Abstract
Viral infections induce Type I interferons (IFN-α and -β) that recruit unexposed cells in a self-amplifying response. We report that the transcription factor MAFB thwarts auto-amplification by a metastable switch behavior. MAFB acts as a weak positive basal regulator of transcription at the IFN-β promoter through activity at AP-1-like sites. Interferon elicitors recruit the transcription factor IRF3 to the promoter, whereupon MAFB acts as a transcriptional antagonist, impairing the interaction of CREB-binding protein (CBP) with IRF3. Mathematical modeling supports the view that prepositioning of MAFB on the promoter allows the system to respond rapidly to fluctuations in IRF3 activity. Elevated expression of MAFB in human pancreatic islet β-cells might increase cellular vulnerability to viral infections associated with the etiology of type I diabetes.
Keywords: MAFB, interferon, diabetes, antiviral, model
Viral infections in mammalian cells elicit responses by strain-nonspecific cellular pattern-recognition receptors, including Toll-like receptors (TLRs), RIG-like helicase, double-strand-specific kinases, and cytosolic DNA receptors1. Engagement of these sensors triggers an intracellular signaling cascade leading to the production of type-I interferon (IFN-I) and proinflammatory cytokines. The cytokines further activate a subset of genes that enforce and propagate an antiviral state throughout the host, thereby activating the first line of defense against viral pathogens. Although rapid and sensitive cellular induction of cytokines upon virus infection is essential for efficient suppression of viral propagation, mammalian cells have also developed multiple mechanisms to prevent autonomous induction and excess production of Type I IFNs2. Despite major advances over the last decade in our understanding of cellular regulation of Type I IFN induction and signaling pathways, the components of the pathways involved have not been fully elucidated.
In this study, a genome-wide screen of potential gene products interacting with the human type I interferon transcriptional response identified multiple candidates with negative action at the interferon β promoter. One of these, MAFB, a member of the family of MAF transcription factors, is the subject of this report. Members of the MAF family of proto-oncogenes mediate both oncogenic transformation and terminal differentiation3,4. MAFB is ubiquitously expressed in human tissues5, but is found at an especially high amounts in myeloid cells, and facilitates the establishment and maintenance of the monocyte-macrophage lineage6. MAFB has also been implicated in the formation of pancreatic islet beta cells7,8. We report here that MAFB is a regulator of Type I IFN transcription with a dual mode of action as both activator and coactivation inhibitor.
Results
MAFB is a negative regulator of Type I IFN
Candidate positive and negative regulators of Type I IFN transcription were identified by a transcriptional reporter screen in which 17184 individual cDNAs encoding human proteins were cotransfected with an IFN-β luciferase reporter into 293ETN cells. Luciferase activity measured at 2 days post-transfection provided a sensitive and reliable measure of transcriptional enhancement or repression. The screen identified known activators and repressors of IFN-β transcription as well as proteins for which no activity had previously been ascribed. Among the latter, the large MAF family protein MAFB consistently and prominently inhibited the interferon transcriptional response and was selected for further study.
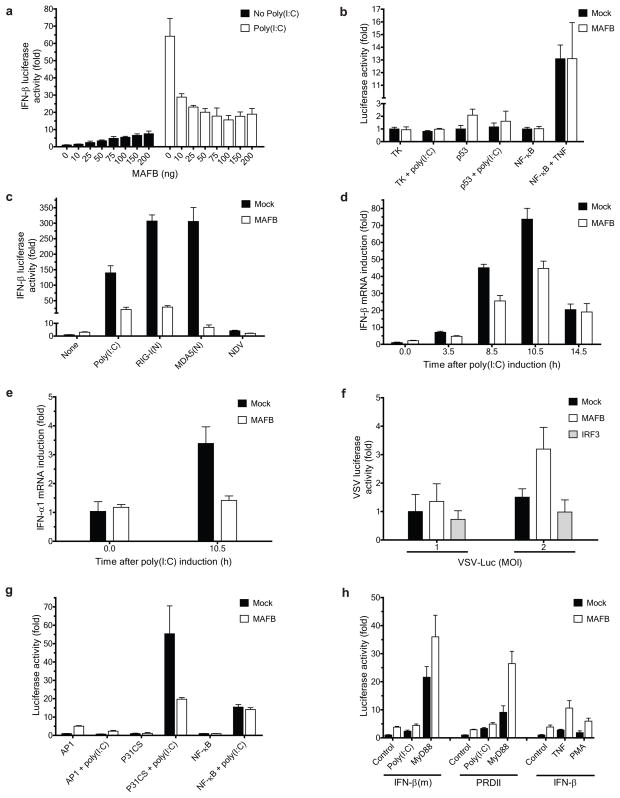
Overexpression of MAFB in 293ETN cells weakly enhanced the basal activity of the IFN-β promoter in a dose-dependent manner (Fig. 1a). By contrast, when the cells were primed with poly(I:C) at 24 h post-transfection, coexpressed MAFB strongly inhibited activation (Fig. 1a). MAFB-mediated inhibition did not reflect a general repression of transcription since the activity of a luciferase expressed under the control of a Herpes simplex thymidine kinase promoter was not affected by MAFB coexpression either in the absence or presence of poly(I:C) (Fig. 1b). In addition, the activities of p53-and NF-κB-promoter luciferase reporters were not suppressed by coexpressed MAFB (Fig. 1b). MAFB strongly inhibited IFN-β activation triggered by other IFN-β inducers, including constitutively active N-terminal forms of RIG-I and MDA59,10, RIG-I(N) and MDA5(N), respectively, and Newcastle disease virus (NDV) (Fig. 1c). Similar results were obtained using HEC1B cells (Supplementary Fig. 1a), which lack a functional type I IFN receptor, indicating that suppression of extracellular feedback mediated by IFN-β secretion is not a required element of the inhibition process. Mafb also inhibited poly(I:C)- and RIG-I(N)-mediated activation of Ifn-β in murine macrophage cell lines (Supplementary Fig. 1b). The action of MAFB on endogenous IFN-α and β promoters recapitulated the results found with the synthetic promoter. In unstimulated 293ETN cells the basal expression of IFN-β and α1 mRNAs was slightly increased by MAFB overexpression, whereas the poly(I:C)-mediated activation was severely impaired (Fig. 1d and e). The luciferase activity of a recombinant vesicular stomatitis virus (VSV-Luc) was enhanced by coexpression of MAFB, whereas expression of IRF3, a positive regulator of Type I IFN induction, suppressed VSV-dependent luciferase expression (Fig. 1f).
Figure 1.
MAFB negatively regulates Type I Interferon induction. (a–c, g, h) Effect of MAFB on viral elicitor-mediated activation of luciferase reporters: (a, c) IFN-β-Luc, (b) TK Renilla, p53-Luc, or NF-κB-Luc, (g) AP1-Luc, P31CS-Luc, or NF-κB-Luc, and (h) IFN-β(m)-Luc (an IRF3 binding deficient mutant of IFN-β), PRDII-Luc (a PRDI, III and IV-deleted mutant of IFN-β), or IFN-β-Luc (10 ng each). Poly(I:C) (25 μg/ml), TNF-α (50 ng/ml), NDV (12 HA units), or PMA (60 ng/ml) was added at 24 h post-transfection. RIG-I(N), MDA5(N) or MyD88 (100 ng each) was transfected at 0 h. Luciferase activity was measured at 48 h (poly(I:C), TNF and PMA) or 32 h (NDV, RIG-I(N), MDA5(N) and MyD88) post-transfection, and is displayed as fold increase relative to the basal level luciferase activity in mock-transfected control without stimulation for each reporter. (d, e) Effect of MAFB on activation of endogenous (d) IFN-β and (e) IFN-α1 promoters. mRNA levels were measured by RT-PCR at the times depicted after poly(I:C) induction. All values were normalized to β-actin, and the values of each gene were further normalized to those before induction. (f) Effect of MAFB on VSV replication. Cells were infected with VSV-Luc at indicated MOIs 24 h after transfection with an indicated vector (25 ng each). VSV replication was assayed by measuring luciferase activity at 9 h post-infection. Data indicate mean ± SD of at least three (d, e) or four (a–c, f–h) within-plate replicates, and results representative of two (b, h) or three (a, c–g) independent experiments are shown. 293ETN cells were used in all panels.
