Abstract
Diagnosis of human bladder cancer in untreated tissue sections is achieved by using imaging data from desorption electrospray ionization mass spectrometry (DESI-MS) combined with multivariate statistical analysis. We use the distinctive DESI-MS glycerophospholipid (GP) mass spectral profiles to visually characterize and formally classify twenty pairs (40 tissue samples) of human cancerous and adjacent normal bladder tissue samples. The individual ion images derived from the acquired profiles correlate with standard histological hematoxylin and eosin (H&E)-stained serial sections. The profiles allow us to classify the disease status of the tissue samples with high accuracy as judged by reference histological data. To achieve this, the data from the twenty pairs were divided into a training set and a validation set. Spectra from the tumor and normal regions of each of the tissue sections in the training set were used for orthogonal projection to latent structures (O-PLS) treated partial least-square discriminate analysis (PLS-DA). This predictive model was then validated by using the validation set and showed a 5% error rate for classification and a misclassification rate of 12%. It was also used to create synthetic images of the tissue sections showing pixel-by-pixel disease classification of the tissue and these data agreed well with the independent classification that uses histological data by a certified pathologist. This represents the first application of multivariate statistical methods for classification by ambient ionization although these methods have been applied previously to other MS imaging methods. The results are encouraging in terms of the development of a method that could be utilized in a clinical setting through visualization and diagnosis of intact tissue.
Keywords: cancer, desorption electrospray ionization, lipidomics, molecular imaging, multivariate statistics, mass spectrometry
Introduction
Mass spectrometry (MS) is a powerful technique in many scientific disciplines, and it is playing an increasingly important role in the field of molecular imaging. Imaging MS allows the direct investigation of the distribution of a variety of endogenous and exogenous compounds in plant and animal tissues with high specificity and without the need for fluorescent or radioactive labeling normally used in histochemical protocols.[1,2] In particular, tissue MS-based molecular imaging has the potential to provide additional histopathological information for the diagnosis and prognosis of many disease states, including cancer. A range of ionization techniques is employed in imaging MS, each with its own advantages and disadvantages. Secondary ion mass spectrometry (SIMS)[3] and matrix-assisted laser desorption ionization (MALDI)[4] are the two most commonly used methods. In addition, laser-based ionization techniques have been used in imaging MS at atmospheric pressure, including electrospray-assisted laser desorption ionization (ELDI)[5] and laser ablation electrospray ionization (LAESI).[2,6] Ambient ionization is a new family of highly simplified ionization methods in which samples are examined in their native state by mass spectrometry at atmospheric pressure. Here, we report on the application of a well-known ambient ionization method, desorption electrospray ionization (DESI),[7] to generate ion images of cancerous and adjacent normal human bladder tissue from the lipid profiles of the samples. Our goal is to develop a method applicable to in situ diagnosis of intact tissue in a clinical setting. One step to achieving this goal is to image tissue sections by DESI-MS in conjunction with multivariate statistical analysis. We use a training set to establish the best predictive features and a validation set to evaluate performance on representative samples. This is the first application of multivariate statistical methods for classification to DESI-MS imaging data although we have previously used principal component analysis (PCA).
In a growing number of reports on imaging MS, images are based on the distribution of lipid species, including glycerophospholipids (GPs). This choice is due to the ease of lipid ionization and their important and diverse roles in cellular processes, including those involved in various forms of cancer.[8] DESI-MS has been applied previously to the analysis of multiple types of human and animal cancers: in canine bladder cancer GP profiles allow distinction between tumor and normal tissues;[9] in human prostate cancer a single molecule, cholesterol sulfate, is elevated in precancerous and cancerous tissue versus normal tissue;[10] in human gliomas GP profiles allow the discrimination of tumor subtypes.[11] Although principal component analysis (PCA) has been applied to single patient DESI-MS data, no statistical analysis has been performed by using the DESI-MS data from a set of human tissues. This study represents for the first time a complete multivariate statistical approach that has been used in combination with DESI-MS data to provide a predictive model for future analysis of human bladder tissue. As a step toward developing a clinical tool for visualization and diagnosis of disease, we use distinctive DESI-MS GP profiles to visually characterize and formally classify twenty pairs of cancerous and adjacent normal human bladder tissue samples. We show that images derived from the acquired profiles correlate with features of standard histological examination by using hematoxylin and eosin (H&E)-stained serial sections. The profiles allow classification of the disease status of the tissue samples with high accuracy; the resulting predictive model can be validated and applied to unknown samples.
