Abstract
Antibodies are critical reagents in biological research and are increasingly being developed as therapeutic agents. They typically exhibit very high affinity and selectivity for their ligands. Synthetic protein-binding agents rarely achieve the combination of high affinity and selectivity for their target protein that is typical of a good antibody. However, significant efforts are underway to develop a new generation of protein ligands with improved properties. This article reviews progress towards this goal and suggests fruitful strategies for future research.
Antibodies are the gold standard of protein-binding agents. A good antibody will bind its target protein with a low nM, or even better, equilibrium dissociation constant and sufficient selectivity to ignore almost every other molecule in the proteome. Moreover, it is often possible to identify an antibody that binds only a particular post-translationally modified form of a protein, for example phosphoprotein-specific antibodies. Many different methods in molecular biology are critically dependent on the existence of a good antibody for the protein of interest. Over the past several years there has also been an explosion in the development of antibodies as therapeutic agents. This is, in part, due to the aforementioned high affinity and selectivity of antibodies, which maximizes potency and minimizes off target effects. But just as important are the long circulating lifetimes of antibodies, which are too large to be filtered through the glomerulus.
While they are remarkable molecules, antibodies have significant limitations. In the therapeutic realm, they must be employed as injectables and are restricted to extracellular targets since they are not cell permeable or orally bioavailable. Under normal circumstances, they cannot traverse the blood brain barrier. While fully humanized antibodies have greatly alleviated the problem of immune responses against them when they are employed as therapeutics, therapeutic antibodies do not entirely escape immune surveillence. Moreover, their manufacture in quantity is expensive and requires considerable skill in the art. With regard to their use as research reagents for protein detection, there is a significant failure rate, particularly for anti-phosphoprotein-specific antibodies and it is not feasible to immunize animals with peptides carrying less stable modifications, such as acetylated serine or threonine [1]. Finally, as folded proteins, they have limited stability and will only function in a relatively narrow window of conditions.
For these reasons, there has been considerable interest in the development of synthetic molecules as antibody surrogates for applications in both biology and medicine [2]. Ideally, these would be molecules with protein-binding parameters that rival those of a good antibody, yet are cell permeable, orally bioavailable and simple and cheap to produce. Progress towards the development of such reagents and some thoughts about the near future of this interesting field are presented in this article.
High affinity protein ligands through multivalency
High-throughput screening of small molecules compound collections based on a functional assay can now be performed in many academic and industrial settings. However, these screens are quite expensive. Moreover, most compound collections have not been assembled with ease of hit optimization in mind, necessitating tedious medicinal chemistry campaigns to achieve high affinity. So as a general source of large numbers of high affinity protein ligands, it is not clear that function-based high-throughput screening of small molecule libraries is a good place to start. An alternative to functional screening to mine one bead one compound (OBOC) libraries for protein ligands using a simple binding assay, [3–6•]. In most such cases, the library will have been made by split and pool synthesis and thus the identity of the compound on any particular bead will not be known a priori. Thus, the nature of the immobilized molecule must allow their direct structural characterization, usually by mass spectrometry, or they must be encoded [7,8]. Encoding, especially using DNA-based methods [9,10], opens the way for the creation of very large libraries of drug-like molecules in an OBOC format.
OBOC libraries are generally screened by incubating a labeled protein with the library and identifying candidate protein ligands by following the label. This is done most conveniently when the library is synthesized on a hydrophilic resin such as TentaGel, which does not bind proteins non-specifically [11,12]. Alternatively, compounds can be cleaved from polystyrene resins and displayed in other formats, such as a small molecule microarray [13,14]. OBOC libraries that are screened when attached to a solid surface have the advantage that one knows where they can be modified without concern for ablating the ability of the molecule to bind the target protein: the point of attachment of the molecule to the surface.
The hits that arise from such screening efforts are almost always micromolar or, at best, high nanomolar ligands [15]. At least a 1000-fold improvement in the affinity of these hits is desirable in order to mimic the binding of a good antibody. Many different strategies have been employed to achieve this jump in affinity, but it is fair to say that no general strategy has yet been demonstrated to be general and capable of high-throughput. This is a critical issue in the field. The following discussion is not intended to be comprehensive, but rather highlight generally interesting methodologies that have the potential to contribute to the eventual design of a convenient and high-throughput system for protein ligand development.
