Abstract
The HIV-1 capsid is modeled as a Fullerene cone that is composed of ~250 hexamers of the viral CA protein and 12 CA pentamers. Structures of CA hexamers have been difficult to obtain because the hexamer-stabilizing interactions are inherently weak, and CA tends to spontaneously assemble into capsid-like particles. Here, we describe a two-step biochemical strategy to obtain soluble CA hexamers for crystallization. First, the hexamer was stabilized by engineering disulfide crosslinks (either A14C/E45C or A42C/T54C) between the N-terminal domains of adjacent subunits. Second, the crosslinked hexamers were prevented from polymerizing further into hyperstable capsid-like structures by mutations (W184A and M185A) that interfered with dimeric association between the C-terminal domains that link adjacent hexamers. The structures of two different HIV-1 CA hexamers were nearly identical, and we combined the non-mutated portions of the structures to generate an atomic-resolution model for the native hexamer. This hybrid approach for structure determination should be applicable to other viral capsomers and protein-protein complexes in general.
Keywords: capsomer, electron microscopy, engineered disulfide bonds, hexamer, HIV-1 CA, virus structure, X-ray crystallography
Introduction
The mature capsid of an infectious retrovirus is a protein shell that is assembled from ~1,500 copies of the virally encoded CA protein and packages and organizes the viral genome for delivery into new host cells [see (1) for a recent review]. On the basis of the mathematical principles of a fullerene shell, the CA subunits form ~250 hexamers arrayed on a variably curved hexagonal lattice, which is closed by incorporation of exactly 12 CA pentamers.2–4 CA proteins of different retroviruses share a conserved tertiary fold composed of two independently folded domains (called the NTD and CTD),2,5–11 despite minimal primary sequence homology and large variations in macroscopic capsid shape. The two domains are primarily α-helical, and are separated by a flexible stretch of ~4 amino acid residues.
The quaternary organization of the CA subunits in the hexagonal capsid lattice was first established by electron microscopy and biochemical studies.3,5,12–17 Within each hexamer, the 6 NTDs form a symmetric ring that is stabilized by a central 18-stranded α-helical barrel, with each subunit contributing 3 helices. The 6 CTDs are arranged as an outer ring surrounding the central NTD ring, with each CTD packed against the NTD from the adjacent subunit. Hexamers assemble into the extended capsid lattice via interactions of the outer ring, with each of the 6 CTDs forming a homodimer with corresponding domains from neighboring hexamers, thereby connecting each hexamer to six others.
The NTD ring and CTD dimer interactions have each been visualized separately in X-ray and NMR structures of the isolated domains,5,9,18–20 but atomic resolution structures of the full-length CA hexamer have been more difficult to obtain. From a practical standpoint, this is presumably due to the intrinsic difficulties in producing discrete, soluble CA oligomers that could be crystallized. Recently, we reported 4 distinct X-ray crystal structures of the CA protein of HIV-1 (human immunodeficiency virus type 1), which revealed the atomic details and conformational states of a full-length retroviral CA hexamer.21 The NTD ring of the hexamer appeared relatively rigid, whereas the interactions between the NTD and CTD rings enabled limited rigid-body motions of the outer ring. This is likely to be an underlying mechanism for generating the variably curved hexagonal lattice observed in authentic HIV-1 capsids.20–22
Our structure determination strategy relied on a biochemical approach that facilitated the efficient production of homogeneous preparations of soluble HIV-1 CA hexamers that could be crystallized. In the first step, the CA hexamer was stabilized by the introduction of exogenous cysteines at amino acid positions 14 and 45. Upon oxidation, the engineered cysteines efficiently formed an intermolecular disulfide bond that covalently linked each CA NTD subunit to its two neighbors in the hexameric ring. In the second step, the crosslinked hexamers were prevented from polymerizing further into an extended lattice by disrupting the CTD-CTD dimer linkages, thereby allowing the production of soluble hexamers. Here, we describe in detail the design and characterization of the disulfide-stabilized CA hexamers. We also report an additional HIV-1 CA hexamer structure, derived from a construct with cysteine substitutions at positions 42 and 54. The new structure recapitulates the previous structures and provides additional validation for the crosslinking strategy. Stitching together the native portions of the two disulfide-stabilized structures allowed us to derive a complete atomic model for the hexameric building block of the HIV-1 capsid.