Results and Discussion
Bladder tumor and adjacent normal tissue sections were interrogated by using DESI-MS in the negative ion mode. Figure 1 and Figure S1 in the Supporting Information show typical negative ion mode full-scan mass spectra, and illustrate how a number of different ions in the spectra change in abundance between typical tumor and adjacent normal samples. The data show significant changes in glycerophosphoinositols (PIs), glycerophosphoserines (PSs) and fatty acids. Although chemical identification of the ions underlying each DESI-MS image is not required for diagnostic purposes, the glycerophospholipid species present in the tissue sections were identified based on collision-induced dissociation (CID) tandem MS experiments and comparison with literature data.[12] The fatty acid were tentatively identified based on the mass of the molecular anion, [M–H]−. Ions in the region from m/z 500–650 were identified as dimers of the free fatty acids.
Figure 1.
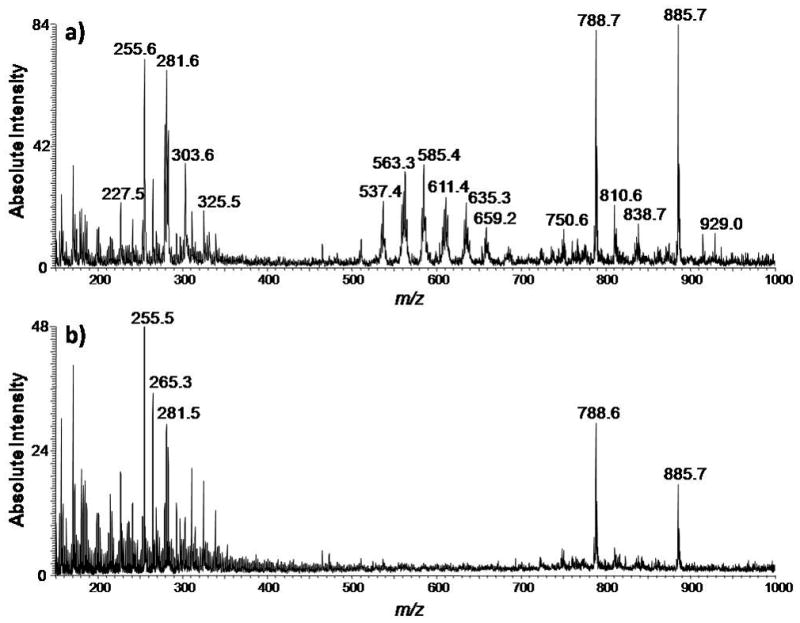
Typical negative ion mode full-scan mass spectra of human bladder cancer tissue and adjacent normal tissue in the range of m/z 150–1000. a) Negative ion mode spectrum of the tumor region of the tissue sample of UH0112-37. b) Negative ion mode spectrum of the normal region of the tissue sample of UH0112-37.
Although individual ions can be used to distinguish tumor from normal tissue, the use of multiple ions should improve the diagnosis. This requires the use of multivariate statistical methods in order to reduce the high-dimensional data we acquire for each sample.[13] A set of twenty tissue pairs was randomly divided in two 10-sample pairs—one a training set, used to develop the predictive models, the second a validation set, used to test the predictive accuracy of the methodology. The use of paired tissue samples ensures that differences between cancerous and normal tissues are not due to differences inherent to individual patients. The training tissue section set was used to manually acquire spectra that excluded non-informative regions. The spectra of the training set were recorded from known tumor and normal regions of the tissue samples, as determined by pathological examination, allowing predictive models to be constructed by using well-understood data. In total, 53 spectra representative of either tumor or normal tissue were acquired for each tissue section in the training set (a total of 20 tissue sections). The spectra were re-sampled to unit resolution, background corrected, and scaled to the median area under the curve. The validation set of ten independent tissue section pairs used the full mass spectral imaging data, with no manual manipulation to construct synthetic images from the statistical models. All synthetic images shown are from the validation set.