To clarify which DNA motif within the IFN-β promoter conveyed the observed MAFB effects, we cotransfected MAFB expression plasmids with luciferase reporters containing multimerized PRDIII-PRDI (P31CS), PRDIV (AP-1) or PRDII (NF-κB) motifs, which bind IRF3 and 7, ATF2 and c-JUN, and NF-κB (p50 and p65) transcription factors, respectively11. In unstimulated cells, the AP1 motif was activated by MAFB coexpression (Fig. 1g), suggesting that this motif is responsible for MAFB-mediated stimulation of IFN-β promoter basal activity. Upon poly(I:C) treatment, the P31CS motif was strongly activated (> 50-fold), and this activation was potently inhibited by MAFB coexpression (Fig. 1g). Coexpression of MAFB with a reporter based on the ISG54 ISRE element (regulated by IRF3 and 7) corroborated these findings (Supplementary Fig. 1c). Mutation of the PRDIII-PRDI motif of the IFN-β promoter that diminishes binding of IRF3 and 712 led to decreased MAFB-mediated inhibition of poly(I:C) induction (Fig. 1h). MAFB had little inhibitory effect on the activation by IFN-β inducers or MyD88 of NF-κB motif-dependent transcription, as exemplified by a mutant IFN-β reporter containing the PRDII motif but lacking PRDIII-PRDI and PRDIV (Fig. 1h) or by multimerized NF-κB (Supplementary Fig. 1d). In addition, MAFB did not inhibit activation of the intact IFN-β promoter by TNF and phorbol myristate acetate (PMA), which act via NF-κB and AP-1 motifs, respectively, and the PRDIII-PRDI mutant IFN-β promoter by MyD88 (Fig. 1h). These results argue against the possibility of nonspecific suppression of IFN-β activation by MAFB coexpression. The RANTES (CCL5) promoter was similarly regulated by MAFB in a signal-dependent manner (Supplementary Fig. 1c), indicating that the regulatory activity of MAFB is not restricted to the IFN-β promoter. The apparent action of MAFB at AP-1 motifs is consistent with the observation that the canonical MAF response element, MARE, contains an AP1 motif as its core (Supplementary Fig. 1c)3,4. Collectively, these data support the view that the stimulatory and inhibitory activities of MAFB at the IFN-β promoter segregate with the AP-1 site and IRF3 and 7 binding site respectively, and that the inhibitory action of MAFB at IRF3 and 7 binding sites is not confined to the IFN-β promoter.
MAFB deficiency facilitates Type I IFN production
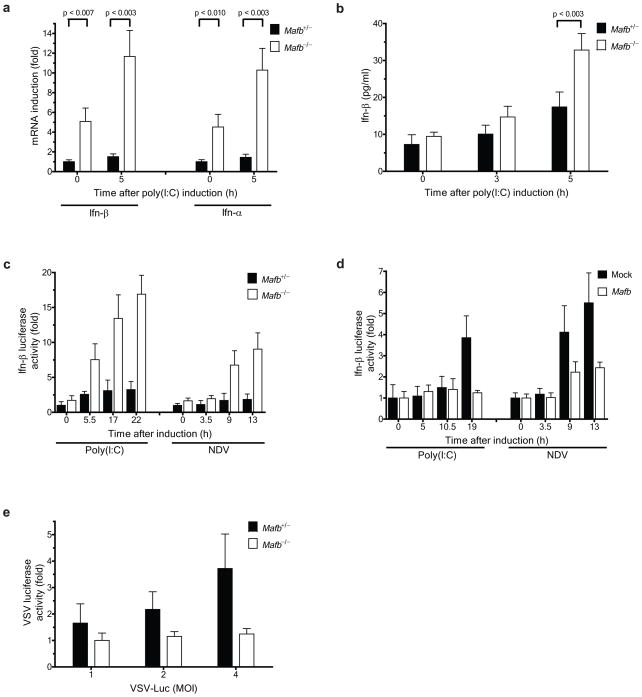
Although Mafb null homozygotes display a perinatal lethal phenotype, the innate antiviral responses of Mafb−/− mouse embryonic fibroblasts (MEFs) could be contrasted with those of MEFs generated from Mafb+/− heterozygous littermates13. In the basal state, Ifn-α and β mRNA abundance was significantly higher in homozygous MEFs compared to heterozygous control MEFs (Fig. 2a). Transcripts of genes induced by Type I IFN signaling, such as Irf7 and Rig-I, were also more highly expressed in homozygous MEFs compared to heterozygous controls (Supplementary Fig. 2a). Poly(I:C) treatment elicited significantly higher quantities of Ifn-α and β mRNA (Fig. 2a) and secreted Ifn-β (Fig. 2b) in homozygous MEFs. Introduction of a plasmid encoding luciferase under the control of the mouse Ifn-β promoter revealed that activation by poly(I:C) treatment or NDV infection was greater in homozygous MEFs than in heterozygous MEFs (Fig. 2c). In addition, reintroduction of mouse Mafb in Mafb null MEFs attenuated Ifn-β activation by poly(I:C) or NDV (Fig. 2d). In accord with the more robust antiviral responses observed in the knockout MEFs, VSV-Luc expression was diminished in homozygous MEFs compared to heterozygous MEFs (Fig. 2e).
Figure 2.
Knockout of Mafb facilitates Type I interferon induction. (a, b) Comparison of activation of endogenous Ifn-α and Ifn-β promoters between Mafb−/− and Mafb+/− MEFs by (a) RT-PCR and (b) ELISA. (a) The mRNA levels and (b) secreted Ifn-β protein in cell supernatants were measured at the times depicted after poly(I:C) induction. (a) All values were normalized to β-actin, and the values of each gene were further normalized to the corresponding gene value from Mafb+/− MEFs at 0 h. (c) Mafb−/− and Mafb+/− MEFs were transfected with a mouse Ifn-β luciferase reporter (mIFN-β-Luc). (d) Mafb−/− MEFs were transfected with vector control or mouse Mafb together with mIFN-β-Luc. (c, d), poly(I:C) or NDV (6 HA units) was added at 24 h post-transfection, and luciferase activity was measured at the indicated times after stimulation as fold increase relative to the basal level luciferase activity in Mafb+/− MEFs (c) or mock-transfected control (d) before stimulation. (e) Mafb−/− and Mafb+/− MEFs were infected with VSV-Luc at indicated MOIs. Luciferase activity was measured at 9 h post-infection, and is displayed as fold increase relative to the activity in Mafb−/− MEFs at MOI = 1. Data indicate mean ± SD of at least three (a) or four (b–e) within-plate replicates, and results representative of three independent experiments are shown.
Homozygous MEFs grew more slowly than heterozygous MEFs, suggesting that the chronic activation of Type I IFN genes might retard growth. Potential MAFB influences on apoptosis induced by viruses or virus-mimetic signals were assessed by measuring caspase 3 activity in homozygous and heterozygous cells (Supplementary Fig. 2b). No difference in caspase 3 activities was observed between null and heterozygous MEFs in the absence of a viral trigger. However, upon poly(I:C) treatment, caspase 3 activity significantly increased in homozygous, but not in heterozygous MEFs (Supplementary Fig. 2b).
The general features of MAFB-mediated regulation of IFN transcription in murine fibroblasts were also found in human cells of diverse origin. Short hairpin or small interfering RNAs targeting Mafb mRNA (Supplementary Fig. 2c) promoted activation of the human IFN-β promoter triggered by poly(I:C) treatment in 293ETN cells (Supplementary Fig. 2d), HeLa and HT-1080 cells (Supplementary Fig. 2e). Thus MAFB is a broadly expressed transcription factor that has an important function in the restraint of IFN-β promoter activity.
