Abstract
Two ring-A-aromatized bile acids, 1 and 2, were isolated from the sponge Sollasella moretonensis, collected from the seabed of northern Queensland. Structures were assigned on the basis of extensive 1D and 2D NMR studies, as well as analysis by HRESIMS. Compound 2 has previously been produced synthetically, though this marks its first isolation from a natural source.
Keywords: marine natural product, sponge, Sollasella moretonensis, ring-A-aromatized bile acids
As part of a collaborative project with the Queensland Museum to investigate the taxonomy, molecular genetics and intraspecific chemical variability of marine sponges sampled from the Great Barrier Reef [1], we studied Sollasella moretonensis collected from the inter-lagoon seabed area in northern Queensland. No chemistry had been reported previously for this species, suggesting this specimen would be an attractive source for chemical investigation. As a result, two ring-A-aromatized bile acids, 1 and 2, were isolated. The isolation and structure elucidation are described herein.
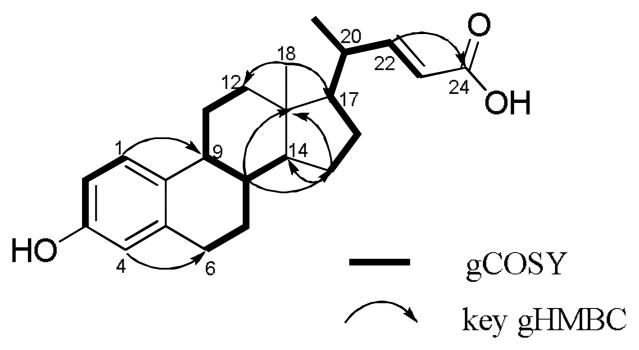
Compound 1 gave the molecular formula C23H30O3, as deduced from HRESIMS ([M + Na]+ m/z 377.2113; Δ -5 ppm). The IR spectrum indicated the presence of a hydroxyl group (3274 cm−1) and an α,β-unsaturated carboxyl group (1685 cm−1). The 1H NMR spectrum exhibited five downfield protons at δH 5.75 (d, J = 15.3 Hz), 6.50 (d, J = 15.3 Hz), 6.46 (d, J = 1.9 Hz), 6.53 (dd, J = 8.5, 1.9 Hz) and 7.06 (d, J = 8.5 Hz), which clearly indicated the existence of a trans double bond and a 1,3,4-trisubstituted benzene ring. The upfield region of the 1H NMR spectrum displayed a singlet methyl at δH 0.77, a doublet methyl at δH 1.12 (d, J = 6.6 Hz) and several methines and methylenes. In addition to the benzene ring system, the COSY experiment revealed two partial structures, CH=CHCH(CH3)CHCH2CH2 and CH2CH2CHCHCH2CH2, corresponding to the C-23, C-22, C-20, C-21, C-17, C-16, C-15 and C-12, C-11, C-9, C-8, C-7, C-6 fragments, respectively (Figure 1). Connectivity between fragments was established 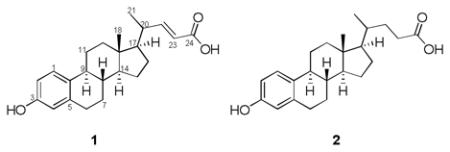 through analysis of the HMBC spectrum (Figure 1). HMBC correlations from H-15 to C-13 and C-14, and from H-8 to C-13 and C-15 allowed connection of these two fragments. HMBC correlations from H3-18 to C-12, C-17 and C-13, and from olefinic protons H-22 and H-23 to C-24 established the locations of Me-18 and the carboxyl group. Finally, HMBC correlations from the aromatic proton H-1 to C-9 and from H-4 to C-6 established the manner of attachment of thex aromatic ring to the alicyclic core, suggesting that compound 1 was a previously undescribed ring-A-aromatized bile acid, namely 3-hydroxy-19-nor-1,3,5(10),22-cholatetraen-24-oic acid.
through analysis of the HMBC spectrum (Figure 1). HMBC correlations from H-15 to C-13 and C-14, and from H-8 to C-13 and C-15 allowed connection of these two fragments. HMBC correlations from H3-18 to C-12, C-17 and C-13, and from olefinic protons H-22 and H-23 to C-24 established the locations of Me-18 and the carboxyl group. Finally, HMBC correlations from the aromatic proton H-1 to C-9 and from H-4 to C-6 established the manner of attachment of thex aromatic ring to the alicyclic core, suggesting that compound 1 was a previously undescribed ring-A-aromatized bile acid, namely 3-hydroxy-19-nor-1,3,5(10),22-cholatetraen-24-oic acid.