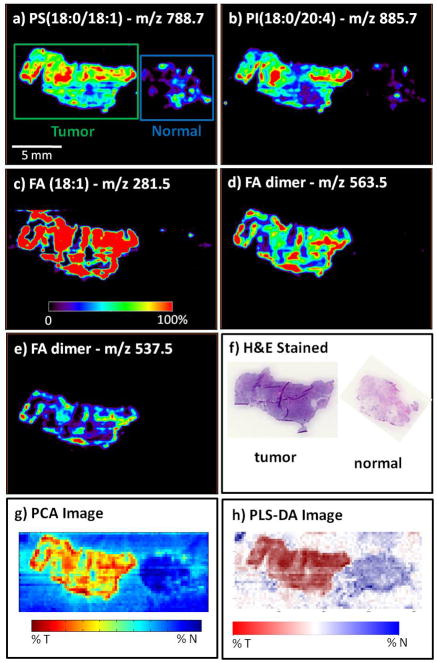
A series of DESI-MS images was used to visually represent the abundance of individual ions in each tissue section, thereby characterizing the distribution of particular molecular species across the set of tissue samples. From among all the lipid species identified, specific ion species were selected from the training dataset to be shown in the DESI-MS ion images of the validation dataset (Figure 2). The selected ions are m/z 788.7 (PS(18:0/18:1)), 885.7 (PI(18:0/20:4)), 281.5 (FA(18:1)), 563.5 (FA dimer), and 537.5 (FA dimer). The images were visually compared to optically scanned images of the H&E-stained sections, which were analyzed by the pathologist.
Figure 2.
Negative ion mode tissue imaging of bladder tissue including areas of cancer and adjacent normal tissue of sample UH0103-23 from the validation set, illustrating individual ion images and the performance of the PCA and partial least-square discriminate analysis (PLS-DA) methods; a) ion image of m/z 788.7, PS(18:0/18:1); b) ion image of m/z 885.7, PI(18:0/20:4); c) ion image of m/z 281.5, FA(18:1); d) ion image of m/z 563.5, FA dimer; e) ion image of m/z 537.5, FA dimer; f) H&E-stained tissue sections of the tumor tissue; and (g) normal PCA-based synthetic image and (h) PLS-DA synthetic image.
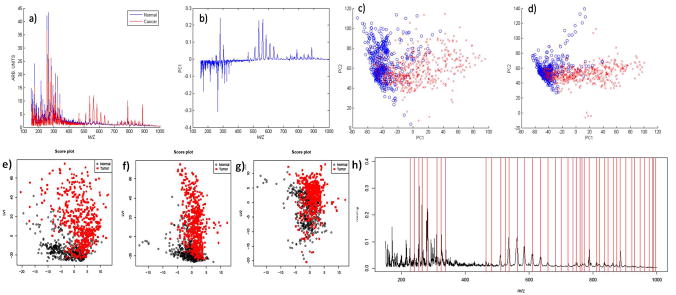
Principal component analysis (PCA) was applied to the spectra acquired from the training dataset to generate a set of principal components (PCs). The principal components are orthogonal linear combinations of DESI-MS peaks, that is, weighted averages of abundances of individual ions with weights maximizing between-tissue variation. All spectra were then projected onto the system of coordinates formed by the PCs, and the scores (i.e., the coordinates of the spectra from the training set on each principal component) were recorded. Figure 3a–d shows the input spectra, the score plots, and two representations of loadings of the first two principal components, respectively. As can be seen in Figure 3b, PC1 accounts for the largest proportion of between-group variation, and fatty acid dimer peaks have large loadings in this PC. Therefore, PC1 was used to produce a synthetic image (termed the PCA image, see for example, Figure 2g) for each tissue in the validation set. This was done by using color-coded pixels to score the underlying spectrum on the coordinate represented by PC1. The image color indicates an approximate strength of prediction based on the first principal component. Values are scaled over the range 0–100% to use the full color map. PCA information from all of the ions summarized in a single image (Figure 2g) is expected and found to perform better than simple visualization of individual ions (Figure 2a–e)).
Figure 3.
