Abstract
Progenitor cells maintain self-renewing tissues throughout life by sustaining their capacity for proliferation while suppressing cell cycle exit and terminal differentiation1,2. DNA methylation3,4,5 provides a potential epigenetic mechanism for the cellular memory needed to preserve the somatic progenitor state through repeated cell divisions. DNA methyltransferase 1 (DNMT1)6,7 maintains DNA methylation patterns after cellular replication. Although dispensable for embryonic stem cell maintenance,8 a clear role for DNMT1 in maintaining the progenitor state in constantly replenished somatic tissues, such as mammalian epidermis, is unknown. Here we show that DNMT1 is essential for epidermal progenitor cell function. DNMT1 protein was found enriched in undifferentiated cells, where it was required to retain proliferative stamina and suppress differentiation. In tissue, DNMT1 depletion led to exit from the progenitor cell compartment, premature differentiation and eventual tissue loss. Genome-wide analysis revealed that a significant portion of epidermal differentiation gene promoters were methylated in self-renewing conditions but were subsequently demethylated during differentiation. Furthermore, we show that UHRF1,9,10 a component of the DNA methylation machinery that targets DNMT1 to hemi-methylated DNA, is also necessary to suppress premature differentiation and sustain proliferation. In contrast, Gadd45A11,12 and B13, which promote active DNA demethylation, are required for full epidermal differentiation gene induction. These data demonstrate that proteins involved in the dynamic regulation of DNA methylation patterns are required for progenitor maintenance and self-renewal in mammalian somatic tissue.
In self-renewing mammalian epithelial tissues, which are the sites of most human malignancies14, the basis for repressed differentiation in somatic stem cells is unclear. Epigenetic gene silencers, such as DNMT1,4 may help preserve progenitor gene expression patterns through repeated cell divisions. Loss of function studies have demonstrated the importance of DNMT1 for imprinting, embryogenesis, and tumorigenesis15. DNMT1 knockout mice display delayed development and lethality by mid-gestation, hindering analysis of the function of DNMT1 in self-renewing epithelia, such as epidermis7,16. In epidermis, undifferentiated progenitor cells residing in the basement membrane-bound basal layer undergo cell cycle arrest, outward migration, and terminal differentiation to generate the cutaneous permeability barrier. Consistent with a role in this process, DNMT1 transcript and protein were repressed upon calcium-induced differentiation of human keratinocytes in culture (Fig. 1a-c, Supplementary Table 1). DNMT1 protein was mainly confined to cells of the basal layer of adult human epidermal tissue and was absent in outer differentiated layers (Fig. 1d). Thus, DNMT1 is expressed in epidermal progenitor-containing cell populations and is lost during differentiation.
Figure 1. DNMT1 is down-regulated during epidermal differentiation.
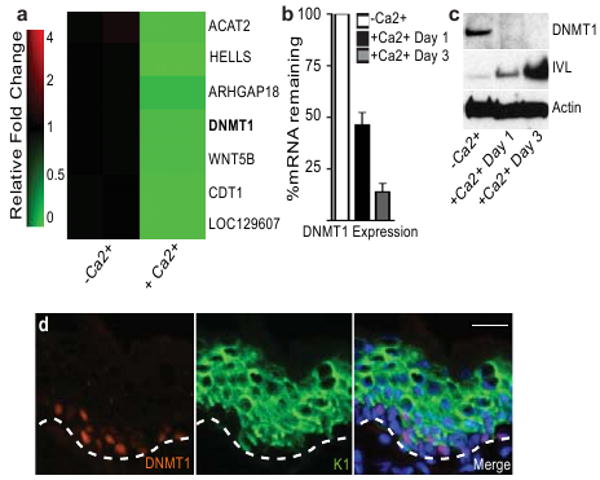
a, Microarray analysis characterizing genes altered during calcium-induced differentiation (genes repressed ≥2 fold are shown in green). b, c, Time course of DNMT1 down-regulation after induction of differentiation on the mRNA and protein level; error bars=s.d., n=3. d, DNMT1 protein distribution in adult human epidermis. DNMT1 (orange), differentiation keratin 1 (K1: green), Hoechst nuclear stain (blue); scale bar=30μm, dotted line denotes basement membrane.