One approach is to join together two modest affinity, non-competitive ligands with an appropriate linker chain to create a bivalent compound, which can exhibit much better affinity than the individual ligands. The simplest strategy is to identify ligand pairs that bind the protein of interest non-competitively, then link them together via a suitable linker arm. A version of this approach, called “SAR by NMR” [16], in which an NMR solution structure of the ligand protein complex was employed to guide linker design, was one of the first successful approaches of this type, but since then, many other strategies for the creation of these bivalent ligands have been reported. [17]
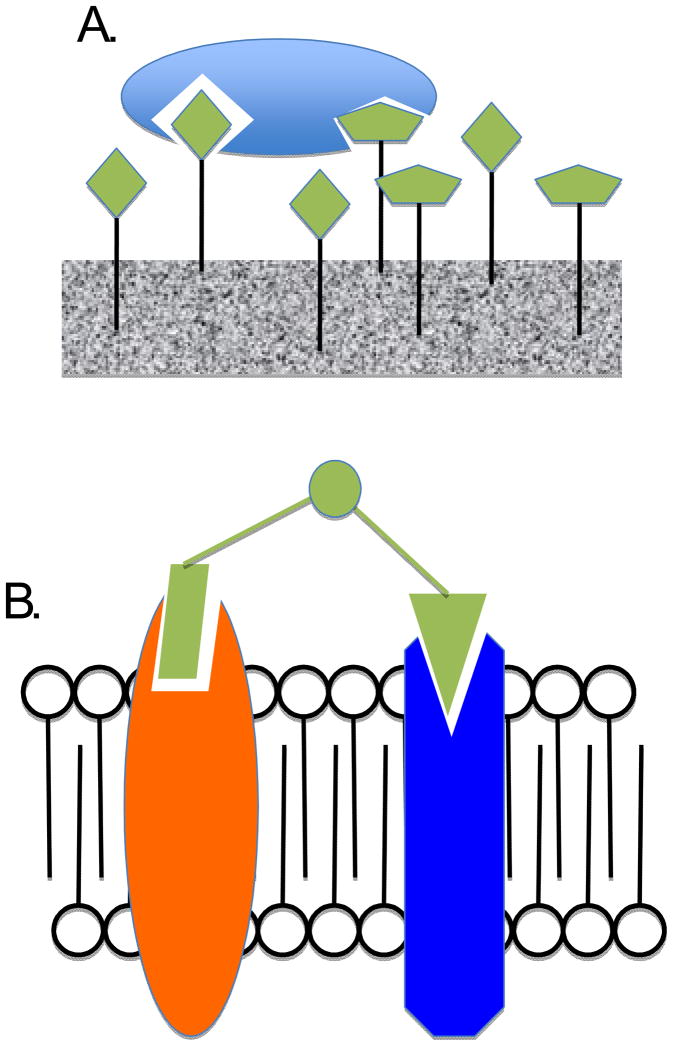
A special case of this type of approach suitable for the creation of high affinity protein capture agents is to simply co-immobilize two non-competitive ligands for the same protein on a densely functionalized surface (Fig. 1A) [18,19]. The idea is that some fraction of the two different immobilized molecules will have the appropriate spatial relationship to engage the target protein in a bivalent fashion, resulting in high affinity. In this case, the surface itself serves as a kind of “library of linkers”, allowing some fraction of the immobilized ligands to act as high affinity bivalent ligands.
Figure 1.
Interactions between a bivalent ligand and a target protein(s) supported by a surface. A. Co-immobilization of two non-competitive ligands for a target protein (light blue oval) can provide a high affinity capture agent by virtue of the fact that some fraction of the two molecules on the surface will have the proper separation to engage the two different binding sites on the protein. B. A bivalent ligand (green) that binds two different integral membrane proteins (orange and blue) can achieve high affinity and cell type selectivity due to the membrane effectively concentrating the two proteins.
A philosophically similar idea, but in reverse, has been reported by Hruby and co-workers as a strategy to create high affinity and selectivity cell surface binding reagents that recognize two different receptors (Fig. 1B) [20,21•]. The idea behind this strategy is that the cell membrane will act as a two-dimensional surface to template binding of covalently linked ligands to two different receptors even if the receptors are not physically associated. [22].
Another approach to bivalent ligands that eliminates the need for linker optimization is to employ a target protein as a template for an uncatalyzed Huisgen cycloaddition between members of a library of alkynes and a library of azides. The cycloaddition of alkynes and azides is highly favorable thermodynamically, but extremely slow kinetically. Thus, when an azide and an alkyne are mixed together, little or no reaction will occur in a reasonable period of time in the absence of a catalyst unless two molecules are brought into close proximity by binding to nearby surfaces of the protein. However, if independent binding of an azide and an alkyne to nearby surfaces of a protein occurs, coupling can take place, creating a high affinity, bivalent ligand in situ. This strategy was used to great effect to find a pM ligand for carbonic anhydrase (CA) [23].