MAFB interferes with IRF3 activity
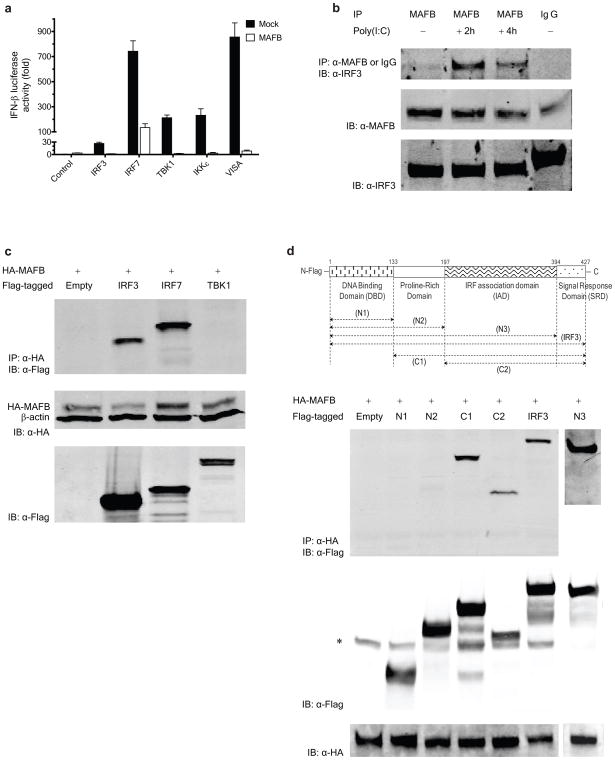
The downstream signaling machinery common to RIG-I, MDA5 and TLR3 includes the kinases TBK1 and IKKε14,15 and the transcription factors IRF3 and IRF716,17. Coexpression of MAFB strongly repressed Type I IFN induction mediated by the upstream effectors of IRF3 and IRF7, as well as IRF3 and IRF7 themselves, in 293ETN cells (Fig. 3a). Similar results were obtained when cells were stimulated using poly(I:C) (Supplementary Fig. 3a). MAFB repressed the IFN-β activation mediated by a constitutively active form of IRF3, IRF3(5D)18,19 (Supplementary Fig. 3a), supporting the interpretation that MAFB functions at a step downstream of IRF3 C-terminal phosphorylation by TBK1 or IKKε. Following transfection, MAFB was found exclusively in the nucleus by immunohistochemistry (Supplementary Fig. 3b) and immunoblotting (Supplementary Fig. 3c), and this localization was unaffected by virus infection, suggesting that the action of MAFB is likely intranuclear.
Figure 3.
MAFB interferes with IRF3 and IRF7 activities. (a) 293ETN cells were transfected with IFN-β-Luc and an expression vector encoding the proteins shown (100 ng each) together with empty control or MAFB (100 ng each). Luciferase activity was measured at 48 h post-transfection (at least in quadruple) as fold increase relative to the basal level luciferase activity in mock-transfected control without MAFB expression. (b) Interaction of endogenous MAFB with endogenous IRF3 in HeLa cells at the indicated times after poly(I:C) stimulation. IP and IB denote immunoprecipitation and immunoblotting, respectively. (c) Interaction of HA-MAFB with Flag-tagged proteins in 293ETN cells (upper panel). At 48 h after transfection, whole cell extracts were immunoprecipitated with anti-HA antibody and then immunoblotted for IRF3, IRF7 and TBK1 with anti-Flag antibody. (d) Interaction of HA-MAFB with Flag-tagged IRF3 deletion mutants in 293 ETN cells. A schematic diagram of full-length IRF3 and its deletion mutants is shown at the top. The bands indicated by * denote nonspecific bands. Immunoblot using 6% (b) or 3% (c, d) of input lysate are also shown. All cell extracts were run on 10% SDS-PAGE.
In untransfected HeLa cells, a weak basal association of endogenous MAFB with IRF3 was found to increase considerably after poly(I:C) stimulation without elevation of MAFB and IRF3 abundance (Fig. 3b). Similar results were obtained using HepG2 cell extracts (Supplementary Fig. 4a), suggesting that the association is not cell type-specific. To evaluate the possibility that MAFB interacts with IRF7, 293ETN cells were transfected with hemagglutinin (HA)-tagged MAFB together with Flag-tagged IRF7. As a specificity control, the interaction of HA-MAFB and Flag-tagged TBK1 was also examined. MAFB associated with IRF7, but not with TBK1, in unstimulated cells (Fig. 3c). Upon poly(I:C) stimulation (Supplementary Fig. 4b), the interaction between MAFB and IRF7 appeared to grow stronger, whereas MAFB remained unassociated with TBK1.
Because MAFB effectively suppresses IFN-β promoter activation by a range of stimuli in HEC1B cells (Supplementary Fig. 1a), in which the inhibitory effect of MAFB is likely mediated through IRF320, we explored potential interactions between IRF3 and MAFB using a series of Flag-tagged IRF3 deletion mutants (Fig. 3d) and HA-tagged MAFB (Supplementary Fig. 4c). Full length MAFB coimmunopreciptated with full length IRF3 and deletion mutants containing the C-terminal IRF association domain (IAD), N3(1-394), C1(134-427) and C2(197-427) fragments, but not with the N-terminal portion of IRF3, N1(1-134) and N2(1-197) (Fig. 3d). This demonstrated that the interaction between MAFB and IRF3 requires the intact IAD of IRF3, whereas the N-terminal DNA binding domain of IRF3 is dispensable for the MAFB–IRF3 interaction. All MAFB deletion mutants could be detected following coimmunoprecipitation with IRF3 (Supplementary Fig. 4c). However, substantial disparities in amounts of MAFB protein expression were present, and an expression-normalized assessment suggests that the C-terminal basic leucine zipper (bZIP) domain of MAFB contributes strongly to the association with IRF3, possibly by facilitation of dimerization. MAFB also exerted inhibitory influences on IRF7-dependent (but IRF3-independent) transcriptional activation (Supplementary Fig. 5a). Efficient binding of MAFB to IRF7 requires the N-terminal portion of IRF7 including the DNA binding domain (Supplementary Fig. 5b).
IRF3 is localized predominantly in the cytoplasm in uninfected cells. Upon virus infection, IRF3 is activated through phosphorylation at its C-terminal serine residues, relieving an intramolecular autoinhibitory association. This conformational change leads to formation of homodimers, accumulation in the nucleus, stimulation of DNA binding, and association with the CBP or p300 coactivator, and thereby activating the Type I IFN genes19–22. Poly(I:C) stimulation substantially increased the formation of heterodimers between Flag-IRF3 and HA-IRF3, but MAFB coexpression only weakly suppressed IRF3 dimer formation (Supplementary Fig. 6a). Similar results were obtained when IRF3 dimerization was analyzed using native PAGE in the presence of deoxycholate20 (Supplementary Fig. 6b).
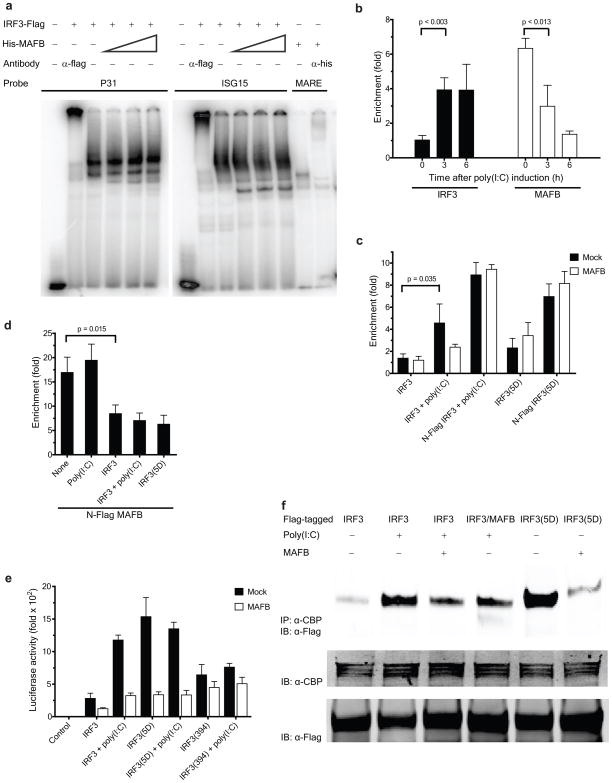
To study the effect of MAFB on IRF3 DNA binding, we carried out electrophoretic mobility shift assays (EMSAs) using a PRDIII-PRDI (P31) oligonucleotide from the IFN-β promoter or an ISRE element from the ISG15 promoter as a probe (Fig. 4a). Addition of in vitro translated His-MAFB did not have an observable effect on the binding of immunopurified IRF3-Flag to the P31 or ISG15 motif (Fig. 4a, lanes 4–6 and 10–12). Similarly, addition of 293ETN-expressed and immunopurified Flag-MAFB did not have any effect on the IRF3 binding (Supplementary Fig. 6c). Equivalent results were obtained using nuclear extracts (NE) prepared from HEC1B cells (Supplementary Fig. 6d), which were transfected with C-terminal Flag-IRF3(5D) in the absence and in the presence of untagged MAFB. Binding of MAFB to the P31 motif could not be detected, although a weak binding of MAFB to the PRDIV motif was detected (Supplementary Fig. 6e).
Figure 4.