To study effects of DNMT1 loss on epidermal progenitor function in vivo, short hairpin RNA constructs targeting DNMT1 (DNMT1i) were generated (Supplementary Fig. 1). DNMT1i and control knockdown cells were used to regenerate human skin on immune-deficient mice by an approach recapitulating features of intact tissue, including epidermal self-renewal kinetics17,18. By two weeks, DNMT1i epidermis was markedly hypoplastic (Fig. 2a) without increased cell death (Supplementary Fig. 2), and only 34% of DNMT1-deficient epidermal tissues survived to week 3 compared to 100% for controls (Fig. 2a,b). DNMT1i tissue displayed ectopic expression of differentiation genes, such as keratin 1 (K1), in the normally undifferentiated basal layer (Fig. 2a), indicating premature differentiation occurs in the absence of DNMT1. Granular layer proteins were not seen in the basal layer of DNMT1i epidermis, possibly because of DNMT1i cell loss from this setting prior to their expression (data not shown). Consistent with premature entry into the post-mitotic differentiation pathway, DNMT1 depletion also diminished cell proliferation by week 3 to <15% of control (Supplementary Fig. 3a, b). These results indicate that epidermal DNMT1 loss leads to premature differentiation within the progenitor-containing compartment and loss of tissue self-renewal.
Figure 2. DNMT1 loss triggers differentiation and failure of self-renewal.
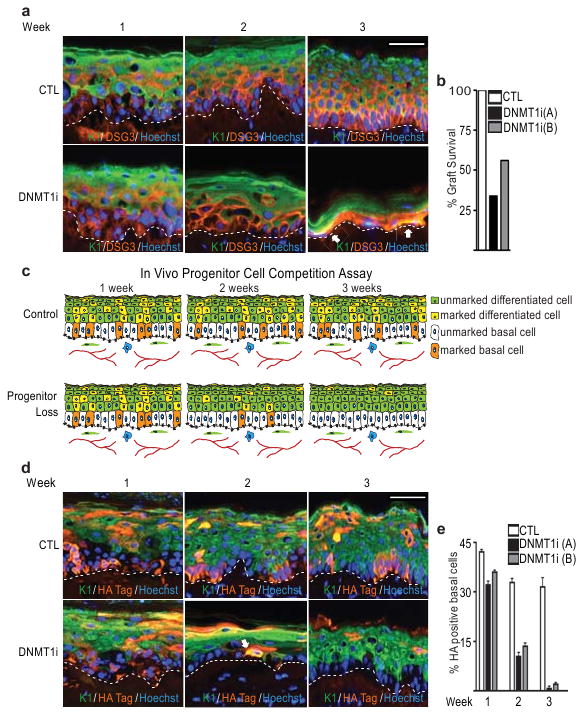
a, Keratinocytes expressing shRNAs for DNMT1 (DNMT1i) or control (CTL) were used to regenerate human epidermis on immune deficient mice. K1 (green) marks differentiated epidermal layers; human species-specific antibody against Desmoglein 3 (DSG3:orange). White arrows denote areas of ectopic basal layer differentiation. Scale bar=50μm, dotted line=basement membrane. b, Graft survival (n=9 grafted mice/group). c, Human epidermal progenitor competition assay. Marked cells with altered gene function are mixed with unmarked controls to regenerate epidermis. Maintenance of marked undifferentiated progenitor cells in the basal layer (orange) as well as in differentiating suprabasal layers (green in control; yellow in marked) over time. The proportion of marked cells expressing control shRNAs remains constant [top panel] whereas the proportion of marked cells depleted of genes required for progenitor persistence decreases over time [bottom panel]. d, Human epidermal progenitor tracking in vivo. shRNA-receiving cells (top panels=control, bottom panels=DNMT1i) marked with HA tag (orange) in tissue mosaic with non-marked cells. White arrow denotes ectopic K1 differentiation protein expression in the basal layer of a marked DNMT1i cell. Scale bar=50μm. e, Quantitation of HA marked basal layer cells. 500 basal nuclei were counted for each time point (n=9 grafted mice/group);error bars=s.d.