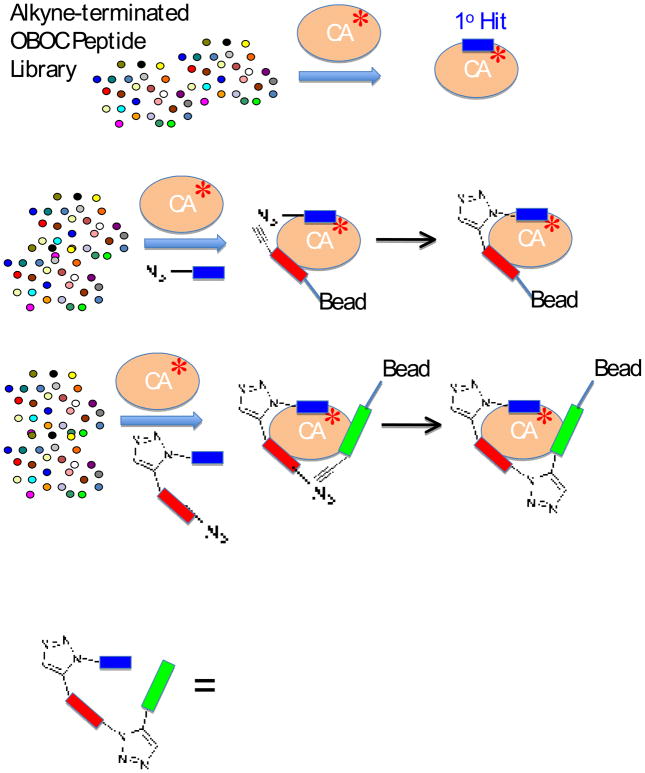
The problem with this clever approach is that the identity of the resultant bivalent compound had to be determined directly from the screening mixture. Only stoichiometric amounts of ligand (with respect to the protein), at best, could be created in one experiment owing to the slow off rate of the bivalent ligand. This meant that very large amounts of protein were required for the screen, making this technique impractical for most protein targets. However, a re-engineering of this system by Heath, Sharpless and co-workers has been reported that alleviates this problem and provides a powerful and much more practical system for multivalent ligand discovery (Fig. 2) [24••]. In this case, an OBOC peptide library was first screened using a standard approach to identify a ligand for CA. This peptide, which exhibited a weak affinity (KD ≈ 500 μM) for CA, was then synthesized with an N-terminal azide appendage. A new OBOC library was then created that terminated with an alkyne-containing residue. The library, the soluble original hit and the labeled target protein, but now at a lower concentration were then mixed and new hits were identified. As before, higher affinity hits were obtained through a protein-templated Huisgen cycloaddition, leading to retention of the labeled protein. In this case however, only a small fraction of the peptide molecules on the resin entered into the reaction. The remainder could easily be sequenced via Edman degradation, allowing the identity of the so-called bi-ligand to be deduced. One of these bi-ligands was found to be improved about 160-fold (KD ≈ 3 μM) over the original hit. Finally, the process was repeated again, this time using the azide-modified biligand as the soluble partner and a tri-ligand was identified with a KD of about 40 nM. This begins to approach the antibody range, yet the mass of the molecule is more than a 100-fold lower than that of an antibody. The important point is that only modest amounts of fluorescently labeled protein are required for this procedure, removing the major drawback of the original assay. Although this study employed peptides, presumably any class of molecules whose sequence could be determined from a single bead or by encoding could be employed in this kind of protocol.
Figure 2.
An iterative, protein-templated Huisgen cycloaddition strategy for protein ligand discovery. Step 1: An OBOC peptide library is screened for a ligand to fluorescently labeled Carbonic Anhydrase (CA). Step 2: The OBOC alkyne-terminated peptide library is again screened, but in the presence of soluble azide-terminated hit from the primary screen. The hope is that a protein-templated cycloaddition will occur. Step 3: Same as step 2, except the bivalent ligand terminated in an azide is employed as the soluble ligand. The hit identified in this step is a trivalent ligand. The structure of the CA ligand reported by Heath and co-workers [24••] is shown at the bottom of the figure.
A related approach, but which did not employ a protein-templated reaction, was reported independently and at about the same time by our laboratory [25]. In this case, a previously identified, low affinity (KD ≈ 300 μM) peptoid ligand for the KIX domain [26] of the transcriptional coactivator CBP was modified with an azide functionality, then incubated with a microarray-displayed peptoid library terminating in an alkyne functionality. A copper catalyst was added to “click” the soluble lead molecule onto each of the immobilized library compounds. The array was then incubated with labeled target protein. This procedure resulted in the identification of a 300-fold improved bivalent ligand.