Mechanisms underlying MAFB-mediated suppression of Type I IFN induction. (a) EMSA analysis using immunopurified IRF3. C-terminal Flag-IRF3 Protein (0.6 μg) was incubated with a 32P-labeled PRDIII-PRDI (P31) or ISG15 probe in the absence and presence of in vitro translated N-terminal His-MAFB (1, 3 and 5 μl for lanes 4(10), 5(11) and 6(12), respectively). Binding activity and specificity of Flag-IRF3 and His-MAFB (1μl) to indicated probes were demonstrated by supershift experiments. (b–d), ChIP assay to determine the recruitment of (b) endogenous IRF3 and MAFB, (c) overexpressed IRF3 and (d) overexpressed MAFB to the IFN-β promoter. The fold enrichment of the IFN-β promoter over IgG control was quantified using qPCR. (b) HeLa cells were cross-linked at the times depicted after poly(I:C) induction. (c, d), 293ETN cells were transfected with depicted forms of IRF3 and MAFB vectors. All samples were cross-linked at 36 h post-transfection. (e) Effect of MAFB on activation of (Gal4)5-Luc by fusion proteins of the Gal4 DBD and IRF3 variants. 293ETN cells were transfected with (Gal4)5-Luc and Gal4-IRF3 vectors together with empty control or MAFB. Luciferase activity was measured at 48 h post-transfection. (f) Effect of MAFB on the interaction of IRF3 with CBP. 293ETN cells were transfected with indicated Flag-tagged vectors. Where indicated, untagged MAFB was cotransfected at 0 h. Cell lysates were prepared at 36 h post-transfection. (c–f), poly(I:C) was added at 24 h post-transfection as indicated.
The effect of MAFB on recruitment of IRF3 to the IFN-β promoter in vivo was explored by chromatin immunoprecipitation (ChIP), followed by qPCR. Enrichment of IFN-β promoter sequences was detected in immunoprecipitates of endogenous IRF3 from HeLa cells (Fig. 4b) or transfected 293ETN cells (Fig. 4c) and the enrichment was significantly enhanced by poly(I:C) stimulation. MAFB coexpression had either limited or no inhibitory effect on IFN-β promoter enrichment by activated IRF3 (Fig. 4c). These results were confirmed by experiments relying on coexpression of IRF3(5D) and its Flag-tagged form (Fig. 4c). In addition, we examined whether MAFB could be directly recruited to the IFN-β promoter. Immunoprecipitation of endogenous MAFB produced a robust enrichment of IFN-β promoter sequences from chromatin prepared from unstimulated HeLa cells, but upon poly(I:C) stimulation the enrichment diminished (Fig. 4b). Similar results were observed for transfected 293ETN cells expressing Flag-MAFB (Fig. 4d): promoter binding was compromised by IRF3 coexpression and particularly by coexpression of constitutively activated forms of IRF3. Taken together, these results indicate that MAFB does not significantly interfere with DNA binding of IRF3. Instead, IRF3 appears to displace MAFB upon activation and hence the action of MAFB must lie subsequent to IRF3 binding. The transcriptional activation of Gal4-IRF3 fusion proteins is suppressed by coexpressed MAFB thus supporting this notion (Fig. 4e). In addition, a DNA binding-defective mutant of MAFB (N248S)23 retained inhibitory capacity at the IFN-β promoter (Supplementary Fig. 7a), although its transcriptional activity at MARE motifs was severely impaired (Supplementary Fig. 7b), suggesting that binding of MAFB to the IFN-β promoter is not a required element of the inhibition process24. Basal transcriptional activity of the Gal4-IRF3(1-394) truncation mutant (lacking the C-terminal activation domain) was somewhat higher than that of Gal4-IRF3 (Fig. 4e), possibly attributable to a conformational change to an open structure19–22. No considerable increase in the transactivation by Gal4-IRF3(1-394) was observed upon poly(I:C) stimulation (Fig. 4e), and MAFB had little effect on transactivation by Gal4-IRF3(1-394), suggesting that MAFB interferes with functions of IRF3 mediated through the C-terminal five Ser-Thr residues.
Because this region of IRF3 can facilitate the recruitment and activation of the CBP or p300 coactivator19–22, we explored the possibility that MAFB directly interferes with IRF3-CBP interactions. A strong association was observed between Flag-IRF3(5D) and CBP (Fig. 4f, lane 5), and between Flag-IRF3 and CBP upon poly(I:C) stimulation (Fig. 4f, lane 2). The introduction of MAFB or its Flag-tagged form severely impaired the association of CBP with activated forms of IRF3 (Fig. 4f, lanes 3, 4 and 6). The relative strengths of IRF3-CBP interactions (Fig. 4f and Supplementary Fig. 7c) displayed a good correlation with the transactivation induced by Gal4-IRF3 fusion proteins (Fig. 4e). Collectively, these results support the view that interference with IRF3-CBP interactions by MAFB is primarily responsible for the observed MAFB-mediated inhibition of IRF3 and its Gal4 fusions (Fig. 3a and 4e and Supplementary Fig. 3a).
Ectopic expression of CBP did not lead to any observable reversal of the MAFB-mediated inhibition of IRF3-triggered IFN-β activation (Supplementary Fig. 7d). MAFB did not synergize with CBP at the IFN-β promoter (Supplementary Fig. 7e), and no interaction between overexpressed CBP and MAFB could be documented. MAFB also did not inhibit transactivation by the peroxisome proliferator-activated receptor gamma (PPARγ) and PPARγ coactivator-1α (PGC1α) (Supplementary Fig. 7f), which can synergize with CBP to activate transcription25. These results argue that MAFB does not compete with IRF3 for binding to CBP, but rather competes with CBP for binding to IRF3.
We also examined whether sumoylation of MAFB was involved in inhibition of Type I IFN induction. All of the single and double sumoylation-deficient mutants of MAFB (K32R, K297R and K32,297R)26 were as effective as wild-type MAFB in repressing poly(I:C)-mediated activation of IFN-β (Supplementary Fig. 7g), indicating that the sumoylation status of MAFB is not linked with its ability to repress IFN-β activation. Taken together, the foregoing results indicate that MAFB acts as a transcriptional antagonist of Type I IFN induction primarily by impairing recruitment of the transcriptional coactivator CBP to IRF3.
Cellular regulation of MAFB expression
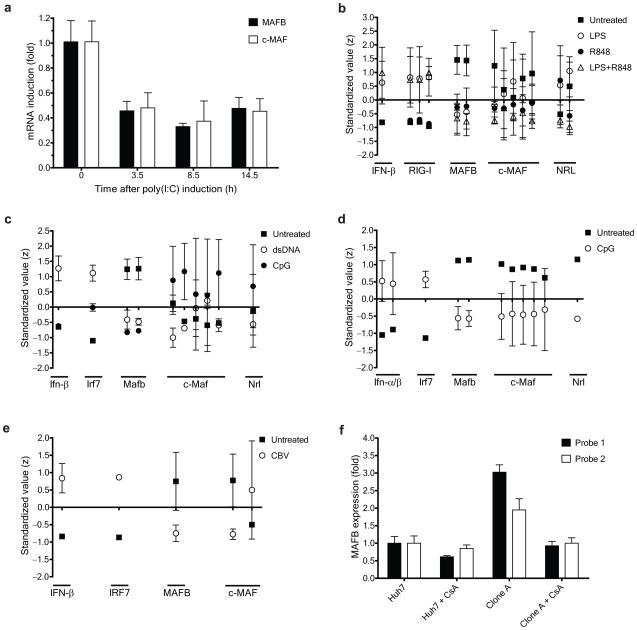
We next investigated how the expression of MAFB varies in response to a viral trigger. MAFB expression in 293ETN cells decreased upon poly(I:C) stimulation (Fig. 5a) and the expression of MAFB over time was inversely correlated with that of IFN-α and β (Fig. 1d and e). Expression of c-MAF, another member of the large MAF family protein, also declined in 293ETN cells (Fig. 5a) in response to poly(I:C) stimulation, and c-MAF suppressed IFN-β activation triggered by a range of Type I IFN inducers including poly(I:C), RIG-I(N) and MDA5(N) (Supplementary Fig. 8a). Expression of the large MAF transcription factors, MAFA and NRL, was modest compared to MAFB and c-MAF in 293ETN cells (data not shown). The change in MAFB abundance differs from that of other reported negative regulators of Type I IFN signaling pathway2, such as A20, DUBA and RNF125, the expression of which is upregulated by viral triggers. Attenuation of MAFB expression upon induction suggests that MAFB acts principally to restrain Type I IFN production in response to low-level cues that might not reflect actual viral infections.
Figure 5.
