Biochim. Biophys. Acta. 2010 doi: 10.1016/j.bbapap.2010.07.012. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hildebrand DP, Burk DL, Maurus R, Ferrer JC, Brayer GD, Mauk AG. Biochemistry. 1995;34:1997–2005. doi: 10.1021/bi00006a021. [DOI] [PubMed] [Google Scholar]
- 36.Egeberg KD, Springer BA, Martinis SA, Sligar SG, Morikis D, Champion PM. Biochemistry. 1990;29:9783–9791. doi: 10.1021/bi00494a004. [DOI] [PubMed] [Google Scholar]
- 37.Ogura H, Nishida CR, Hoch UR, Perera R, Dawson JH, Ortiz de Montellano PR. Biochemistry. 2004;43:14712–14721. doi: 10.1021/bi048980d. [DOI] [PubMed] [Google Scholar]
- 38.Bruce GS. M.S. Thesis. University of South Carolina; 1987. [Google Scholar]
- 39.Bandara I. Ph.D. Dissertation. University of South Carolina; 2010. [Google Scholar]
- 40.Sun J, Loehr TM, Wilks A, Ortiz de Montellano PR. Biochemistry. 1994;33:13734–13740. doi: 10.1021/bi00250a026. [DOI] [PubMed] [Google Scholar]
- 41.Kerr EA, Mackin HC, Yu N-T. Biochemistry. 1983;22:4373–4379. doi: 10.1021/bi00288a005. [DOI] [PubMed] [Google Scholar]
- 42.Pluym M, Muryoi N, Heinrichs DE, Stillman MJ. J. Inorg. Biochem. 2008;102:480–488. doi: 10.1016/j.jinorgbio.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 43.Myer YP, Pande A, Dolphin D, editors. The Porphyrins, Vol. III, Physical Chemistry, Part A. New York: Academic Press; 1978. pp. 271–322. [Google Scholar]