Results and Discussion
Design of double cysteine mutants of HIV-1 CA
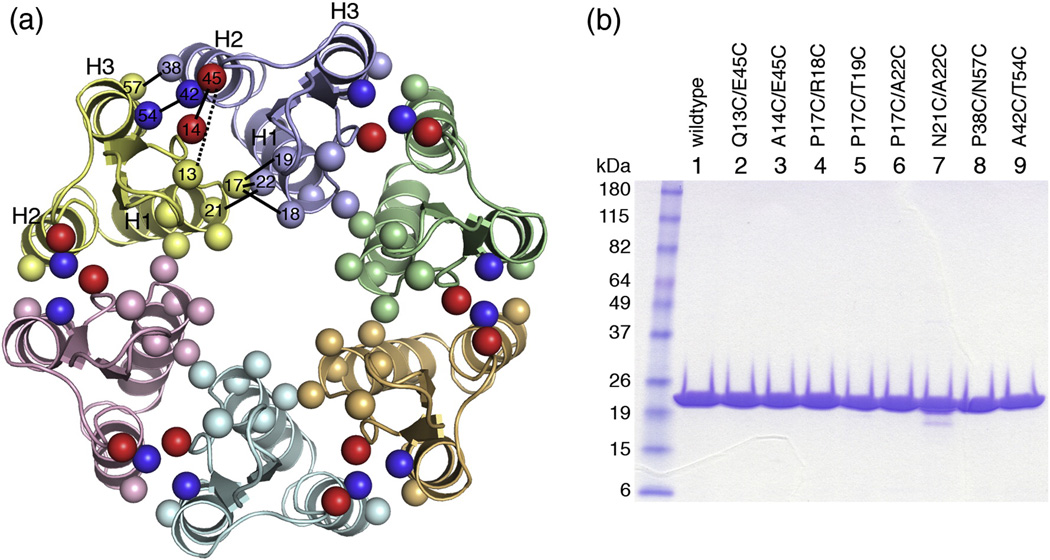
We used the 9-Å cryoEM-based model of the HIV-1 CA hexamer (PDB code 3dik)15 to identify residues that appeared to be in close contact across the hexamerization interface. We restricted our list of candidates to those within the NTD ring of the hexamer because this was the best-defined region in the cryoEM structure. Residues that appeared to mediate interactions between the NTD and CTD rings were also avoided in order to obtain an unbiased view of the molecular interactions in this region. The Cβ-Cβ distance in native disulfide bonds is ~4 Å.23,24 Since the cryoEM model only gave an approximate position for each residue, we used a simple distance filter and selected seven pairs of residues whose Cβ atoms were separated by ≤6 Å (listed in Table 1; amino acid positions in the cryoEM model are shown in Fig. 1a). As a negative control, we also mutated an eighth pair, Gln13/Glu45, which had a predicted Cβ-Cβ distance of 10.4 Å and would not be expected to form a disulfide bond. Two native cysteines in the CTD, Cys198 and Cys218, were left in place because the cryoEM model indicated that these residues were unlikely to interfere with the crosslinking reactions. All constructs could be expressed in milligram quantities Escherichia coli, could be purified to homogeneity (Fig. 1b), and were generally well behaved in reducing solution conditions.
Table 1.
Residue pairs selected for cysteine substitution
| Residue 1 | Residue 2 | Cβ-Cβ distances (Å) |
||
|---|---|---|---|---|
| cryoEMa | CC1b | CC2c | ||
| Gln13 | Glu45 | 10.45 | 8.93 | 9.42 |
| Ala14 | Glu45 | 5.16 | 3.65* | 4.21 |
| Pro17 | Arg18 | 5.47 | 5.56 | 5.35 |
| Pro17 | Thr19 | 5.16 | 4.56 | 5.31 |
| Pro17 | Ala22 | 5.93 | 4.64 | 4.69 |
| Asn21 | Ala22 | 5.78 | 6.71 | 6.37 |
| Pro38 | Asn57 | 4.20 | 3.85 | 3.73 |
| Ala42 | Thr54 | 4.51 | 4.68 | 3.91* |
Predicted distances in the cryoEM model; PDB code 3dik was rebuilt into a polyalanine model prior to measuring distances
Measured distances in the actual CC1 structure; PDB code 3h47
Measured distances in the actual CC2 structure; PDB code 3mge
Distances refer to disulfide-bonded cysteine substitutions
Figure 1.