Figure 1.
gCOSY and key gHMBC correlations of 1.
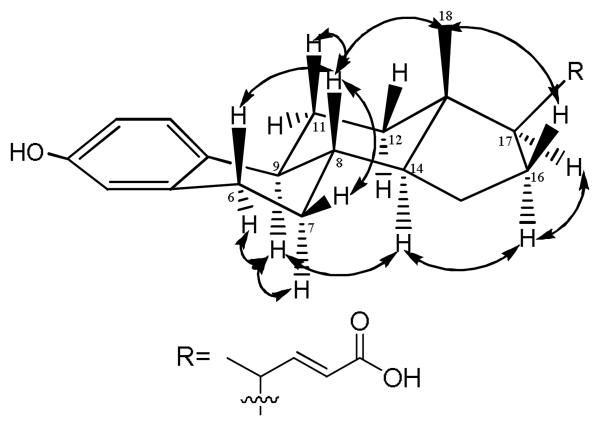
The relative configuration of 1 was determined using NOESY (Figure 2). H-8 displayed a NOESY correlation with H3-18, and H-9 showed a correlation with H-14. Similarly, H-9 exhibited correlations with H-12β and H-7β, whereas H-8 showed correlations with H-11α and H-6α. These data indicated that the B/C and C/D ring junctures were both trans with H-8, H3-18, H-11α, and H-6α on one face of the ring system and H-9, H-12β, H-7β, and H-14 on the opposite face. In addition, NOESY correlations from H-16β to both H-14 and H-17 suggested that the latter is located on the same face as H-9.
Figure 2.
Key NOESY correlations for 1.
Compound 2 showed a pseudomolecular ion peak at m/z 357.2426 ([M + H]+; Δ +1 ppm) in the HRESIMS, suggesting a molecular formula of C23H32O3, which differs from 1 only by the addition of two hydrogen atoms. The 1H NMR spectrum was very similar to that of 1, the major difference being the absence of the olefinic protons at δH 5.75 (d, J = 15.3 Hz), and 6.50 (d, J = 15.3 Hz). In combination with the difference in the molecular formula, this suggested that 2 was the dihydro analogue of 1, as confirmed by detailed analysis by 2D NMR (gCOSY, gHSQC and gHMBC). The relative configuration of 2 was determined to be the same as 1 based on analysis of the NOESY spectrum, thus identifying it as 3-hydroxy-19-nor-1,3,5(10)-cholatrien-24-oic acid, a compound that has been produced synthetically [2,3], though this is the first report of its isolation from a natural source.
Early interest in ring-A aromatized bile acids arose when they were discovered as metabolites produced by microbes in the human intestinal flora from bile acids [4-7], raising the question of whether they might play a role in development of colon cancer [8]. However, synthetic compound 2 was found to lack mutagenic activity on its own [3], thus discounting a significant role for these compounds in carcinogenesis in the colon [9].
Although sterols with aromatic A rings are common in terrestrial plants and in some animals as hormones (for example, estradiol), few have been reported from marine organisms. To our knowledge, only geodisterol [10] and two sulfated congeners [11] have been reported from marine sponges. One unusual feature of geodisterol among ring-A aromatized sterols is that the side chain is fully intact, whereas it is more common for the side chain to have undergone carbon-carbon bond cleavage, as in 1 and 2. It is interesting to note that 2, a compound thought to be an intermediate in the metabolism of bile acids by human gut bacteria [3], is apparent in the extract of a sponge.