Regulation of MAFB expression in response to pathogen triggers. (a) The levels of MAFB and c-MAF mRNAs in 293ETN cells were measured by RT-PCR at the indicated times after poly(I:C) induction. Values were normalized to β-actin expression, and further normalized to the corresponding value before poly(I:C) induction. (b–e), Regulatory patterns of expression of MAFB and other large MAF transcription factors in response to pathogen-mimetic stimulation in a variety of cell types. (b) Human monocyte-derived dendritic cells in response to LPS or the synthetic imidazoquinoline resiquimod R84827, a TLR7/8 agonist, (c) murine bone marrow-derived dendritic cells in response to lipid-transfected double-stranded DNA (dsDNA) or CpG oligonucleotides28, a TLR9 agonist, (d) murine plasmacytoid dendritic cells (pDC) in response to CpG oligonucleotides31, and (e) human pancreatic islet cells in response to coxsackievirus32. Microarray expression values were standardized (to Z-values) for each probe set separately, and data are expressed as mean ± SD of each treatment for each indicated probe (x-axis). (f) Expression patterns of MAFB in naïve and HCV-replicon-containing (Clone A) Huh7 cells. Microarray expression values (measured in triplicate)33 were normalized to the values of naïve Huh7.
We also analyzed regulatory patterns of MAFB expression in a variety of cell types in response to pathogen-mimetic stimulation using publicly available microarray datasets including the GEO database (Fig. 5b–e, Supplementary Fig. 8b–e)27–32. In most cases, we observed that MAFB expression decreased in response to a pathogen-mimetic stimulus, whereas expression of IFN-α and β and genes regulated by IFN-α and β signaling increased. No systematic correlations could be detected for c-MAF and NRL, and in many cases their expression levels were negligible compared to that of MAFB. These results, combined with a broader tissue distribution of MAFB expression compared to other large MAF family members (http://biogps.gnf.org), suggest a general, lineage-nonspecific role for MAFB. However, some exceptions have been encountered. For example, in HeLa cells, MAFB expression increased slightly upon poly(I:C) stimulation, and induction of Type I IFN was much weaker than in 293ETN cells (Supplementary Fig. 8f). Expression of MAFB (Fig. 5f), and c-MAF (Supplementary Fig. 8g), was higher in an HCV-replicon-containing cell line that was selected to be highly permissive for HCV RNA replication33. The elevation in MAFB expression may be virus-dependent since inhibition of HCV replication by treating the cells with a cyclophilin inhibitor, Cyclosporin A (CsA), strongly reduced MAFB expression but had little effect on the uninfected parental line. These results raise the possibility that viruses induce MAFB as a strategy for suppressing Type I IFN responses.
Mathematical modeling of IFN-I induction
MAFB exhibits both stimulatory and inhibitory activities on the IFN-β promoter depending on experimental conditions (Fig. 1a). To provide a quantitative framework for understanding MAFB-mediated regulation of Type I IFN induction we developed a biochemical mass-action kinetic model, as outlined in the Supplementary Information. Using this model, we identified several key factors influencing the potency of MAFB as an inhibitor of the IFN-β induction: high order interactions of MAFB and IRF3 on the promoter and in the nucleoplasm, and a low order interaction of MAFB and IFN-β promoter, which affect the IFN-β promoter activity in opposing directions (Supplementary Fig. 9b–g and text therein). The model suggests that MAFB, even at a relatively low level, exerts a considerable inhibitory effect on IFN-β induction, primarily by a high order interaction of MAFB with IRF3 on the promoter and resulting inhibition of IRF3-CBP pre-initiation complex formation (Supplementary Fig. 9e and f). These findings are in agreement with the experimental observations in genetically deficient MEFs. Rapid and strong induction of IFN-β in 293ETN cells in response to a viral elicitor can be ascribed to a substantially enhanced processing rate and higher efficiency of a pre-initiation complex for generating a productive Pol II elongation complex (Supplementary Fig. 9h and text therein). The complex between MAFB and IRF3 in the nucleoplasm provides a mechanism to effectively buffer unwarranted low-level activation of IFN-β, and at the same time, combined with MAFB downregulation (Supplementary Fig. 9i), to counteract this buffering role of MAFB in order to facilitate the IFN-β activation upon virus infection (Supplementary Fig. 9j–n and text therein).
MAFB and enterovirus infection of pancreatic β cells
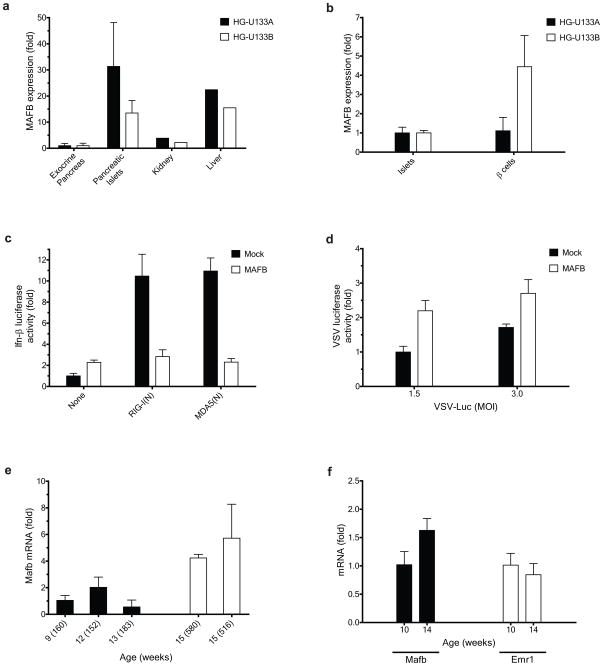
The finding that MAFB expression might predispose cells to viral infection suggests that cells highly expressing MAFB could be vulnerable to viruses. For example, Enteroviruses such as coxsackievirus are capable of infecting pancreatic tissues and such infection might be a trigger for the development of type I diabetes (T1D)34,35. Enterovirus infection is specific to endocrine pancreatic islet cells, but not to exocrine pancreas, in humans35,36. The known enterovirus tropism shows a good correlation with the pancreatic expression pattern of MAFB in humans (Fig. 6a)37. Further, MAFB expression is higher in purified human β cells (Fig. 6b)38, which constitute over 60% of the islet population32, than in islets themselves, suggesting that in humans MAFB expression is maintained at a relatively high level in mature β cells. By contrast, in mice, enterovirus infection of healthy islets is limited even in lethal cases35,39, consistent with the relatively low level of Mafb expression in adult mouse islets7. Mafb expression is restricted to α cells in the adult mouse although the expression is observed both in α and β cells in the mouse embryo. To examine whether the differential MAFB expression might play a role in the observed cell tropism and species selectivity of enterovirus infection, human MAFB was ectopically expressed in a murine pancreatic β cell line, MIN6, and cellular antiviral responses were monitored. Coexpressed MAFB strongly inhibited activation of the murine Ifn-β promoter triggered by RIG-I(N) and MDA5(N) (Fig. 6c). In addition, MAFB expression considerably enhanced replication of VSV-Luc compared to a mock-transfected control (Fig. 6d). Because Type I interferon might play an essential role in preventing β cell destruction induced by coxsackievirus infection40, it is possible that elevated MAFB expression in human islet cells might predispose these cells to coxsackievirus infection or persistence, thereby increasing susceptibility to type I diabetes. Mafb is a candidate gene within a T1D susceptibility locus, Idd13, in non-obese diabetic (NOD)/Lt mice41.
Figure 6.
Roles of MAFB in pancreatic β cells. (a, b). Microarray-based expression patterns of MAFB in human pancreatic tissues. (a) Islet versus exocrine pancreas37 and (b) Purified β cells versus total islet cells38. For each microarray platform, MAFB expression values were normalized to the value of (a) exocrine pancreas and (b) total islet cells. Data indicate (a) mean ± SD (for pancreatic tissues) and a single pooled value (for kidney and liver) of at least two donors, and (b) mean ± SD of three donors. (c, d), Effect of human MAFB overexpression on (c) mIFN-β-Luc activation and (d) VSV replication in a murine β cell line. MIN6 cells transfected with empty control or MAFB were (c) cotransfected with mIFN-β-Luc together with RIG-I(N) or MDA5(N) at 0 h or (d) infected with VSV-Luc 24 h later. Luciferase activities were measured at 34 h post-transfection. (e, f) Age-dependent changes in Mafb expression in islet tissue of NOD mice. (e) beta cell-rich core of islets collected using LCM and (f) whole islets isolated by collagenase perfusion. Mafb and Emr1 mRNA levels were measured by RT-PCR at the age indicated. (e) The blood glucose level for each mouse (mg/dl) is shown in parenthesis. (f) For each age group, two mice were used to obtain pooled total RNA (with blood glucose (mg/dl) of 115 and 143 for 10-week-old and 550 and 579 for 14-week-old). Values were normalized to GAPDH expression, and further normalized to the corresponding values from the youngest mouse (group).