Principal component analysis: a) average tumor (red) and normal (blue) spectra from the training set that were used as inputs for PCA. The spectra are re-sampled to unit resolution, background corrected, and scaled to the median area under the curve for all spectra. b) First principal component. c) PC1/PC2 score plot for training data set. d) PC1/PC2 score plot for validation data set projected onto the training set principal component eigenvectors. e)–g) Score plots for PLS-DA of the training set. The two axes LV1 and LV2 correspond to the scores of the first two components for each sample after applying PLS. A good separation between cancer ( ) and normal (○) samples as observed in e) indicates that the first two components carry sufficient disease-related information, f) and g) show the scores of the first and second components with the third component, which is unnecessary for disease classification. h) Loading plot for the first component in PLS-DA for the training set. The plot illustrates the m/z values and their relative importance for the first component in PLS-DA. The large peaks are detected and marked by vertical lines in the plot, which should correspond to the isotopic patterns. These visible peaks are the primary effects contributing to the separation of normal and tumor samples and subject to further close investigation.
) and normal (○) samples as observed in e) indicates that the first two components carry sufficient disease-related information, f) and g) show the scores of the first and second components with the third component, which is unnecessary for disease classification. h) Loading plot for the first component in PLS-DA for the training set. The plot illustrates the m/z values and their relative importance for the first component in PLS-DA. The large peaks are detected and marked by vertical lines in the plot, which should correspond to the isotopic patterns. These visible peaks are the primary effects contributing to the separation of normal and tumor samples and subject to further close investigation.
Although PCA images show the variation between tissues, they do not use information concerning the disease state of the samples and do not yield a rule for classification of either individual spectra or entire tissues. A second statistical method allowed a formal diagnostic rule to be derived based on the acquired DESI-MS profiles. To this end, 53 spectra from the tumor and normal regions of ten pairs of samples were used as the training set for orthogonal projection to latent structures (O-PLS)[14] treated partial least-square discriminate analysis.[15] PLS is a supervised multivariate procedure, which removes linear combinations of peaks in the spectra that do not contribute to the separation of the disease state of the samples, thereby yielding a model with better interpretability compared with regular PLS. PLS-DA allowed us to formally classify the disease status of individual spectra (i.e., image pixels).
Figure 3e–g shows the PLS score plots of the first three components, indicating that they carry sufficient disease-related information and provide a separation between cancerous and normal tissues. Figure 3h shows the loading plot of the first component, and the relative importance of various m/z values. The visible peaks are the primary effects contributing to the separation of cancerous and normal tissues. The classifier was developed on the training set. However the true disease status of each pixel was not determined for each H&E-stained section, as only the overall disease state for each tissue section could be reliably determined by the pathologist. We therefore evaluated the performance of the method by classifying entire sections as cancerous or normal by using a simple “majority rule”, where the entire tissue sample is classified as cancer if the majority of the spectra are classified as cancerous according to the model. The result was then compared to the pathologist3s diagnosis. This approach allowed a determination of classification error rates. The PLS-DA model with ten components achieved the best prediction performance (10-fold cross-validated Q2=0.727), and produces a 0.052 error rate for classification. We then applied the model obtained from the training set to the ten tissue pairs in the validation set, and obtained a misclassification error rate of 0.119. The classification rule was visualized by using synthetic images, where color intensity of each pixel shows the strength of evidence for cancerous tissue, with white being the cut-off value between tumor and normal. This represents the first application of PLS-DA to full imaging data and its first diagnostic use in imaging mass spectrometry.
For complete visualization, DESI-MS-generated ion images were compared to the synthetic images from the validation set resulting from the PCA and PLS-DA analysis. The synthetic images combine the qualitative information obtained from all of the ions seen in the mass spectral GP profiles. Figures 2 and 4 show the overall results of the PCA and PLS-DA visualization procedures. Figure 2a–e shows the negative ion mode imaging of tissue samples for patient UH0103-23, whereas Figure 2f shows the corresponding H&E-stained sections. Figure 2g–h shows the synthetic images. The tumor tissue appears on the left side and the normal tissue on the right side of the images.
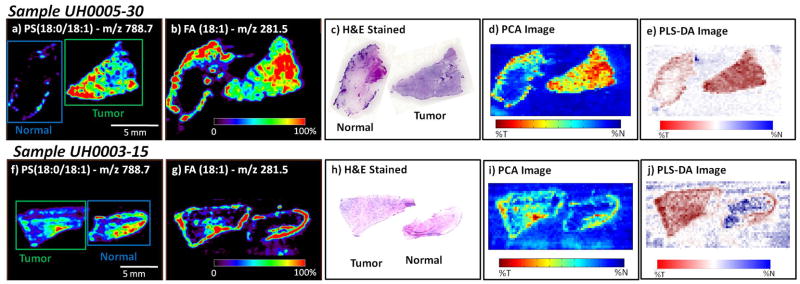
Figure 4.