High affinity protein ligands through hit optimization
An inevitable drawback of all of the fragment condensation strategies is that they accept increased molecular mass as the price one pays for higher affinity. In general, an increase in mass makes it less likely a molecule will be cell permeable and orally bioavailable. Therefore, it would be of great interest to develop alternative strategies that do not mandate an increase in ligand mass to achieve high affinity. The tried and true solution to this problem is classical medicinal chemistry, where a large number of derivatives of a hit are created synthetically and tested for increased efficacy. The problem with this approach is that it has a low throughput and requires considerable manpower skilled in organic synthesis.
Affinity maturation of lead compounds is an area where DNA-encoded libraries of proteins, peptides or RNAs have a great advantage over synthetic libraries. This is because mutagenesis protocols allow the rapid creation of a “derivative library”, i.e., a large collection of compounds that differ modestly from the lead by a few substitutions. For example, given an RNA aptamer that has been pulled out of a primary library, error-prone RT-PCR can be employed to generate a large collection of amplicons in which 0–5 of the positions of a 50mer can be changed to a different base. This derivative library of duplex DNAs can be amplified, transcribed and the resultant RNA library screened under more demanding conditions to isolate an improved ligand. The process can then be repeated several times until a high affinity ligand is obtained [27]. The same kind of approach can be employed with DNA-encoded peptide libraries. Unfortunately, synthetic libraries cannot be amplified and so it is more difficult to apply this kind of derivative library scheme to the rapid affinity maturation of non-natural protein ligands. This is too bad, because the less than optimal pharmacokinetic (PK) properties of peptides and nucleic acids limit the applications for which they can be used.
Given these facts, some researchers have explored a “best of both worlds” compromise in which the ribosome is coaxed to accept unnatural molecules, allowing the creation of DNA-encoded libraries of non-natural molecules with improved PK characteristics [28]. Perhaps the most powerful such example described to date involves the ribosome-mediated synthesis of N-alkylated peptides [29•]. N-alkylation of peptides renders them highly resistant to proteases and far more cell permeable [30,31]. Apparently, elimination of the highly polar N-H bonds in the main chain reduces the sphere of hydration of the compound, making it far easier to cross the phospholipid membrane. It is noteworthy that many bioactive depsipeptides and related compounds such as cyclosporin and FK-506 feature N-methylated amino acids prominently. Suga and colleagues had previously reported the development of two key technologies that were combined to allow the incorporation of N-methyl amino acids at multiple positions in a ribosome-synthesized peptide. The first is the “Flexizyme” system [32]. Flexizyme is a ribozyme that efficiently mediates the charging of almost any tRNA with virtually any amino acid desired. The second is a highly purified in vitro translation system derived from E. coli. The advantage of a completely purified system over extracts is that one must supply the system with aminoacyl-t-RNAs to be employed by the ribosome. By substituting engineered aminoacyl-t-RNAs for native aminoacyl-t-RNA, the desired unnatural amino acids can be incorporated precisely at multiple positions in the ribosome-synthesized peptide. Using this approach, Suga and coworkers demonstrated the ribosome-catalyzed synthesis of peptides with up to 10 consecutive methylated residues as well as cyclic N-methylated peptides in which the ring was closed via a physiologically stable thioether bond. To the best of my knowledge, this relatively new technology has not yet been used to create a large library of cyclic methylated peptides for screening purposes, but it seems likely that such studies will be conducted soon. If successful, this represents a potentially powerful approach to the discovery of bioactive molecules.
But what about synthetic molecules that are not constructed biologically? To avoid difficult and tedious synthesis and testing of derivatives, i.e., classical SAR campaigns, it seems clear that the compound libraries that are employed in screening campaigns will have to be designed carefully with ready optimization in mind [15]. One of several potential strategies to this end is to develop libraries of oligomeric compounds made from pieces that can easily be swapped out or modified, but which, unlike peptides, retain good cell permeability and other desirable PK properties. Another potential advantage of modest-sized oligomers is that they will probably have sufficient “wingspan” to bind to the rather large, shallow protein surfaces that typically engage in protein-protein interactions. Disruption of such interactions is a challenging, but potentially rich, area of pharmacology [33].