Coxsackieviruses replicates in islet tissue of older prediabetic NOD mice42. To explore whether Mafb plays a role in age-dependent susceptibility of NOD mice to enterovirus infection, we monitored Mafb expression in islet tissue. To avoid potential confounding effects of macrophage infiltration in islets, we selectively collected the β cell-rich core of islets using laser-capture microdissection (LCM). Elevated Mafb transcript abundance was detected in older NOD mice with severe hyperglycemia (blood glucose > 500 mg/dl) compared to younger NOD mice before the onset of hyperglycemia (Fig. 6e). The expression level of a macrophage-specific marker Emr1 (encoding the F4/80) was below the limit of detection in all samples collected by LCM (data not shown). Similar results were obtained when whole islets were isolated from NOD mice by a conventional collagenase perfusion technique (Fig. 6f)43, in which two age groups exhibited similar levels of macrophage infiltration. These results support the notion that the age-dependent accumulation of Mafb contributes to the observed susceptibility of older prediabetic NOD mice to enterovirus infection42. The extent to which endemic enteroviruses contribute to the etiology of NOD strain diabetes is presently unknown, but the variable course and age dependence are consistent with the influence of a stochastic environmental event. Species- and strain-specific, lineage-dependent MAFB expression can be predicted to contribute to susceptibility of islet β-cells to viral infection, a susceptibility that might contribute to the human susceptibility to type 1 diabetes.
Discussion
Results from studies on MEFs from Mafb deficient embryos in this work support the view that the low level of expression of MAFB found in nearly all mammalian tissues has functionally important consequences for antiviral responses. There is ample support for the view that cells, even in the absence of virus infection, are constitutively exposed to low-level activation signals capable of initiating Type I IFN production, as evidenced by the low-level constitutive phosphorylation observed for IRF3 and IRF720,21. Spontaneous induction of Type I IFN should be tightly regulated in uninfected cells since chronic activation diminishes host fitness, facilitates autoimmune disease and induces tissue injury. We propose that as a constitutive inhibitor of the Type I IFN pathway, MAFB buffers cells against unwarranted induction of Type I IFN. In uninfected cells, MAFB proteins might be localized near the IFN-α and β promoters or other IRF3 regulated genes, an affiliation presumably mediated by nearby AP-1 or MARE motifs, and exhibits a weak transcriptional activity. Upon adventitious binding of activated IRF3 to the promoter, MAFB masks IRF3 from CBP by binding to the C-terminal IAD domain of IRF3, inhibiting interaction of IRF3 and CBP and thereby preventing the formation of functional pre-initiation complexes. MAFB might also bind to IRF3 in the nucleoplasm, interfering with cofactor-mediated recruitment of IRF3 to relevant promoters. Upon virus infection, the fluxes of activated IRF3 to the nucleus and recruitment of IRF3 to relevant promoters significantly increase. Further, IRF3 binding to MAFB in the nucleoplasm might counteract the MAFB-mediated inhibition by reducing the pool size of free MAFB as recruitment of IRF3 to the promoters increases, accelerating the production of Type I IFN, which is further amplified by its positive feedback loop. MAFB expression might be downregulated, depending on cell type, to facilitate IRF3-dependent transactivation. In this way, MAFB could create an inhibitory threshold for active Type I IFN induction, with the exact nature of this threshold be determined by cell-type specific MAFB expression and its regulation. The dual role of MAFB as activator and coactivation inhibitor might be intrinsically linked, co-locating MAFB in the chromatin neighborhood of highly sensitive promoters and thereby preventing stochastic fluctuation from provoking runaway amplification.
The mechanism of inhibitory action of MAFB is distinct from that of previously described repressors since, unlike IRF244, MAFB directly binds to IRF3 and IRF7, and does not directly inhibit NF-κB-dependent transactivation. In addition, unlike PIN145 that targets IRF3 for ubiquitination and subsequent proteasome-mediated degradation, MAFB coexpression does not reduce IRF3 protein abundance. The dual mode of action of MAFB as activator and coactivation inhibitor is distinguishable from that of glucocorticoid receptor, a well-documented dual-regulator of transcription46, since the MAFB activity does not depend strongly on its direct association with CBP.
In addition to its effects on IRF3, MAFB might also exert inhibitory influences on IRF7-dependent transcriptional activation under physiological conditions. For example, in cell types with high constitutive IRF7 expression, such as plasmacytoid dendritic cells and macrophages, cells should also restrain ectopic induction of IRF7-dependent transcription. Our results combined with evidence of elevated expression of MAFB in these cells compared to other cell types (http://biogps.gnf.org), suggest that MAFB might also be important in this process. However, the mode of action of MAFB on IRF7 might differ to that observed for IRF3 since virus infection induces significant recruitment of CBP to IRF3, but not IRF718–21, and efficient binding of MAFB to IRF7 requires the DNA binding domain of IRF7, raising the possibility that MAFB interferes with IRF7 DNA binding. Further, our mathematical model suggests that in cell types with elevated expression of MAFB, regulation of recruitment and binding of an interferon-inducing transcription factor to a relevant promoter might constitute a primary mode of action of MAFB for controlling the IFN-β activation (Supplementary Information), supporting the view that MAFB functions as a constitutive inhibitor of IRF7-dependent transactivation in these cells. We also found that, unlike TBK1, MAFB showed significant binding to IKKε, which, combined with its inhibitory effect on IRF7, raises the possibility that MAFB also functions to inhibit late phase interferon signaling.
Regulation of MAFB expression in response to pathogen triggers, combined with the unstable nature of MAFB protein47, its weak transforming activity47 and its ability to strongly repress Type I IFN induction, might enable host cells to respond rapidly to viral pathogens. MAFB exhibits a myeloid expression pattern with prominent transcription in monocytes and macrophages5,6, implying that it broadly shapes the innate immune potential of the organism toward an antibacterial (i.e. monocytic or macrophage) instead of an antiviral (dendritic cell) capability. Because MAFA is highly expressed in mature β cells and might play a fundamental role in regulating survival and function of these cells7,8, MAFB might be dispensable or at least not central for islet function and hence therapeutic inhibition of MAFB with the intent of clearing islet infection might not have adverse consequences for glucose homeostasis. Collectively, our results raise the possibility that MAFB expression and regulation has important consequences for Type I IFN responses and predicts a lineage dependence of viral susceptibility that could have important consequences for diseases of viral or autoimmune etiology.
Methods
Plasmids and mutagenesis
Human and mouse expression plasmids were from Open Biosystems (Huntsville, AL) or OriGene (Rockville, MD) unless otherwise specified. Full-length or truncated open reading frames from plasmids were subcloned into pCMV-HA (Clontech), which was modified to remove the HA-tag, or to add an N- or C-terminal Flag-tag. The IRF3 nuclear localization sequence, amino acids 71 to 89, was added to the N-terminal end of the C-terminal IRF3 and IRF7 deletion mutants (IRF3(134-427), IRF3(197-427), IRF7(151-503) and IRF7(305-503)). Constitutively active IRF318 and DNA binding- and sumoylation-deficient mutants of MAFB23,26 were generated by site-directed mutagenesis PCR. For in vitro translation, N-terminal His-tagged MAFB was subcloned into the pET-28 vector (EMD Biosciences). Gal4-IRF3 fusion plasmids were constructed using the pM vector (Clontech). Transcriptional reporters were generated as summarized in Supplementary Table 1.
Cell culture, transfection and luciferase assay
Cell lines were cultured in DMEM, and MEFs lacking Mafb were cultured in IMDM. Both media were supplemented with 10% calf serum plus iron and 15 μg/ml gentamycin, and further with 50 μM β-mercaptoethanol for IMDM. 293ETN and HEC1B cells were transfected using TransFectin (BioRad). HeLa, HT-1080, MIN6 and RAW 264.7 cells were transfected using Lipofectamine LTX (Invitrogen). MEFs lacking Mafb were transfected using the Amaxa nucleofection system (LONZA). For luciferase assays, cell lines were plated in either 96- or 48-well plates at 4 × 104 cells per well. Unless otherwise specified, cells were transfected with 100 ng of an expression or empty control plasmid together with 10 ng of a luciferase reporter, and 10 ng of pRL-TK Renilla (Promega). For MEFs lacking Mafb, 106 cells were transfected with 2–3 μg of an expression plasmid and 500 ng of a luciferase reporter, and then transfected cells were plated on 48-well plates at 4 × 104 cells per well. For stimulation, 25 μg/ml of poly(I:C) (GE Healthcare), 50 ng/ml of TNFα (R&D Systems) or 60 ng/ml of PMA (Sigma) were added to media at 24 h post-transfection. RIG-I(N), MDA5(N) or MyD88 expression plasmid (100 ng each) was transfected at 0 h. Cells were infected with NDV (LaSota strain, Charles River Laboratories) or VSV-Luc (luciferase-inserted Indiana strain of VSV48) at the indicated hemagglutination (HA) units or multiplicity of infection (MOI) 24 h post-transfection. At various times after transfection, Photinus luciferase activity in total cell lysates was measured using Steady-Lite HTS luciferase substrate (Perkin Elmer), or both Photinus and Renilla luciferase activities were measured using the Dual Luciferase Assay System (Promega).