To confirm that the inability of DNMT1-deficient tissue to persist was due to a cell-intrinsic loss of progenitor function as opposed to a global tissue failure process, an in vivo epidermal progenitor cell competition assay was developed using tissue mosaicism (Fig. 2c). Epidermal cells marked with a construct expressing a hemagglutinin (HA) tag were infected with DNMT1i or control vectors and mixed with unmarked cells. These cells were then used to regenerate mosaic human epidermis in vivo. Initially, there were a similar percentage of DNMT1i and control cells contributing to all layers (Fig. 2d,e). By week 2, however, there was a significant decrease in the number of DNMT1i cells in the basal layer, with up to 11% of retained cells displaying ectopic expression of differentiation genes (Supplementary Fig. 4; Fig. 2d,e). In contrast, marked cells in control shRNA epidermis remained stable without signs of ectopic differentiation (Fig. 2d,e; Supplementary Fig.4). By week 3, DNMT1i cells generated with multiple DNMT1 shRNA constructs comprised <2% of the basal layer compared to >30% for controls, a process paralleled by progressive loss of DNMT1i cells from suprabasal layers (Fig. 2e). Thus, DNMT1 loss leads to a cell-intrinsic failure of epidermal progenitor maintenance and premature differentiation.
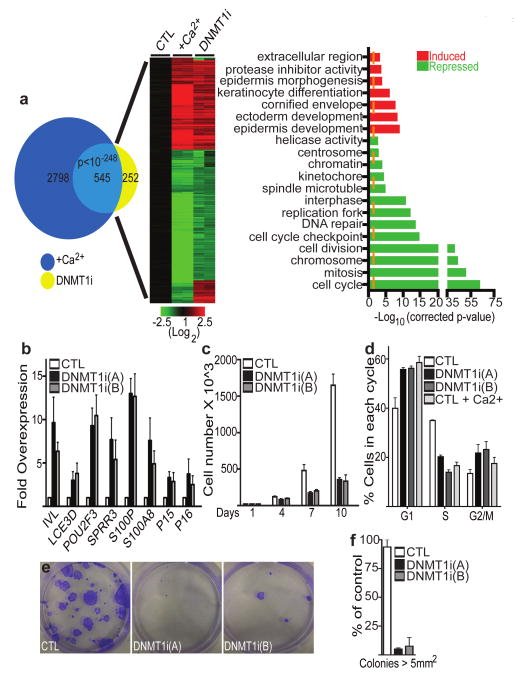
Global gene expression profiles in DNMT1-deficient cells cultured in growth conditions was next compared to that of control cells induced to differentiate with calcium. 3,343 genes (1,366 induced; 1,977 repressed) changed during calcium-induced differentiation and 797 genes (450 induced; 347 repressed) were altered by DNMT1 loss (Supplementary Tables 1 and 2). Comparing these two gene sets revealed a statistically significant overlap of 545 genes, the vast majority of which were regulated in the same direction (Fig. 3a, b; Supplementary Table 3). Up-regulated genes in this shared set were enriched for differentiation gene ontology (GO) terms, while repressed genes were enriched for proliferation GO terms (Fig. 3a). DNMT1 thus controls an epidermal gene expression program compatible with a role in sustaining proliferation and repressing differentiation.
Figure 3. DNMT1 is required to repress differentiation and sustain proliferation.
a, Venn diagram (left) illustrating overlap between changes identified with DNMT1 loss and calcium-induced differentiation (+Ca2+). Heat map of the 545 genes shared between the two profiles (P-value<1×10-248; Fisher's exact test); red (induced) and green (repressed), log2-based scale. GO analysis (right) (P-values represent a Bonferroni-corrected EASE score). b, QRT-PCR verification of array data. Error bars=s.d., n= 3. c, Cell proliferation; error bars=s.d., n= 2. d, DNMT1 depletion results in G1 cell cycle arrest, flow cytometry profiles; error bars=s.d., n= 2. e, Clonogenic assays of control shRNA and DNMT1i keratinocytes plated at limiting dilution. f, Quantitation of colonies >5 mm2 (n=3/group); error bars=s.d.
To study the basis for the impaired proliferation seen with DNMT1 loss, we examined the proliferative stamina of epidermal progenitors. DNMT1i cells prematurely exited S phase into G1 arrest, similar to cells undergoing calcium-induced differentiation (Fig. 3c,d). In clonogenic assays, single DNMT1i cells failed to proliferate into larger colonies (>5mm2), producing only small, abortive colonies (Fig. 3e,f). Similar to in vivo, decreased cell numbers were not due to increased apoptosis (Supplementary Fig. 5). Rather, DNMT1 loss resulted in the upregulation of cyclin dependent kinase (Cdk) inhibitors, p15INK4B and p16INK4A (Fig. 3b; Supplementary Table 2), implicated in inhibiting stem cell self-renewal19,20,21. In keratinocytes, the effects of p15INK4B and p16INK4A can be bypassed by co-expression of Cdk4 and cyclin D122. This G1 Cdk-cyclin pair partially rescued DNMT1i cell proliferation (Supplementary Fig. 6a,b). Wild-type DNMT1, but not a catalytically inactive point mutant23, reversed the proliferation defects as well as the Cdk inhibitor induction observed with DNMT1 loss (Supplementary Fig. 7), indicating that DNMT1 DNA methylation capacity is required to repress p15INK4B/p16INK4A to permit progenitor proliferation. These data suggest that DNMT1 sustains progenitor proliferation by inhibiting expression of Cdk inhibitors.