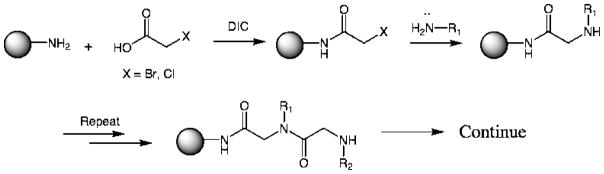
Over the last few years, we have explored peptoids (N-substituted oligoglycines) for this purpose. Peptoids [34], originally created by Zuckerman and co-workers at Chiron are made readily using a “sub-monomer” route in which a single unit of the peptoid oligomer is created from an α-haloacetate and a primary amine (Fig. 3) [35,36]. Because thousands of primary amines are commercially available or synthesized easily and because OBOC libraries of several million peptoids can be made in a week or so, this is an almost ideal chemistry for generating molecular diversity. We have also reported effective protocols with which to screen these libraries for ligands to either soluble proteins or integral membrane receptors [6•,37,38][39••][40].
Figure 3.
The sub-monomer synthesis of peptoids [35].
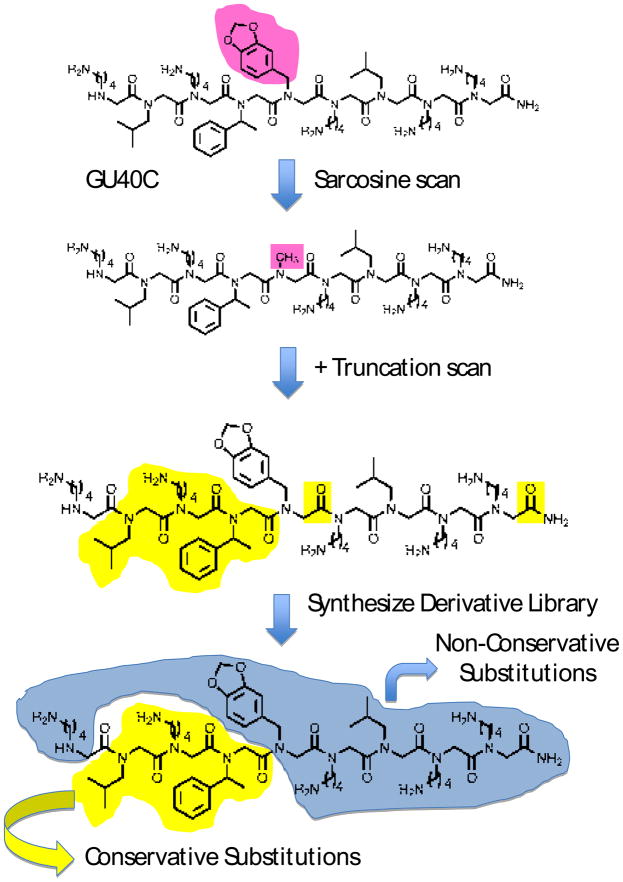
The utility and current limitations of this technology are illustrated by a screen that provided a ligand for the VEGF Receptor 2 (VEGFR2) called GU40C (Fig. 4) [39••]. The KD of the GU40C•VEGFR2 complex was about 3 μM, respectable for a primary screening hit, but far off the affinity typical of a good antibody. A high affinity dimer of this compound was created that exhibited a KD of ≈ 25 nM for the extracellular domain of VEGFR2 and exhibited potent inhibition of VEGFR2-mediated angiogenic signaling in cultured cells as well as living animals. But the dimer was not cell permeable and was introduced into the bloodstream of mice via an implanted pump. It would be of interest to develop an orally bioavailable VEGFR2 inhibitor, so we have also begun to attempt to optimize the monomeric peptoid without increasing it’s mass.
Figure 4.
Work flow for the optimization of a protein-binding peptoid using GU40C4 as an example. First, the side chains important for binding the protein are identified by synthesizing derivatives of the hit in which a methyl group is substituted for one of the side chains (for example at the position highlighted in pink). These derivatives are tested for binding to the protein target. Once the critical positions are identified, a putative minimal pharmacophore consisting of only these residues is synthesized and tested for binding (not shown). If it does not bind, truncated derivatives of the hit are made and tested for binding. In this way, the side chains and main chain residues of GU40C highlighted in yellow were found to be important for VEGFR2 binding. To identify improved derivatives of GU40C, new libraries will be created using amines different than those employed in the synthesis (see Fig. 4) of the original library. At the positions found to be important for binding, amines with structures similar, but different to that found in GU40C will be employed in an attempt to optimize the fit of these side chains with the protein pocket they contact. At positions found not to be important for binding, amines drastically different than those employed in the synthesis of the original library will be employed in an attempt to pick up new, productive peptoid-protein contacts. The derivative library will be screened again under more demanding conditions.