RNA interference
Lentiviral shRNA vector and pre-designed siRNAs for knockdown of human MAFB were purchased from Sigma (shRNA (TRCN0000017681)), Ambion (α-MAFB-1 (s19279), α-MAFB-2 (s19280) and α-MAFB-3 (a pool of s19279 and s19280)) and Dharmacon (α-MAFB-4 (L-009018)). Empty pLKO.1puro vector or non-targeting siRNAs from the corresponding manufacturers were used as controls. For luciferase assays, cells were transfected with either 100 ng of the shRNA or 5 pmol of a siRNA together with luciferase reporter.
Real-time PCR and ELISA
Total RNA was isolated from cells using the RNeasy RNA extraction kit (Qiagen), and cDNA was prepared using the iScript cDNA Synthesis Kit (Bio-Rad). Real-time PCR was performed using an iQ5 RT-PCR Detection System (Bio-Rad) and iQ SYBR Green Supermix (Bio-Rad). Primers are summarized in Supplementary Table 2. Secreted Ifn-β protein in cell supernatants was measured using murine Ifn-β ELISA Kit (PBL Biomedical).
Immunoblotting
For immunoprecipitation, cell lysates were incubated either with Anti-Flag M2 or Anti-HA Affinity Gel (Sigma), or with an appropriate antibody (anti-CBP (sc-369), Santa Cruz; anti-MAFB (MAB3810), R&D Systems), followed by protein A/G agarose (Sigma). Immunoblotting used the following antibodies: anti-IRF3 (sc-9082, Santa Cruz), anti-HA (MMS-101P, Covance), anti-β-actin (ab8226, Abcam), Goat anti-mouse and anti-Rabbit IRDye 800CW (LI-COR) and IRDye800 anti-Flag and anti-HA (Rockland). Immunocomplexes were visualized using an Odyssey Infrared Imaging System (LI-COR).
EMSA and chromatin immunoprecipitation
Electrophoretic mobility shift assays were based on methods described previously19,20. Flag-tagged IRF3 and MAFB were purified using M2 Affinity Gel. In vitro translated His-MAFB was produced using the TNT Coupled Reticulocyte Lysate System (Promega). All nuclear extracts were treated with 1% deoxycholate to reduce CBP effects on IRF3 DNA binding. Supershift assays were performed using Anti-Flag M2 (Sigma) and anti-His (A00186, GenScript) antibodies. Probes used in the study are summarized in Supplementary Table 3. Chromatin immunoprecipitation was performed according to the manufacturer’s recommendations (Magna ChIP A kit, MILLIPORE). Immunoprecipitation was performed using an appropriate antibody (described earlier) and Protein A Magnetic Beads (MILLIPORE). Purified DNA samples were quantified by real-time PCR using the following primers: 5′-tgacataggaaaactgaaagggag (forward) and 5′-gtcctttctccatgggtatgg (backward).
Native PAGE and caspase 3 assay
Native PAGE was performed as described49. Caspase 3 activity was measured using the Caspase-Glo 3/7 Assay kit (Promega).
NOD mice and islet isolation
Female NOD/ShiLtJ mice were obtained from The Jackson Laboratory. Mice were maintained under SPF conditions in accordance with US Government and local institutional guidelines. Glucose levels were measured weekly using blood samples collected from tail veins. Test groups of mice were sacrificed within 24 hours of glucose measurement. Whole islets were isolated from mice as previously described43.
Laser capture microdissection
Whole mouse pancreatic tissue was harvested and frozen immediately in OCT® (Sigma). 8 micron cryo-sections were mounted on microscopy slides. Each tissue section was lightly fixed in 70% ethanol, rinsed with RNAse free diH2O, and incubated in a toluidine blue solution for 1 min. The stained sections were then dehydrated in increasingly concentrated ethanol 70–100% into xylene. The central core of islets was microdissected and captured onto Arcturus Macro Cap using Veritas Microdissection System (Molecular Devices). The collecting cap was incubated at 42°C for 30 minutes in Arcturus GITC-containing extraction buffer, and RNA was isolated using the Arcturus picopure RNA isolation system (Molecular Devices).
Microarray analysis, mathematical modeling and statistics
Microarray datasets were obtained either from the GEO database (http://www.ncbi.nlm.nih.gov/geo) or directly from the literature, and processed using R. Expression profiles were summarized using the GCRMA method without log2 transformation as implemented in Bioconductor (http://www.bioconductor.org). Expression values were further standardized (to Z-values) for each probe set. Mathematical modeling (Supplementary Information) was performed following a similar approach to ref 50. Two-tailed Student’s t-test or one-way analysis of variance (ANOVA) was applied to compare two or more than two data sets, respectively, using a significance level of 0.05.
Supplementary Material
Acknowledgments
We thank John Darga for assistance with automated assays and high throughput screens, and Rasma Niedra, Naifang Lu and Weihua Zhou for experimental help. We appreciate the contributions of the Seed and Xavier groups in sharing reagents and information. We are grateful to Satoru Takahashi, Michito Hamada and Doug Engel for KO Mafb MEFs and discussion, Sean Whelan for VSV-Luc, Takashi Fujita for the CBP construct and Jun-ichi Miyazaki and Donald F Steiner for MIN6 cells. We thank Charles Vanderburg for assistance with LCM. We also thank Marc Wathelet, Rongtuan Lin, Sunmi Han and Horim Lee for helpful discussions.
Footnotes
Author Contributions
H.K. designed and performed experiments and wrote the manuscript; B.S. designed and supervised research and wrote the manuscript.