Negative ion mode tissue imaging of bladder tissues including areas of cancer and adjacent normal tissue (note the tissue orientation is different between the two samples). Both samples are from the validation set, illustrating the performance of the methods on future samples; a) UH0005-30 ion image of m/z 788.7, PS(18:0:18:1); b) UH0005-30 ion image of m/z 281.5, FA(18:1); c) UH0005-30 H&E-stained tissue sections of the normal and tumor tissue; d) UH0005-30 PCA-based synthetic image; e) UH0005-30 PLS-DA-based synthetic image. The normal tissue appears to contain a border of carcinoma in situ on the upper left hand side of the tissue section; f) UH0003-15 ion image of m/z 788.7, PS(18:0:18:1); g) UH0003-15 ion image of m/z 281.5, FA(18:1); h) UH0003-15 H&E-stained tissue sections of the tumor and normal tissue; i) UH0003-15 PCA-based synthetic image; j) PLS-DA-based synthetic image.
The tumor tissue exhibits increased intensity for the ions at m/z 788.7 (PS(18:0/18:1)), 885.7 (PI(18:0/20:4)), 281.5 (FA(18:1)), 563.5 (FA dimer), and 537.5 (FA dimer) when compared to the normal tissue. These increased intensities of fatty acids and fatty acid dimers could represent the presence of lipid droplets in the tissue, with the co-localization of increased amounts of fatty acids contributing to dimer formation. These lipid droplets are known to be increased in inflammation and cancer.[16] Both PCA and PLS-DA output a continuous score, with a larger value indicating stronger evidence for tumor. PCA does not have a natural score cutoff for classification. For PLS-DA, the color scheme was calibrated so that “white” corresponds to the cutoff value, red indicates a stronger presence of tumor and blue indicates greater presence of normal tissue.
Figure 4 shows analogous images for two more tissue pairs, UH0005-30 and UH0003-15. In the DESI-MS and PCA images both samples appear to have a border of tumor on the normal tissue. In one sample, UH0005-30, the normal tissue does contain a border of carcinoma in situ, as determined by pathological examination of the H&E-stained tissue. Therefore, the correct diagnosis can be made on the basis of the lipid signals as seen in the DESI-MS images. In the other sample, UH0003-15, pathological examination of the H&E-stained tissue revealed that the border seen in the DESI and PCA images is that of normal urothelial cells, which make up the transitional epithelium or urothelium layer lining the inside wall of the bladder. These urothelial cells are the site of origin for transitional cell carcinoma (TCC), which accounts for approximately 97% of bladder cancer cases and all but one cancerous sample in this work. As the cancer progresses these urothelial cells proliferate into the cell layers beyond the lining of the bladder, into the muscle of the bladder, to the surrounding fatty tissue, and to other sites in the body. Seemingly normal urothelial cells were detected as cancerous by DESI-MS, however, these samples are from a patient with TCC. It is probable that the tissue appearing to be normal has some malignancy-associated changes that are not apparent when visually examined by a pathologist. Early changes in the tissue in accordance with the field effect theory on bladder cancer tumorigenesis are most likely present resulting in increased signal for the normal tissue in the DESI-MS images.[17] Further studies are necessary to investigate this possibility, including studies involving truly normal urothelium from patients without bladder cancer.
In addition to the three sample pairs presented in Figures 2 and 4, Figures S2–S5 in the Supporting Information present four more tissue pairs and their corresponding DESI-MS, PLS-DA, PCA, and H&E-stained images. Figure S5 presents a single case in which the DESI-MS images do not agree with the pathological diagnosis: based on the ion images the normal tissue would be diagnosed as tumor. For this case the statistically generated synthetic images show a more accurate picture with only select pixels in the normal tissue being classified as tumor. This represents a much more likely scenario where the adjacent normal tissue from a cancer patient contains a small number of tumor cells.
Conclusion
In total twenty sample pairs were visually characterized and statistically classified. For fifteen matched tissue pairs the DESI-MS and PCA images agree with the pathological diagnosis of cancer and normal tissue as determined from the H&E-stained tissue. There is excellent agreement between the H&E stains, individual ion images, principal component synthetic images (PC1), and PLS-DA synthetic images. In one case (Figure 4a–e) a border of tumor was detected on the normal tissue, demonstrating the utility of DESI-MS for determination of surgical margins of the tumor. Overall, we successfully classified regions and whole tissue sections as containing cancerous or normal tissue.