Model-based design of disulfide-stabilized HIV-1 CA hexamers. (A) Top view ribbons representation of the 18-helix barrel comprising the NTD-NTD hexamerization interactions, as seen in the cryoEM model (3dik).15 Each subunit is colored differently. The locations of mutated residues are indicated by spheres, and are labeled within a single interface (between yellow and blue subunits). Pairs are connected by black lines (dashed for the Q13/E45 negative control). The A14/E45 and A42/T54 pairs are colored red and blue, respectively. (B) SDS-PAGE profile of purified HIV-1 CA double cysteine mutants. Ten µg protein was loaded in each lane. Molecular weight markers are labeled on the left.
Assembly and crosslinking of double cysteine CA mutants
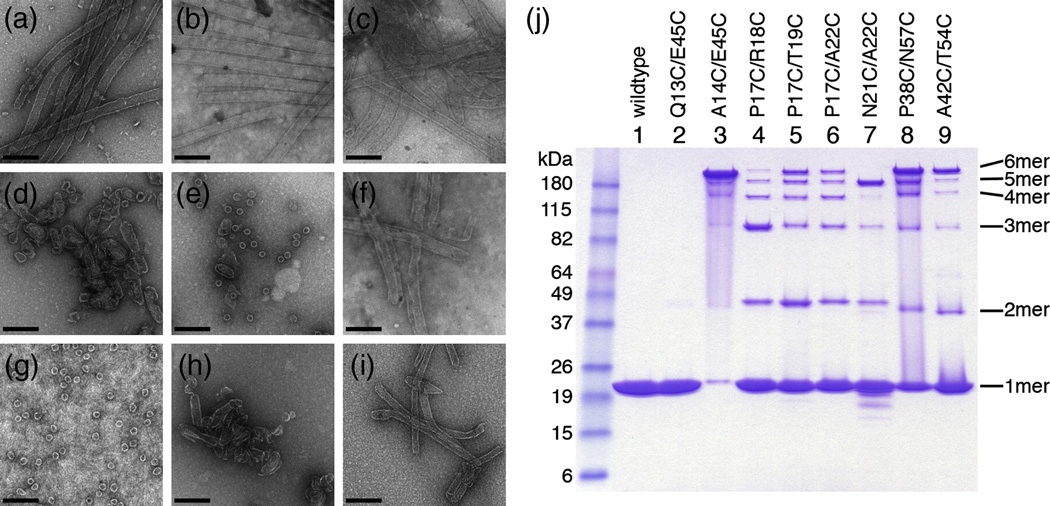
Pure recombinant wildtype HIV-1 CA protein spontaneously assembles into long, hollow tubes upon incubation in buffer containing 1 M NaCl at near neutral pH (Fig. 2a).14,25–28 These in vitro tubes are organized as helical arrays of CA hexamers,3,20 and a variety of studies from several groups have shown that the tubes faithfully recapitulate the hexameric lattice interactions within authentic capsids.12–14,27,29–31 The double cysteine mutants were therefore assembled in vitro to identify constructs that retained the ability to form tubes. Four mutants assembled into tubes that appeared similar in morphology to the wildtype tubes (Q13C/E45C, Fig. 2b; A14C/E45C, Fig. 2c; P17C/A22C, Fig. 2f; and A42C/T54C, Fig. 2i), implying that these pairs of engineered cysteines did not disrupt the native assembly interfaces. The remainder of the constructs formed highly curved particles (i.e., spheres, small rods, cones, and spirals) similar to the previously described R18A and R18L HIV-1 CA mutants.14,15 Note that these constructs contain mutations at or near Arg18 (Fig.1a and Table 1), and therefore the highly curved and closed particles presumably arose by the incorporation of pentamers into the assembling hexagonal lattices, in analogy to the R18A and R18L mutants.14 We speculate that the engineered cysteines may have induced pentamer incorporation by directly stabilizing the pentamer, and/or indirectly by increasing the curvature of the hexagonal lattice.
Figure 2.
Assembly and crosslinking of double cysteine CA mutants. (A–I) Representative negatively stained EM images of in vitro assembled (A) Wildtype HIV-1 CA, (B) Q13C/E45C, (C) A14C/E45C, (D) P17C/R18C, (E) P17C/T19C, (F) P17C/A22C, (G) N21C/A22C, (H) P38C/N57C, and (I) A42C/T54C. The assembly buffer contained 10 mg/mL protein in 50 mM Tris, pH 8, 1 M NaCl, 20 mM βME. Scale bar = 200 nm. (J) Non-reducing SDS-PAGE profiles of the assembly reactions. Molecular weight markers are labeled on the left, and the positions of crosslinked n-mers are indicated on the right.