The DNMT1 requirement in maintaining the progenitor state suggests that DNA methylation changes during differentiation. Methylated DNA immunoprecipitations (MeDIP)24,25 were thus performed in undifferentiated and differentiated keratinocytes followed by hybridization to promoter tiling arrays covering 24,659 human genes (Supplementary Table 4 and 5). Methylated DNA enrichment was found on 5,999 gene promoters in undifferentiated keratinocytes (Supplementary Table 4), with an overabundance of differentiation genes (Fig. 4a). 364 of the 1,366 induced differentiation gene set in Fig. 3 had methylated DNA promoters in undifferentiated cells (Fig.4b; Supplementary Table 6), >50% of which lost methylation during differentiation (Fig. 4b; Supplementary Table 7). Of the 232 differentially methylated genes, 75% contained a CpG island within 2kb upstream of their transcription start site (Supplementary Table 7), a proportion similar to that in the epidermal differentiation gene set above as well as in the human genome. Among the 232 genes, a significant portion exhibited methylation in undifferentiated cells in regions outside of CpG islands known as “shores” (Supplementary Fig.8), which exhibit tissue-specific methylation patterns correlated with gene expression26. These methylation marks were erased from these low CpG shores during epidermal differentiation. Because of variability in distance of shores from CpG islands, some of which are >2kb away26, the shores for all genes were not encompassed within the tiled region of the arrays. Thus, the genes regulated in this manner during epidermal differentiation may be underestimated. MeDIP-coupled QPCR confirmed that differentiation genes, such as LCE3D and S100P, lost DNA methyl marks ≥3-fold during differentiation whereas the stably imprinted gene, KCNQ1, remained unaltered (Fig. 4c). Bisulfite sequencing of S100P corroborated MeDIP profiling (Supplementary Fig.9). Several transcription factors regulating epidermal differentiation27,28,29,30, including POU2F3, MAFF, and SP1 (Fig. 4c; Supplementary Table 7) also lost methylation during differentiation, corresponding with increased expression (Supplementary Table 1). Interestingly, 372 of the 1,977 genes repressed during differentiation became de novo methylated during differentiation, possibly via the de novo DNMTs, 3A and 3B, which are both expressed in keratinocytes (Supplementary Table 8). DNA methylation thus undergoes dynamic alterations during epidermal differentiation.
Figure 4. DNA methylation during epidermal differentiation.
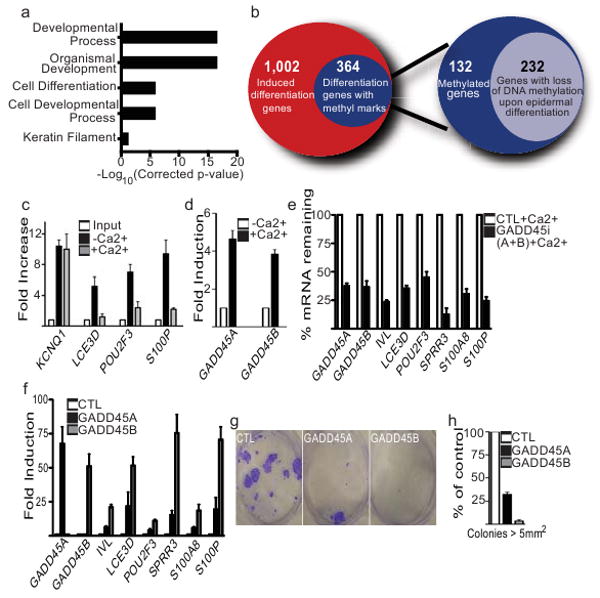
a, GO analysis of gene promoters enriched for DNA methylation in undifferentiated keratinocytes. The x-axis represents the P-values for each GO category shown that contains enrichment for DNA methylation (P-values=Bonferroni-corrected EASE score). b, Venn diagrams of epidermal differentiation genes enriched for promoter methylation. The left diagram depicts the number of up-regulated epidermal differentiation genes with methylated DNA promoters (blue:364 genes) in undifferentiated cells. The right diagram shows the number of DNA methylation enriched differentiation genes that lose their methyl marks upon differentiation (light blue: 232 genes). c, DNA methylation enrichment ratios on individual gene promoters in the absence (-Ca2+) or presence (+Ca2+) of calcium-induced epidermal differentiation, error bars=s.d., n= 3. d, Gadd45A and B mRNA is induced during epidermal differentiation; error bars=s.d., n= 3. e, Combined Gadd45A/B loss impairs epidermal differentiation; error bars=s.d., n= 3. f, Forced expression of Gadd45A or B induces premature epidermal differentiation. Error bars=s.d., n= 3. g, Effects of expression of Gadd45 A or B on clonogenic growth. h, Quantitation of clonogenic assays with colonies > 5 mm2 (n=3/group); error bars=s.d.