Experimental
General experimental procedures
Optical rotations were measured on a JASCO DIP-370 digital polarimeter. UV spectra were acquired in spectroscopy grade methanol using a Hewlett-Packard 8452A diode array spectrophotometer. IR spectra were recorded on a JASCO FT/IR-420 spectrophotometer. NMR data were collected using a Varian INOVA 500 (1H 500 MHz, 13C 125 MHz) NMR spectrometer with a 3 mm Nalorac MDBG probe with a z-axis gradient and utilized residual solvent signals for referencing (δH 3.30 ppm, δC 49.00 ppm for CD3OD). High-resolution ESIMS analyses were obtained using a Micromass Q-Tof Micro mass spectrometer. Analytical and semi-preparative HPLC was accomplished utilizing a Beckman System Gold 126 solvent module equipped with a 168 PDA detector.
Sponge material
Sollasella moretonensis van Soest, Hooper, Beglinger and Erpenbeck, 2006 was collected from an inter-lagoon seabed area of Torres Strait, far North Queensland (−19° 37′ 30.00″ N, 148° 8′ 6.00″ E) under permit [1]; a voucher specimen of G329277 is maintained at the Queensland Museum.
Extraction and isolation
The frozen sponge (150 g wet wt) was exhaustively extracted with MeOH to yield 6.0 g of crude extract that was then separated on HP20SS resin using a gradient of H2O to IPA in 25% steps, and a final wash of 100% MeOH, yielding 5 fractions. The third fraction (50/50 H2O/IPA) was further fractionated on Sephadex LH-20 with 1:1 CH3Cl: MeOH to give 6 fractions (Fr. 3.1-3.6). Fr. 3.5 was chromatographed by HPLC using a Phenomenex Luna C18 column (250 × 10 mm) employing 80% CH3CN/H2O at 4 mL/min to yield compound 1 (0.9 mg, tR = 4.5 min) and compound 2 (1.2 mg, tR = 5.2 min).
3-Hydroxy-19-nor-1,3,5(10),22-cholatetraen-24-oic acid (1)
Colorless solid
[α]D: +27° (c 0.1, MeOH).
IR (film): 3274, 2927, 2866, 1685, 1652, 1557, 1505, 1456, 1403, 1287, 1254, 1209, 1181, 1140, 1029, 985, 928, 868, 844, 813, 786, 725 cm−1.
UV/Vis λmax (MeOH) nm (log ε): 206 (4.77), 222 (4.36), 284 (3.94).
1H NMR and 13C NMR: Table 1.
Table 1.
NMR data for 1 (1H 500 MHz, 13C 125 MHz δ ppm, J in Hz) in CD3OD.
| position | δ C | δH (J in Hz) | COSY | HMBC | NOESY |
|---|---|---|---|---|---|
| 1 | 127.0, CH | 7.06, d (8.5) | H-2 | C-3, C-5, C-9 | H-2, H-9, H-11α, H-11β, H-12β |
| 2 | 113.5, CH | 6.53, dd (8.5,1.9) | H-1 | C-3, C-4, C-10 | H-1 |
| 3 | 156.0, C | ||||
| 4 | 115.8, CH | 6.46, d (1.9) | H-6 | C-2, C-3, C-6, C-10 | H-6α, H-6β |
| 5 | 138.9, C | ||||
| 6α | 30.6, CH2 | 2.76, m | H-4, H-7 | C-4, C-5, C-7, C-8, C-10 | H-4, H-6β, H-7α, H-8 |
| 6β | 1.30, m | H-4 | H-4, H-6α, H-9 | ||
| 7α | 28.9, CH2 | 1.86, m | H-6, H-8 | C-8, C-9 | H-6α, H-7β |