As a first step, the core pharmacophore of the molecule was characterized [41•]. First, a “sarcosine scan” was conducted that identified three side chains as being important for binding to the receptor (Fig. 4). At all of the other positions, substitution of a methyl group for the side chain present in GU40C did not have a major effect upon receptor binding. However, when this trimer was synthesized, it exhibited no detectable affinity for VEGFR2. The characterization of truncated GU40C derivatives revealed that the main chain carbonyl groups in residues 1 and 5 were also important contributors to binding. With the core pharmacophore of GU40C4 now well defined, efforts are under way to obtain an improved molecule. The scheme that will be employed is shown schematically in Fig. 4. A derivative library will be made in which each of the important side chains is altered modestly by using amines at that position that more or less resemble the amine used to create the GU40C parent compound in an attempt to optimize the fit of the residue at this position in the protein pocket it recognizes. In contrast, at positions identified as unimportant by the sarcosine scan, amines will be used in the derivative library synthesis that are quite different structurally than those used in the construction of the original library in the hopes of picking up new interactions that will lead to high affinity. These results will be reported in due course.
A significant limitation of peptoids is their lack of conformational constraints. Therefore, we, and others, are exploring libraries of macrocyclic peptoids as well as mixed libraries that contain small peptoid units connected via stiff linker elements. One can also imagine other types of oligomer libraries that are comprised of conformationally constrained backbones. These new libraries may provide higher affinity protein ligands than simple linear peptoids. At the moment however, a high-throughput alternative to traditional SAR experiments remains a key, but so far elusive, goal in the field.
Acknowledgments
The work from my own laboratory described in this review was funded by a contract from the NHLBI (NO1-HV-28185) and the NIH Director’s Pioneer Award (DP10D00066301).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mukherjee S, Keitany G, Li Y, Wang Y, Ball HL, Goldsmith EJ, Orth K. Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science. 2006;312:1211–1214. doi: 10.1126/science.1126867. [DOI] [PubMed] [Google Scholar]
- 2.Kodadek T, Reddy MM, Olivos HJ, Bachhawat-Sikder K, Alluri PG. Synthetic molecules as antibody replacements. Acc Chem Res. 2004;37:711–718. doi: 10.1021/ar030145l. [DOI] [PubMed] [Google Scholar]
- 3.Lam KS, Salmon SE, Hersh EM, Hruby VJ, Kazmierski WM, Knapp RJ. A new type of synthetic peptide library for identifying ligand-binding activity. Nature. 1991;354:82–84. doi: 10.1038/354082a0. [DOI] [PubMed] [Google Scholar]
- 4.Blackwell HE, Perez L, Stavenger RA, Tallarico JA, Eatough AEC, Foley MA, Schreiber SL. A one-bead, one-stock solution approach to chemical genetics: part 1. Chem & Biol. 2001;8:1167–1182. doi: 10.1016/s1074-5521(01)00085-0. [DOI] [PubMed] [Google Scholar]
- 5.Clemons PA, Koehler AN, Wagner BK, Sprinings TG, Spring DR, King RW, Schreiber SL, Foley MA. A one-bead, one-stock solution approach to chemical genetics: part 2. Chem & Biol. 2001;8:1183–1195. doi: 10.1016/s1074-5521(01)00086-2. [DOI] [PubMed] [Google Scholar]
- 6•.Astle JMS, Impson LS, Huang Y, Reddy MM, Wilson R, Connell S, Wilson J, Kodadek T. Seamless bead to microarray screening: Rapid identification of the highest affinity protein ligands from large combinatorial libraries. Chem & Biol. 2010;17:38–45. doi: 10.1016/j.chembiol.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohlmeyer MH, Swanson RN, Dillard LW, Reader JC, Asouline G, Kobayashi R, Wigler M, Still WC. Complex synthetic chemical libraries indexed with molecular tags. Proc Natl Acad Sci USA. 1993;90:10922–10926. doi: 10.1073/pnas.90.23.10922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu R, Marik J, Lam KS. A novel peptide-based encoding system for “one-bead one-compound” peptidomimetic and small molecule combinatorial libraries. J Amer Chem Soc. 2002;124:7678–7680. doi: 10.1021/ja026421t. [DOI] [PubMed] [Google Scholar]