The SET and RING finger-associated (SRA) domain of UHRF19,10, binds DNMT1 and recruits it to hemi-methylated DNA. Similar to DNMT1, UHRF1 was predominately detected in the epidermal basal layer and was down-regulated during differentiation (Supplementary Fig. 10a-c). UHRF1 depletion (Supplementary Fig. 10d) de-repressed differentiation genes in progenitors (Supplementary Fig. 10e) and dramatically reduced their proliferative potential (Supplementary Fig. 10f,g). DNMT1-UHRF1 preservation of progenitor function predicted that mediators of DNA demethylation might induce epidermal differentiation. In support of this, Gadd45A11,12 and B13, which interact with components of both base excision (BER) and nucleotide excision repair (NER) DNA demethylase complexes to demethylate DNA, were induced during epidermal differentiation (Fig. 4d; Supplementary Table 1). Gadd45 depletion inhibited calcium induction of genes shown to be de-methylated during differentiation and impaired erasure of S100P promoter DNA methylation (Fig. 4e; Supplementary Fig. 9; Supplementary Table 7). Furthermore, increasing Gadd45 expression induced premature differentiation and inhibited progenitor growth (Fig. 4f,g,h). DNMT1 and Gadd45 impacts in epidermal progenitors suggest they may function similarly in other tissues. Consistent with this, DNMT1 expression is down-regulated during muscle differentiation (Supplementary Fig.11a) and DNMT1 depletion or enforced Gadd45 expression results in the increased expression of skeletal muscle differentiation genes such as skeletal actin, tropomyosin, and myosin heavy chain (Supplementary Fig.11b-d).
Taken together, these data suggest that tissue self-renewal may be controlled by a dynamic antagonism between regulators of DNA methylation. In this model, the progenitor state is sustained by methylation-induced silencing of the differentiation program via the concerted actions of DNMT1 and UHRF1; their retention in some lower suprabasal epidermal cells suggests their action may be partially retained early in differentiation. The differentiated cell fate, in contrast, is promoted by Gadd45 DNA demethylation-enhancing proteins. The basis for the reciprocal expression of DNMT1-UHRF1 in progenitors and Gadd45 in differentiation, as well as whether this balance is disrupted in epithelial cancers, represents avenues for future study.
Methods Summary
Tissue culture
Primary human keratinocytes were derived from fresh foreskin. Cells were grown in KSF-M (GIBCO-BRL) supplemented with epidermal growth factor (EGF) and bovine pituitary extract (BPE). Cells were induced to differentiate by the addition of 1.2 mM calcium for 1 and 3 days in full confluence. Amphotrophic phoenix cells were maintained in DMEM and 10% fetal bovine serum.
A full methods section accompanies this paper. (See Supplementary Methods)
Supplementary Material
Acknowledgments
We thank M. P. Scott, G.R. Crabtree, M.T. Fuller, J. Wysocka, H.Y. Chang, A.E. Oro, T.W. Ridky, B.J. Zarnegar, M. Kretz, D. Webster, R. Flockhart for critical comments and pre-submission review, X. Bao for help with figures, and G. Lai for informatics.
Footnotes
Supplemental Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions G.L.S. performed and designed the experiments and wrote the manuscript. J.A.R. analyzed the bioinformatic data and wrote the manuscript. D.E.W. analyzed the bioinformatic data. L.Z. performed experiments. P.A.K. supervised the project, discussed the results and wrote the manuscript.