| 7β | 1.32, m | H-6, H-8 | H-7α, H-9 | ||
| 8 | 40.4, CH | 1.33, m | H-7, H-9, H-14 | C-13, C-15 | H-6α, H-11α, H-18 |
| 9 | 45.1, CH | 2.14, m | H-8, H-11 | C-7, C-11, C-14 | H-1, H-6β, H-7β, H-11β, H-12β, H-14 |
| 10 | 132.9, C | ||||
| 11α | 27.9, CH2 | 2.26, m | H-9, H-12 | C-8, C-13 | H-1, H-8, H-11β, H-12α, H-18 |
| 11β | 1.44, m | H-9, H-12 | H-1, H-9, H-11α | ||
| 12α | 41.1, CH2 | 2.12, m | H-11 | C-9, C-11, C-18 | H-11α, H-12β, H-18, H-21 |
| 12β | 1.44, m | H-11 | H-1, H-9, H-14, H-12α, H-18 | ||
| 13 | 44.1, C | ||||
| 14 | 56.7, CH | 1.28, m | H-8 | H-9, H-12β, H-16β | |
| 15α | 29.3, CH2 | 1.34, m | H-16 | C-13, C-14, C-16 | H-15β, H-18 |
| 15β | 1.79, m | H-16 | H-15α | ||
| 16α | 24.9, CH2 | 1.19, m | H-15, H-17 | C-13, C-15, C-17 | H-16β, H-18 |
| 16β | 1.68, m | H-15, H-17 | H-14, H-16α, H-17 | ||
| 17 | 56.9, CH | 1.34, m | H-16, H-20 | C-14, C-15 | H-16β, H-20, H-22, H-23 |
| 18 | 12.6, CH3 | 0.77, s | C-12, C-13, C-17 | H-8, H-11α, H-12, H-15α, H-16α, H-20, H-21 | |
| 20 | 40.7, CH | 2.24, m | H-17, H-21, H-22 | H-17, H-18, H-21, H-22, H-23 | |
| 21 | 20.2, CH3 | 1.12, d (6.6) | H-20 | C-17, C-20, C-22 | H-12α, H-18, H-20, H-22, H-23 |
| 22 | 150.1, CH | 6.50, d (15.3) | H-20, H-23 | C-17, C-20, C-21, C-24 | H-17, H-20, H-21, H-23 |
| 23 | 126.3, CH | 5.75, d (15.3) | H-22 | C-20, C-24 | H-17, H-20, H-21, H-22 |
| 24 | 175.9, C |
HRESIMS: m/z [M + Na+] calcd for C23H30O3Na: 377.20926; found: 377.2113.
3-Hydroxy-19-nor-1,3,5(10)-cholatrien-24-oic acid (2)
Colorless solid
[α]D: +29° (c 0.1, MeOH).
IR (film): 3274, 2925, 2866, 1731, 1558, 1499, 1455, 1445, 1415, 1378, 1286, 1240, 121, 1182, 1142, 1041, 928, 867, 843, 805, 786, 724 cm−1.
UV/Vis λmax (MeOH) nm (log ε): 206 (4.82), 222 (4.40), 284 (4.05).
1H NMR (500 MHz, CD3OD): 7.04 (1H, d, J = 8.4 Hz, H-1), 6.51 (1H, dd, J = 8.4,1.6 Hz, H-2), 6.45 (1H, d, J =1.6 Hz, H-4), 2.74 (1H, m, H-6α), 1.28 (1H, m, H-6β), 1.85 (1H, m, H-7α), 1.30 (1H, m, H-7β), 1.30 (1H, m, H-8), 2.10 (1H, m, H-9), 2.22 (1H, m, H-11α), 1.41 (1H, m, H-11β), 2.12 (1H, m, H-12α), 1.39 (1H, m, H-12β), 1.29 (1H, m, H-14), 1.29 (1H, m, H-15α), 1.86 (1H, m, H-15β), 1.17 (1H, m, H-16α), 1.69 (1H, m, H-16β), 1.20 (1H, m, H-17), 0.73 (3H, s, H3-18), 1.46 (1H, m, H-20), 0.98 (3H, d, J = 6.7 Hz, H3-21 ), 1.80 (1H, m, H-22α), 1.30 (1H, m, H-22β), 2.22 (1H, m, H-23α), 2.05 (1H, m, H-23β).
13C NMR (125 MHz, CD3OD): 127.0 (C-1), 113.5 (C-2), 156.7 (C-3), 115.9 (C-4), 139.7, (C-5), 30.8 (C-6), 28.9 (C-7), 40.4 (C-8), 45.2 (C-9), 133.5 (C-10), 27.8 (C-11), 41.3, (C-12), 44.6 (C-13), 57.0 (C-14), 29.2 (C-15), 24.8 (C-16), 57.4 (C-17), 12.4 (C-18), 37.2 (C-20), 18.9 (C-21), 34.0 (C-22), 36.3 (C-23), 167.5 (C-24).