DESI-MS imaging shows great promise as a molecular pathology technique that uses the GP profiles of tissues to visualize and diagnose cancerous and normal tissue. This is the first large scale DESI-MS analysis of GP profiles of human tissue and it supports previous literature in showing that alterations in the GP composition occur in cancerous tissues. The study follows up and expands on the previous bladder studies conducted in animals.[9] Most significantly, this is the first DESI-MS imaging study to include complete multivariate statistical analysis and classification. Expansion of the current work will include searches for other disease markers, examination of additional clinical samples to further validate this classification method and to examine the possibility of assigning a stage to the disease and making a prognosis based on DESI-MS data. An advantage to using a spray-based ionization method is the ability to perform chemically specific reactions at the surface prior to detection.[18] This advantage will be exploited in the future in order to more thoroughly explore the wealth of chemical information present in each sample.
Experimental Section
All tissue samples were handled in accordance with approved institutional review board (IRB) protocols at the Indiana University School of Medicine. All patients, from whom tissue was collected, had voluntarily signed the most current informed consent and HIPAA documents prior to study participation in accordance to the IRB protocol. The samples were flash frozen in liquid nitrogen and stored at −80°C until sliced into 15 μm thick sections and thaw-mounted onto glass slides. The slides were stored at −80°C; prior to analysis the samples were allowed to come to room temperature and then dried under nitrogen in a dessicator for approximately 20 min. All twenty pairs of tissue samples were subjected to DESI-MS imaging analysis. For multivariate statistical analysis, 53 representative, individual spectra were acquired from known tumor and normal regions of each of the ten samples in the training set, excluding the background regions of glass. All experiments were carried out by using a commercial LTQ linear ion trap mass spectrometer controlled by XCalibur 2.0[19] software and a lab-built prototype DESI ion source.[20] Biomap software (freeware, http://www.maldi-msi.org) was used to generate single and overlaid ion images.
Supplementary Material
Acknowledgments
We thank Timothy Ratliff, Director, Purdue University Center for Cancer Research for assistance in obtaining the human bladder cancer samples. This work was supported by the National Institutes of Health (Grant 1 R21 EB00 9459-01).
Footnotes
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/chem.201001692.
References
- 1.Lane AL, Nyadong L, Galhena AS, Shearer TL, Stout EP, Parry RM, Kwasnik M, Wang MD, Hay ME, Fernandez FM, Kubanek J. Proc Natl Acad Sci USA. 2009;106:7314. doi: 10.1073/pnas.0812020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nemes P, Barton AA, Vertes A. Anal Chem. 2009;81:6668. doi: 10.1021/ac900745e. [DOI] [PubMed] [Google Scholar]
- 3.Ostrowski SG, Van Bell CT, Winograd N, Ewing AG. Science. 2004;305:71. doi: 10.1126/science.1099791. [DOI] [PMC free article] [PubMed] [Google Scholar]; Colliver TL, Brummel CL, Pacholski ML, Swanek FD, Ewing AG, Winograd N. Anal Chem. 1997;69:2225. doi: 10.1021/ac9701748. [DOI] [PubMed] [Google Scholar]; Fletcher JS, Lockyer NP, Vaidyanathan S, Vickerman JC. Anal Chem. 2007;79:2199. doi: 10.1021/ac061370u. [DOI] [PubMed] [Google Scholar]
- 4.Caprioli RM, Farmer TB, Gile J. Anal Chem. 1997;69:4751. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]; Chaurand P, Schwartz SA, Caprioli RM. Anal Chem. 2004;76:86A. doi: 10.1021/ac0351264. [DOI] [PubMed] [Google Scholar]