Within the assembled tubes, if the cysteine pairs were at the appropriate distance and geometry then they should spontaneously oxidize and form disulfide bonds. Note that the assembly reactions were performed in the presence of a 50-fold molar excess of β-mercaptoethanol (βME, 20 mM) relative to the protein concentration (10 mg/mL or 0.4 mM). This highly reducing assembly environment discouraged formation of spuriously crosslinked species, and promoted disulfide bonding only if the local geometry was favorable. To assess disulfide bond formation, the assembled particles were denatured in the presence of a thiolcapping reagent (to prevent further crosslinking), and then analyzed by using non-reducing SDS-PAGE. The different constructs migrated as ladders of crosslinked n-mers, with n = 1–6 (Fig. 2j). Note that we observed little or no n > 6 species. This indicates that disulfide bond formation was being driven by the non-covalent intersubunit interactions within the assembled particles and not by random, non-specific association of soluble protein, which would lead to higher molecular weight aggregates.
As expected, the negative control, Q13C/E45C, migrated as a single band corresponding to the non-crosslinked monomer, consistent with the prediction based on the cryoEM model (Fig. 2j, lane 2). Of the constructs that assembled into wildtype-like tubes, A14C/E45C (Fig. 2j, lane 3) and A42C/T54C (lane 9) had enriched 6-mer bands, indicating that these cysteine pairs were within favorable disulfide bonding distance. A14C/E45C crosslinked with at least 90% efficiency and was therefore selected for further analysis (see below). The N21C/A22C construct was a notable outlier: its SDS-PAGE profile showed an enriched 5-mer band, with essentially no 6-mer band (Fig. 2j, lane 7). This construct assembled predominantly into spheres approximately 35 nm in diameter (Fig. 2g), which matches the expected dimensions of a T=3 icosahedral particle composed of 20 hexamers and 12 pentamers. Although the N21C/A22C spheres lacked strict icosahedral symmetry, this result supports the finding that retroviral CA hexamers and pentamers are quasi-equivalent, i.e., the same (or overlapping) protein surfaces are used to create both capsomers.22 Substituting Asn21 and Ala22 with cysteines apparently created an artificial switch that shifted the equilibrium between the two capsomers in favor of the pentamer. We speculate that within the N21C/A22C particles, the engineered cysteines may be partly oxidized in the hexamers and completely oxidized in the pentamers.
Characterization of CA A14C/E45C hexamers
We first focused on the A14C/E45C construct, because it assembled into wildtype-like tubes and crosslinked efficiently into 6-mers. A series of follow-up experiments showed that: (1) the in vitro crosslinking efficiency of A14C/E45C could be driven essentially to completion by performing the assembly reactions in stoichiometric amounts of βME (data not shown; but see Fig. 3b); (2) the projection structure of two-dimensional crystals of the crosslinked hexamer was essentially identical to the wildtype lattice at 15 Å resolution (data not shown); (3) the crosslinked tubes bound to human cyclophilin A, a known interactor of the HIV-1 capsid32 (data not shown); and (4) a proviral clone containing the two cysteine substitutions produced mature virions and cores, albeit non-infectious (D. Christensen and W.I. Sundquist, personal communication). Taken together, these results indicated that the engineered disulfide bond stabilized the A14C/E45C hexamer both in vitro and in authentic virions, and did not disrupt the native architecture or surface properties of the hexameric capsid lattice.
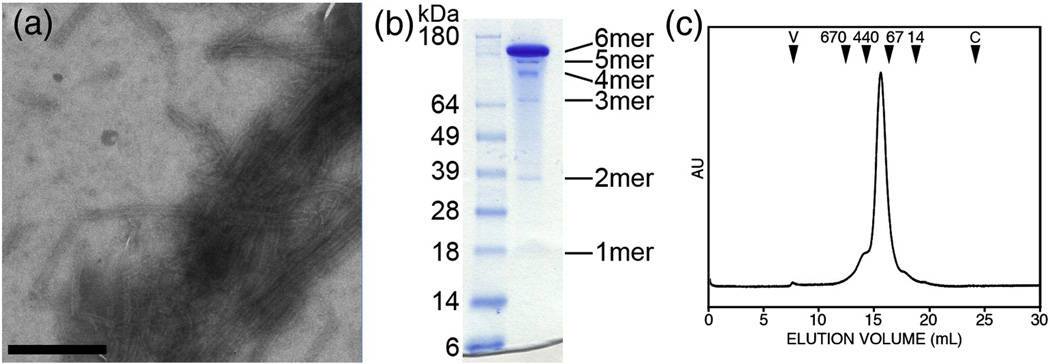
Figure 3.