References
- 1.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 2.Yoneyama M, Fujita T. RNA recognition and signal transduction by RIG-I-like receptors. Immunol Rev. 2009;227:54–65. doi: 10.1111/j.1600-065X.2008.00727.x. [DOI] [PubMed] [Google Scholar]
- 3.Motohashi H, O’Connor T, Katsuoka F, Engel JD, Yamamoto M. Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene. 2002;294:1–12. doi: 10.1016/s0378-1119(02)00788-6. [DOI] [PubMed] [Google Scholar]
- 4.Eychene A, Rocques N, Pouponnot C. A new MAFia in cancer. Nat Rev Cancer. 2008 doi: 10.1038/nrc2460. [DOI] [PubMed] [Google Scholar]
- 5.Wang PW, et al. Human KRML (MAFB): cDNA cloning, genomic structure, and evaluation as a candidate tumor suppressor gene in myeloid leukemias. Genomics. 1999;59:275–281. doi: 10.1006/geno.1999.5884. [DOI] [PubMed] [Google Scholar]
- 6.Kelly LM, Englmeier U, Lafon I, Sieweke MH, Graf T. MafB is an inducer of monocytic differentiation. Embo J. 2000;19:1987–1997. doi: 10.1093/emboj/19.9.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Artner I, et al. MafB: an activator of the glucagon gene expressed in developing islet alpha- and beta-cells. Diabetes. 2006;55:297–304. doi: 10.2337/diabetes.55.02.06.db05-0946. [DOI] [PubMed] [Google Scholar]
- 8.Nishimura W, et al. A switch from MafB to MafA expression accompanies differentiation to pancreatic beta-cells. Dev Biol. 2006;293:526–539. doi: 10.1016/j.ydbio.2006.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 10.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 11.Maniatis T, et al. Structure and function of the interferon-beta enhanceosome. Cold Spring Harb Symp Quant Biol. 1998;63:609–620. doi: 10.1101/sqb.1998.63.609. [DOI] [PubMed] [Google Scholar]
- 12.Escalante CR, Nistal-Villan E, Shen L, Garcia-Sastre A, Aggarwal AK. Structure of IRF-3 bound to the PRDIII-I regulatory element of the human interferon-beta enhancer. Mol Cell. 2007;26:703–716. doi: 10.1016/j.molcel.2007.04.022. [DOI] [PubMed] [Google Scholar]
- 13.Moriguchi T, et al. MafB is essential for renal development and F4/80 expression in macrophages. Mol Cell Biol. 2006;26:5715–5727. doi: 10.1128/MCB.00001-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fitzgerald KA, et al. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 15.Sharma S, et al. Triggering the interferon antiviral response through an IKK-related pathway. Science. 2003;300:1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- 16.Hiscott J. Triggering the innate antiviral response through IRF-3 activation. J Biol Chem. 2007;282:15325–15329. doi: 10.1074/jbc.R700002200. [DOI] [PubMed] [Google Scholar]
- 17.Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol. 2008;26:535–584. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- 18.Lin R, Heylbroeck C, Pitha PM, Hiscott J. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol Cell Biol. 1998;18:2986–2996. doi: 10.1128/mcb.18.5.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin R, Genin P, Mamane Y, Hiscott J. Selective DNA binding and association with the CREB binding protein coactivator contribute to differential activation of alpha/beta interferon genes by interferon regulatory factors 3 and 7. Mol Cell Biol. 2000;20:6342–6353. doi: 10.1128/mcb.20.17.6342-6353.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wathelet MG, et al. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-beta enhancer in vivo. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 21.Yoneyama M, et al. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. Embo J. 1998;17:1087–1095. doi: 10.1093/emboj/17.4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar KP, McBride KM, Weaver BK, Dingwall C, Reich NC. Regulated nuclear-cytoplasmic localization of interferon regulatory factor 3, a subunit of double-stranded RNA-activated factor 1. Mol Cell Biol. 2000;20:4159–4168. doi: 10.1128/mcb.20.11.4159-4168.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cordes SP, Barsh GS. The mouse segmentation gene kr encodes a novel basic domain-leucine zipper transcription factor. Cell. 1994;79:1025–1034. doi: 10.1016/0092-8674(94)90033-7. [DOI] [PubMed] [Google Scholar]
- 24.Sieweke MH, Tekotte H, Frampton J, Graf T. MafB is an interaction partner and repressor of Ets-1 that inhibits erythroid differentiation. Cell. 1996;85:49–60. doi: 10.1016/s0092-8674(00)81081-8. [DOI] [PubMed] [Google Scholar]
- 25.Mizukami J, Taniguchi T. The antidiabetic agent thiazolidinedione stimulates the interaction between PPAR gamma and CBP. Biochem Biophys Res Commun. 1997;240:61–64. doi: 10.1006/bbrc.1997.7602. [DOI] [PubMed] [Google Scholar]
- 26.Tillmanns S, et al. SUMO modification regulates MafB-driven macrophage differentiation by enabling Myb-dependent transcriptional repression. Mol Cell Biol. 2007;27:5554–5564. doi: 10.1128/MCB.01811-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Napolitani G, Rinaldi A, Bertoni F, Sallusto F, Lanzavecchia A. Selected Toll-like receptor agonist combinations synergistically trigger a T helper type 1-polarizing program in dendritic cells. Nat Immunol. 2005;6:769–776. doi: 10.1038/ni1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 29.Pruett SB, Schwab C, Zheng Q, Fan R. Suppression of innate immunity by acute ethanol administration: a global perspective and a new mechanism beginning with inhibition of signaling through TLR3. J Immunol. 2004;173:2715–2724. doi: 10.4049/jimmunol.173.4.2715. [DOI] [PubMed] [Google Scholar]
- 30.Hammer M, et al. Dual specificity phosphatase 1 (DUSP1) regulates a subset of LPS-induced genes and protects mice from lethal endotoxin shock. J Exp Med. 2006;203:15–20. doi: 10.1084/jem.20051753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao W, et al. Toll-like receptor-mediated induction of type I interferon in plasmacytoid dendritic cells requires the rapamycin-sensitive PI(3)K-mTOR-p70S6K pathway. Nat Immunol. 2008;9:1157–1164. doi: 10.1038/ni.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ylipaasto P, et al. Global profiling of coxsackievirus- and cytokine-induced gene expression in human pancreatic islets. Diabetologia. 2005;48:1510–1522. doi: 10.1007/s00125-005-1839-7. [DOI] [PubMed] [Google Scholar]
- 33.Gaither LA, et al. Multiple cyclophilins involved in different cellular pathways mediate HCV replication. Virology. 2010;397:43–55. doi: 10.1016/j.virol.2009.10.043. [DOI] [PubMed] [Google Scholar]
- 34.Hyoty H, Taylor KW. The role of viruses in human diabetes. Diabetologia. 2002;45:1353–1361. doi: 10.1007/s00125-002-0852-3. [DOI] [PubMed] [Google Scholar]
- 35.Ylipaasto P, et al. Enterovirus infection in human pancreatic islet cells, islet tropism in vivo and receptor involvement in cultured islet beta cells. Diabetologia. 2004;47:225–239. doi: 10.1007/s00125-003-1297-z. [DOI] [PubMed] [Google Scholar]
- 36.Foulis AK, et al. A search for the presence of the enteroviral capsid protein VP1 in pancreases of patients with type 1 (insulin-dependent) diabetes and pancreases and hearts of infants who died of coxsackieviral myocarditis. Diabetologia. 1990;33:290–298. doi: 10.1007/BF00403323. [DOI] [PubMed] [Google Scholar]
- 37.Maffei A, et al. Identification of tissue-restricted transcripts in human islets. Endocrinology. 2004;145:4513–4521. doi: 10.1210/en.2004-0691. [DOI] [PubMed] [Google Scholar]
- 38.Marselli L, et al. Gene expression of purified beta-cell tissue obtained from human pancreas with laser capture microdissection. J Clin Endocrinol Metab. 2008;93:1046–1053. doi: 10.1210/jc.2007-0931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.See DM, Tilles JG. Pathogenesis of virus-induced diabetes in mice. J Infect Dis. 1995;171:1131–1138. doi: 10.1093/infdis/171.5.1131. [DOI] [PubMed] [Google Scholar]
- 40.Chehadeh W, et al. Persistent infection of human pancreatic islets by coxsackievirus B is associated with alpha interferon synthesis in beta cells. J Virol. 2000;74:10153–10164. doi: 10.1128/jvi.74.21.10153-10164.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Serreze DV, Prochazka M, Reifsnyder PC, Bridgett MM, Leiter EH. Use of recombinant congenic and congenic strains of NOD mice to identify a new insulin-dependent diabetes resistance gene. J Exp Med. 1994;180:1553–1558. doi: 10.1084/jem.180.4.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Drescher KM, Kono K, Bopegamage S, Carson SD, Tracy S. Coxsackievirus B3 infection and type 1 diabetes development in NOD mice: insulitis determines susceptibility of pancreatic islets to virus infection. Virology. 2004;329:381–394. doi: 10.1016/j.virol.2004.06.049. [DOI] [PubMed] [Google Scholar]
- 43.Carter JD, Dula SB, Corbin KL, Wu R, Nunemaker CS. A Practical Guide to Rodent Islet Isolation and Assessment. Biol Proced Online. 2009 doi: 10.1007/s12575-009-9021-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Senger K, et al. Gene repression by coactivator repulsion. Mol Cell. 2000;6:931–937. doi: 10.1016/s1097-2765(05)00081-x. [DOI] [PubMed] [Google Scholar]
- 45.Saitoh T, et al. Negative regulation of interferon-regulatory factor 3-dependent innate antiviral response by the prolyl isomerase Pin1. Nat Immunol. 2006;7:598–605. doi: 10.1038/ni1347. [DOI] [PubMed] [Google Scholar]
- 46.Revollo JR, Cidlowski JA. Mechanisms generating diversity in glucocorticoid receptor signaling. Ann N Y Acad Sci. 2009;1179:167–178. doi: 10.1111/j.1749-6632.2009.04986.x. [DOI] [PubMed] [Google Scholar]
- 47.Pouponnot C, et al. Cell context reveals a dual role for Maf in oncogenesis. Oncogene. 2006;25:1299–1310. doi: 10.1038/sj.onc.1209171. [DOI] [PubMed] [Google Scholar]
- 48.Whelan SP, Ball LA, Barr JN, Wertz GT. Efficient recovery of infectious vesicular stomatitis virus entirely from cDNA clones. Proc Natl Acad Sci U S A. 1995;92:8388–8392. doi: 10.1073/pnas.92.18.8388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iwamura T, et al. Induction of IRF-3/-7 kinase and NF-kappaB in response to double-stranded RNA and virus infection: common and unique pathways. Genes Cells. 2001;6:375–388. doi: 10.1046/j.1365-2443.2001.00426.x. [DOI] [PubMed] [Google Scholar]
- 50.Kim H, Perelson AS. Viral and latent reservoir persistence in HIV-1-infected patients on therapy. PLoS Comput Biol. 2006;2:e135. doi: 10.1371/journal.pcbi.0020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.