HRESIMS: m/z [M + H+] calcd for C23H33O3: 357.2430; found: 357.2426.
Supplementary Material
Acknowledgments
Funding for the Varian INOVA 500 MHz NMR spectrometer was provided through NIH grant RR06262. This work was supported by NIH grant CA36622 (C.M.I). Samples were provided by the Torres Strait Seabed Mapping & Characterisation Project [1], a collaboration between the Commonwealth Scientific and Industrial Research Organisation (CSIRO), the Queensland Museum (QM) and the Queensland Department of Primary Industries & Fisheries (QDPI&F); funded by the CRC Torres Strait Program and the National Oceans Office of Australia. We thank the crews of the FRV Gwendoline May (QDPI&F) and RV James Kirby (James Cook University) and acknowledge the traditional inhabitants of the Torres Strait Islands and Papua New Guinea for access to their sea territories.
References
- 1.Pitcher CR, Haywood M, Hooper JNA, Coles R, Bartlett C, Browne M, Cannard T, Carini G, Carter A, Cheers S, Chetwynd D, Colefax A, Cook S, Davie P, Ellis N, Fellegara I, Forcey K, Furey M, Gledhill D, Hendriks P, Jacobsen I, Johnson J, Jones M, Last P, Marks S, McLeod I, Sheils J, Sheppard J, Smith G, Strickland C, Vander Geest C, Venables W, Wassenberg T, Yearsley G. Mapping and Characterisation of key biotic and physical attributes of the Torres Strait Ecosystem. CSIRO/QM/QDPI CRC Torres Strait Task Final Report. 2007:145. http://www.rrrc.org.au/crctorres/index/crctspublications.html
- Tsuda K, Nozoe S, Ohata K. Steroid studies. XLIII. An aromatization reaction of a cross conjugated dienone system with zinc. Chemical & Pharmaceutical Bulletin. 1963;11:1265–1270. doi: 10.1248/cpb.11.1265. [DOI] [PubMed] [Google Scholar]
- Namba T, Hirota T, Hayakawa S. Synthesis and mutagenicity of a ring-A-aromatized bile acid, 3-hydroxy-19-nor-1,3,5(10)-cholatrien-24-oic acid. Journal of Lipid Research. 1988;29:809–814. [PubMed] [Google Scholar]
- Goddard P, Hill MJ. The dehydrogenation of the steroid nucleus by human-gut bacteria. Biochemical Society Transactions. 1973;1:1113–1115. [Google Scholar]
- Drasar BS, Hill MJ. Human Intestinal Flora. Academic Press; London: 1974. p. 139.p. 140. [Google Scholar]
- Goddard P, Hill MJ. Degradation of steroids by intestinal bacteria. IV. The aromatization of ring A. Biochimica et Biophysica Acta. 1972;280:336–342. doi: 10.1016/0005-2760(72)90101-4. [DOI] [PubMed] [Google Scholar]
- Goddard P, Hill MJ. The in vivo metabolism of cholesterol by gut bacteria in the rat and guinea-pig. Journal of Steroid Biochemistry. 1974;5:569–572. doi: 10.1016/0022-4731(74)90106-x. [DOI] [PubMed] [Google Scholar]
- Hill MJ. The role of colon anaerobes in the metabolism of bile acids and steroids, and its relation to colon cancer. Cancer. 1975;36:2387–2400. doi: 10.1002/1097-0142(197512)36:6<2387::aid-cncr2820360618>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Chaplin MF. Bile acids, fibre and colon cancer: the story unfolds. The Journal of the Royal Society for the Promotion of Health. 1998;118:53–61. doi: 10.1177/146642409811800111. [DOI] [PubMed] [Google Scholar]
- Crews P, Wang GYS. Geodisterol, a novel polyoxygenated sterol with an aromatic A ring from the tropical marine sponge Geodia sp. Tetrahedron Letters. 1996;37:8145–8146. [Google Scholar]
- DiGirolamo JA, Li XC, Jacob MR, Clark AM, Ferreira D. Reversal of fluconazole resistance by sulfated sterols from the marine sponge Topsentia sp. Journal of Natural Products. 2009;72:1524–1528. doi: 10.1021/np900177m. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.