- 5.Shiea J, Huang MZ, HSu HJ, Lee CY, Yuan CH, Beech I, Sunner J. Rapid Commun Mass Spectrom. 2005;19:3701. doi: 10.1002/rcm.2243. [DOI] [PubMed] [Google Scholar]
- 6.Nemes P, Barton AA, Li Y, Vertes A. Anal Chem. 2008;80:4575. doi: 10.1021/ac8004082. [DOI] [PubMed] [Google Scholar]; Nemes P, Vertes A. Anal Chem. 2007;79:8098. doi: 10.1021/ac071181r. [DOI] [PubMed] [Google Scholar]
- 7.Takats Z, Wiseman JM, Gologan B, Cooks RG. Science. 2004;306:471. doi: 10.1126/science.1104404. [DOI] [PubMed] [Google Scholar]; Wiseman JM, Ifa DR, Song Q, Cooks RG. Angew Chem. 2006;118:7346. doi: 10.1002/anie.200602449. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2006;45:7188. [Google Scholar]
- 8.Aboagye EO, Bhujwalla ZM. Cancer Res. 1999;59:80. [PubMed] [Google Scholar]; Glunde K, Jie C, Bhujwalla ZM. Cancer Res. 2004;64:4270. doi: 10.1158/0008-5472.CAN-03-3829. [DOI] [PubMed] [Google Scholar]; Yamaji-Hasegawa A, Tsujimoto M. Biol Pharm Bull. 2006;29:1547. doi: 10.1248/bpb.29.1547. [DOI] [PubMed] [Google Scholar]; Utsugi T, Schroit AJ, Connor J, Bucana CD, Fidler IJ. Cancer Res. 1991;51:3062. [PubMed] [Google Scholar]; Zwaal RFA, Comfurius P, Bevers EM. Cell Mol Life Sci. 2005;62:971. doi: 10.1007/s00018-005-4527-3. [DOI] [PubMed] [Google Scholar]
- 9.Dill AL, Ifa DR, Manicke NE, Costa AB, Ramos-Vara JA, Knapp DW, Cooks RG. Anal Chem. 2009;81:8758. doi: 10.1021/ac901028b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eberlin LS, Dill AL, Costa AB, Ifa DR, Cheng L, Masterson T, Koch M, Ratliff TL, Cooks RG. Anal Chem. 2010;82:3430. doi: 10.1021/ac9029482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eberlin LS, Dill AL, Golby AJ, Ligon KL, Wiseman JM, Cooks RG, Agar NYR. Angew Chem. 2010;122:6089. doi: 10.1002/anie.201001452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manicke NE, Wiseman JM, Ifa DR, Cooks RG. J Am Soc Mass Spectrom. 2008;19:531. doi: 10.1016/j.jasms.2007.12.003. [DOI] [PubMed] [Google Scholar]; Pulfer M, Murphy RC. Mass Spectrom Rev. 2003;22:332. doi: 10.1002/mas.10061. [DOI] [PubMed] [Google Scholar]; Hsu FF, Turk J. J Am Soc Mass Spectrom. 2008;19:1681. doi: 10.1016/j.jasms.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hsu FF, Turk J. J Am Soc Mass Spectrom. 2005;16:1510. doi: 10.1016/j.jasms.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 13.Clarke R, Ressom HW, Wang AT, Xuan JH, Liu MC, Gehan EA, Wang Y. Nat Rev Cancer. 2008;8:37. doi: 10.1038/nrc2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trygg J, Wold S. J Chemom. 2002;16:119. [Google Scholar]
- 15.R. D. C. Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: 2009. [Google Scholar]
- 16.Bozza PT, Viola JPB. Prostaglandins, Leukotrienes Essent Fatty Acids. 2010;82:243. doi: 10.1016/j.plefa.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 17.Lopez-Beltran A, Cheng L, Andersson L, Brausi M, de Matteis A, Montironi R, Sesterhenn I, van der Kwast T, Mazerolles C. Virchows Arch. 2002;440:3. doi: 10.1007/s00428-001-0577-6. [DOI] [PubMed] [Google Scholar]
- 18.Wu C, Ifa DR, Manicke NE, Cooks RG. Anal Chem. 2009;81:7618. doi: 10.1021/ac901003u. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chen H, Cotte-Rodriguez I, Cooks RG. Chem Commun. 2006:597. doi: 10.1039/b515122h. [DOI] [PubMed] [Google Scholar]; Huang G, Chen H, Zhang X, Cooks RG, Ouyang Z. Anal Chem. 2007;79:8327. doi: 10.1021/ac0711079. [DOI] [PubMed] [Google Scholar]
- 19.XCalibur 2.1. Thermo Fisher Scientific; San Jose: 2009. [Google Scholar]
- 20.Ifa DR, Wiseman JM, Song QY, Cooks RG. Int J Mass Spectrom. 2007;259:8. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.