Author Information Profiling and MeDIP-chip data have been deposited with the GEO database under accession code GSE18590. Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Morrison SJ, Spradling AC. Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell. 2008;132:598–611. doi: 10.1016/j.cell.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orford KW, Scadden DT. Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat Rev Genet. 2008;9:115–128. doi: 10.1038/nrg2269. [DOI] [PubMed] [Google Scholar]
- 3.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 4.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 5.Fuks F. DNA methylation and histone modifications: teaming up to silence genes. Curr Opin Genet Dev. 2005;15:490–495. doi: 10.1016/j.gde.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 6.Bestor T, Laudano A, Mattaliano R, Ingram V. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J Mol Biol. 1988;203:971–983. doi: 10.1016/0022-2836(88)90122-2. [DOI] [PubMed] [Google Scholar]
- 7.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 8.Fouse SD, et al. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell. 2008;2:160–169. doi: 10.1016/j.stem.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bostick M, et al. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 10.Sharif J, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 11.Rai K, et al. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barreto G, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- 13.Ma DK, et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.(SEER Incidence) Surveillance, Epidemiology, and End Results (SEER) Program. National Cancer Institute, DCCPS, Surveillance Research Program, Cancer Statistics Branch; SEER*Stat Database: Incidence - SEER 9 Regs, Nov 2002 Sub (1973-2000) www.seer.cancer.gov. released April 2003, based on the November 2002 submission. [Google Scholar]
- 15.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 16.Lei H, et al. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122:3195–3205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
- 17.Khavari PA. Modelling cancer in human skin tissue. Nat Rev Cancer. 2006;6:270–280. doi: 10.1038/nrc1838. [DOI] [PubMed] [Google Scholar]
- 18.Robbins PB, et al. In vivo restoration of laminin 5 beta 3 expression and function in junctional epidermolysis bullosa. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(9):5193–5198. doi: 10.1073/pnas.091484998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maurelli R, et al. Inactivation of p16INK4a (inhibitor of cyclin-dependent kinase 4A) immortalizes primary human keratinocytes by maintaining cells in the stem cell compartment. Faseb J. 2006;20:1516–1518. doi: 10.1096/fj.05-4480fje. [DOI] [PubMed] [Google Scholar]
- 20.Carlson ME, Hsu M, Conboy IM. Imbalance between pSmad3 and Notch induces CDK inhibitors in old muscle stem cells. Nature. 2008;454:528–532. doi: 10.1038/nature07034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He J, Kallin EM, Tsukada Y, Zhang Y. The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15(Ink4b) Nat Struct Mol Biol. 2008;15:1169–1175. doi: 10.1038/nsmb.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lazarov M, et al. CDK4 coexpression with Ras generates malignant human epidermal tumorigenesis. Nat Med. 2002;8:1105–1114. doi: 10.1038/nm779. [DOI] [PubMed] [Google Scholar]
- 23.Damelin M, Bestor TH. Biological functions of DNA methyltransferase 1 require its methyltransferase activity. Mol Cell Biol. 2007;27:3891–3899. doi: 10.1128/MCB.00036-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weber M, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 25.Weber M, Schubeler D. Genomic patterns of DNA methylation: targets and function of an epigenetic mark. Curr Opin Cell Biol. 2007;19:273–280. doi: 10.1016/j.ceb.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 26.Irizarry RA, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andersen B, et al. Functions of the POU domain genes Skn-1a/i and Tst-1/Oct-6/SCIP in epidermal differentiation. Genes Dev. 1997;11:1873–1884. doi: 10.1101/gad.11.14.1873. [DOI] [PubMed] [Google Scholar]
- 28.Eckert RL, Crish JF, Banks EB, Welter JF. The epidermis: genes on - genes off. J Invest Dermatol. 1997;109:501–509. doi: 10.1111/1523-1747.ep12336477. [DOI] [PubMed] [Google Scholar]
- 29.Motohashi H, Katsuoka F, Engel JD, Yamamoto M. Small Maf proteins serve as transcriptional cofactors for keratinocyte differentiation in the Keap1-Nrf2 regulatory pathway. Proc Natl Acad Sci U S A. 2004;101:6379–6384. doi: 10.1073/pnas.0305902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakamura Y, et al. The combination of ubiquitous transcription factors AP-1 and Sp1 directs keratinocyte-specific and differentiation-specific gene expression in vitro. Exp Dermatol. 2007;16:143–150. doi: 10.1111/j.1600-0625.2006.00528.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.