Assembly and analysis of A14C/E45C/W184A/M185A (CC1 construct). (A) Negatively-stained EM image of tubes assembled by dialyzing 30 mg/mL protein in 50 mM Tris, pH 8, 1 M NaCl, 2 mM βME. Scale bar = 500 nm. (B) Non-reducing SDS-PAGE profile of crosslinked soluble CC1 hexamers. Molecular weight markers are labeled on the left, and the expected positions of crosslinked n-mers are indicated on the right. (C) Size exclusion chromatographic profile of soluble crosslinked CC1. The elution volumes of protein standards are indicated by black arrowheads (numbers indicate molecular weight in kDa, V = void volume, C = column volume). The A42C/T54C/W184A/M185A mutant (CC2 construct) behaved similarly, but only crosslinked into hexamers with ~50% efficiency (data not shown).
Tubes composed of crosslinked CA A14C/E45C were hyperstable
Having identified a pair of cysteines that efficiently crosslinked CA hexamers, we expected that the crosslinked tubes would now be hyperstable compared to wildtype tubes. Indeed, intact crosslinked tubes were observed even after 10,000-fold dilution of assembly reactions into water, from 0.4 mM to 40 nM CA, or after extended incubation with cyclophilin A (data not shown). Under these conditions, wildtype CA tubes rapidly disintegrate. The A14C/E45C construct also assembled readily even at low nanomolar concentrations (data not shown). Thus, it was not possible to disassemble the crosslinked tubes and crystallize the A14C/E45C hexamers. We note, however, that the enhanced stability of the crosslinked tubes makes them attractive reagents for biochemical studies of capsid-binding factors such as cyclophilin A32 and TRIM5α,33 which tend to induce rapid disassembly of wildtype CA assemblies.
Addition of dimer interface mutations to A14C/E45C gave soluble hexamers
CA hexamers are linked into an extended lattice by homodimerization of the CTD, via a hydrophobic interface that includes helix 9.9,13,14,29 To obtain discrete, soluble A14C/E45C hexamers for crystallization, we disrupted the CTD-CTD linkages by introducing the W184A and M185A mutations into helix 9, thereby preventing further polymerization of the crosslinked hexamers into tubes or other capsid-like particles. Because the CTD-CTD interface does not directly stabilize the hexamer, the W184A and M185A mutations were not expected to affect formation of the crosslinked A14C/E45C hexamers.
We tested the W184A and M185A mutations singly and in combination. In contrast to the wildtype background, addition of these mutations to A14C/E45C produced constructs that remained competent for assembly. The A14C/E45C/W184A construct assembled into mixtures of tubes and flat sheets, which crosslinked into 6-mers with ~100% efficiency (data not shown). Many of these assemblies were single-layered (data not shown), suggesting that the W184A mutation may have altered the native curvature of the hexameric lattice, by changing the packing geometry at the dimerization interface. The A14C/E45C/M185A mutant was prone to aggregation (but also assembled; data not shown) and was therefore not analyzed further.
The A14C/E45C/W184A/M185A construct (hereinafter referred to as CC1) also formed tubes (Fig. 3a), but with considerably lower efficiency. Specifically, CC1 tubes were observed only when the protein concentration was at least 30 mg/mL (0.8 mM), and we also observed more amorphous particles. Tubes were more prevalent when the assembly reactions contained lower amounts of βME (Fig. 3a). Of greater importance, CC1 crosslinked into 6-mers with almost 100% efficiency in the presence of stoichiometric amounts of βME (Fig. 3b), even at protein concentrations wherein tubes were not readily observed. This result indicated that the protein was forming soluble hexamers. Indeed, size exclusion chromatography of crosslinked CC1 (solubilized in low salt buffer) displayed a nearly symmetric peak centered at the appropriate elution volume for a hexamer (Fig. 3c).