- 9.Wrenn SJ, Harbury PB. Chemical evolution as a tool for molecular discovery. Annu Rev Biochem. 2007;76:331–349. doi: 10.1146/annurev.biochem.76.062205.122741. [DOI] [PubMed] [Google Scholar]
- 10.Scheuermann J, Neri D. DNA-encoded chemical libraries: a tool for drug discovery and for chemical biology. Chembiochem. 2010;11:931–937. doi: 10.1002/cbic.201000066. [DOI] [PubMed] [Google Scholar]
- 11.Alluri PG, Reddy MM, Bacchawat-Sikder K, Olivos HJ, Kodadek T. Isolation of protein ligands from large peptoid libraries. J Amer Chem Soc. 2003;125:13995–14004. doi: 10.1021/ja036417x. [DOI] [PubMed] [Google Scholar]
- 12.Peng L, Liu R, Marik J, Wang X, Takada Y, Lam KS. Combinatorial chemistry identifies high-affinity peptidomimetics against alpha4beta1 integrin for in vivo tumor imaging. Nature Chem Biol. 2006;2:381–389. doi: 10.1038/nchembio798. [DOI] [PubMed] [Google Scholar]
- 13.MacBeath G, Koehler AN, Schreiber SL. Printing small molecules as microarrays and detecting protein-ligand interactions en masse. J Amer Chem Soc. 1999;121:7967–7968. [Google Scholar]
- 14.MacBeath G, Schreiber S. Printing proteins as microarrays for high-throughput function determination. Science. 2000;289:1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 15.Kodadek T. Rethiking screening. Nature Chem Biol. 2010;6:162–165. doi: 10.1038/nchembio.303. [DOI] [PubMed] [Google Scholar]
- 16.Shuker SB, Hajduk PJ, Meadows RP, Fesik SW. Discovering high-affinity ligands for proteins: SAR by NMR. Science. 1996;274:1531–1534. doi: 10.1126/science.274.5292.1531. [DOI] [PubMed] [Google Scholar]
- 17.Congreve M, Chessari G, Tisi D, Woodhead AJ. Recent developments in fragment-based drug discovery. J Med Chem. 2008;51:3661–3680. doi: 10.1021/jm8000373. [DOI] [PubMed] [Google Scholar]
- 18.Bachhawat-Sikder K, Kodadek T. Mixed element capture agents (MECAs): A simple strategy for the construction of synthetic, high affinity protein capture ligands. J Amer Chem Soc. 2003;125:9550–9551. doi: 10.1021/ja034912n. [DOI] [PubMed] [Google Scholar]
- 19.Roska RL, Lama TG, Hennes JP, Carlson RE. Small molecule-based binding environments: combinatorial construction of microarrays for multiplexed affinity screening. J Am Chem Soc. 2009;131:16660–16662. doi: 10.1021/ja9046944. [DOI] [PubMed] [Google Scholar]
- 20.Xu L, Vagner J, Josan J, Lynch RM, Morse DL, Baggett B, Han H, Mash EA, Hruby VJ, Gillies RJ. Enhanced targeting with heterobivalent ligands. Mol Cancer Ther. 2009;8:2356–2365. doi: 10.1158/1535-7163.MCT-08-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21•.Vagner J, Xu L, Handl HL, Josan JS, Morse DL, Mash EA, Gillies RJ, Hruby VJ. Heterobivalent ligands crosslink multiple cell-surface receptors: the human melanocortin-4 and delta-opioid receptors. Angew Chem Int Ed Engl. 2008;47:1685–1688. doi: 10.1002/anie.200702770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Josan JS, Sankaranarayanan R, Handl HL, Fernandes S, Xu L, Vagner J, Gillies RJ, Hruby VJ. Heterobivalent ligands crosslink multiple cell-surface receptors--a step towards personal medicine. Adv Exp Med Biol. 2009;611:413–414. doi: 10.1007/978-0-387-73657-0_178. [DOI] [PubMed] [Google Scholar]
- 23.Lewis WG, Green LG, Grynszpan F, Radic Z, Carlier PR, Taylor P, Finn MG, Sharpless KB. Click chemistry in situ: acetylcholineesterase as a reaction vessel for the selective assembly of a femtomolar inhibitor from an array of building blocks. Angew Chem Intl Ed. 2002;41:1053–1057. doi: 10.1002/1521-3773(20020315)41:6<1053::aid-anie1053>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 24••.Agnew HD, Rohde RD, Millward SW, Nag A, Yeo WS, Hein JE, Pitram SM, Tariq AA, Burns VM, Krom RJ, et al. Iterative In Situ Click Chemistry Creates Antibody-like Protein-Capture Agents. Angew Chem Int Ed Engl. 2009 doi: 10.1002/anie.200900488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim HS, Reddy MM, Xiao X, Wilson J, Wilson R, Connell S, Kodadek T. Rapid identification of improved protein ligands using peptoid microarrays. Bioorg Med Chem Lett. 2009;19:3866–3869. doi: 10.1016/j.bmcl.2009.03.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao X, Yu P, Lim HS, Sikder D, Kodadek T. A cell-permeable synthetic transcription factor mimic. Angew Chem Int Ed Engl. 2007;46:2865–2868. doi: 10.1002/anie.200604485. [DOI] [PubMed] [Google Scholar]
- 27.Hesselberth J, Robertson MP, Jhaveri S, Ellington AD. In vitro selection of nucleic acids for diagnostic applications. Rev Mol Biotechnol. 2000;74:15–25. doi: 10.1016/s1389-0352(99)00005-7. [DOI] [PubMed] [Google Scholar]
- 28.Forster AC, Tan Z, Nalam MN, Lin H, Qu H, Cornish VW, Blacklow SC. Programming peptidomimetic syntheses by translating genetic codes designed de novo. Proc Natl Acad Sci U S A. 2003;100:6353–6357. doi: 10.1073/pnas.1132122100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29•.Kawakami T, Murakami H, Suga H. Messenger RNA-programmed incorporation of multiple N-methyl-amino acids into linear and cyclic peptides. Chem Biol. 2008;15:32–42. doi: 10.1016/j.chembiol.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 30.Yu P, Liu B, Kodadek T. A high-throughput assay for assessing the cell permeability of combinatorial libraries. Nature Biotech. 2005;23:746–751. doi: 10.1038/nbt1099. [DOI] [PubMed] [Google Scholar]
- 31.Kwon YU, Kodadek T. Quantitative evaluation of the relative cell permeability of peptoids and peptides. J Amer Chem Soc. 2007;129:1508–1509. doi: 10.1021/ja0668623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohuchi M, Murakami H, Suga H. The flexizyme system: a highly flexible tRNA aminoacylation tool for the translation apparatus. Curr Opin Chem Biol. 2007;11:537–542. doi: 10.1016/j.cbpa.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 33.Gadek T. Strategies and methods in the identification of antagonists of protein-protein interactions. Biotechniques. 2003;(Supplement):21–24. [PubMed] [Google Scholar]
- 34.Simon RJ, Kania RS, Zuckermann RN, Huebner VD, Jewell DA, Banville S, Ng S, Wang L, Rosenberg S, Marlowe CK, et al. Peptoids: a modular approach to drug discovery. Proc Natl Acad Sci U S A. 1992;89:9367–9371. doi: 10.1073/pnas.89.20.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zuckermann RN, Kerr JM, Kent SBH, Moos WH. Efficient method for the preparation of peptoids [Oligo(N-substituted glycines)] by submonomer solid-phase synthesis. J Am Chem Soc. 1992;114:10646–10647. [Google Scholar]
- 36.Figliozzi GM, Goldsmith R, Ng SC, Banville SC, Zuckermann RN. Synthesis of N-substituted glycine peptoid libraries. Methods Enzymol. 1996;267:437–447. doi: 10.1016/s0076-6879(96)67027-x. [DOI] [PubMed] [Google Scholar]
- 37.Lim H-S, Archer CT, Kodadek T. Identification of a peptoid inhibitor of the proteasome 19S regulatory particle. J Amer Chem Soc. 2007;129:7750–7751. doi: 10.1021/ja072027p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiao X, Yu P, Lim H-S, Sikder D, Kodadek T. Design and synthesis of a cell permeable synthetic transcription factor mimic. J Comb Chem. 2007;9:592–600. doi: 10.1021/cc070023a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39••.Udugamasooriya DG, Dineen SP, Brekken RA, Kodadek T. A peptoid “antibody surrogate” that antagonizes VEGF receptor 2 activity. J Amer Chem Soc. 2008;130:5744–5752. doi: 10.1021/ja711193x. [DOI] [PubMed] [Google Scholar]
- 40.Qi X, Astle J, Kodadek T. Rapid identification of of orexin receptor binding ligands using cell-based screening accelerated with magnetic beads. Mol BioSystems. 2009;5:102–107. doi: 10.1039/b915611a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41•.Udugamasooriya DG, Dunham G, Ritchie C, Brekken RA, Kodadek T. The pharmacophore of a peptoid VEGF receptor 2 antagonist includes both side chain and main chain residues. Bioorg Med Chem Lett. 2008;18:5892–5894. doi: 10.1016/j.bmcl.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]