Structure of crosslinked HIV-1 CA A14C/E45C/W184A/M185A (CC1)
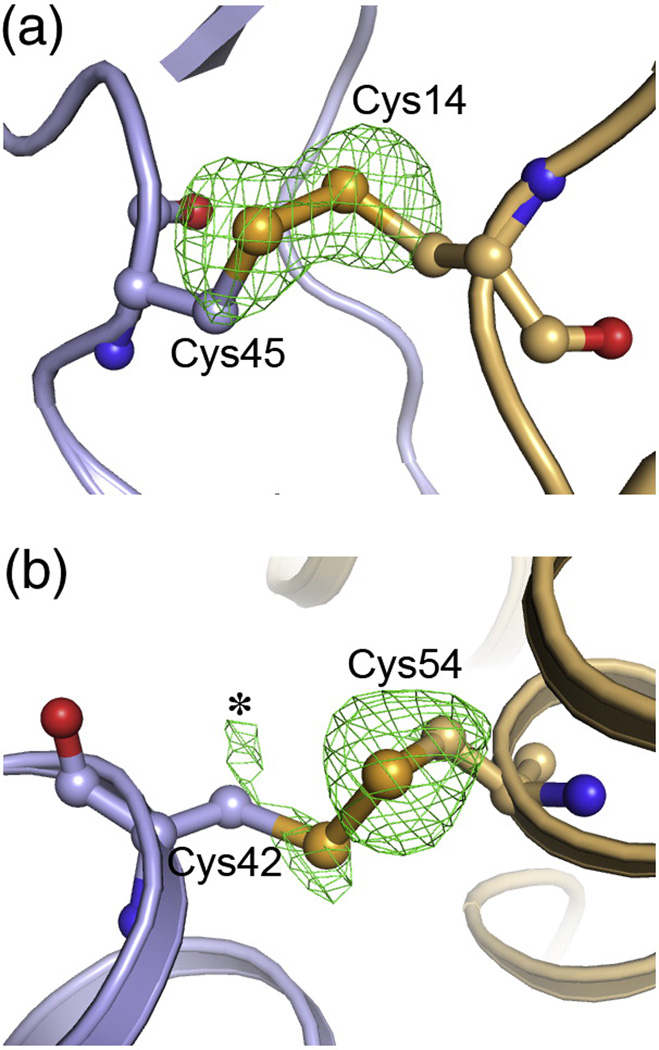
Unlike wildtype HIV-1 CA, CC1 readily crystallized under a variety of conditions. Two crystal forms diffracted to atomic resolution: a hexagonal form (~2 Å resolution), and an orthorhombic form (~2.7 Å).21 In the orthorhombic crystal form, the hexamers packed in a biologically non-relevant configuration, with two independent hexamers stacked head-to-head in the asymmetric unit. In the hexagonal crystal form, the hexamers packed on a flat hexagonal lattice,21 similar to the two-dimensional crystals solved by cryoEM.15 However, the 3D and 2D hexagonal lattices were not exactly matched, likely due to the presence of the CTD-CTD interface mutations in CC1. The crystallographically distinct CC1 hexamers superimposed very well with each other,21 with the cryoEM structure,15 and also with a non-crosslinked CA hexamer structure that was stabilized by fusion with CcmK4, a protein that forms stable hexamers in solution.21,34 Taken together with our biochemical data, the above analysis strongly indicates that the crosslinked construct captured the correct, native hexamer conformation. The engineered Cys14/Cys45 disulfide bond adopted a left-handed configuration, with favorable dihedral angles. Densities for the sulfur atoms indicated a single configuration for both cysteines, i.e., they were ~100% disulfide bonded in the crystal (Fig. 4a). Thus, the X-ray structures of CC1 support the conclusion from our biochemical analyses, that the engineered Cys14/Cys45 disulfide bond was comfortably accommodated within the hexamerization interface.
Figure 4.
Conformations of the engineered disulfide bonds. (A) Cys14 and Cys45 in the CC1 construct, shown in ball-and-stick representation, with the rest of the structure in ribbons. Green mesh show omit mFo-DFc densities for the sulfur atoms contoured at 3σ. The omit map was obtained by setting the sulfur occupancies to zero, introducing random shifts to the remaining atoms (mean displacement = 0.5 Å), and refining the resulting model with simulated annealing. Note that the sulfur Cys14 and Cys45 sulfur density peaks have equal magnitude and are ~100% oxidized. (B) Disulfide-bonded conformation of Cys42 and Cys54 in the CC2 construct. Note that the sulfur density for Cys54 is clear, whereas Cys42 is weaker and more diffuse. The asterisk indicates density ascribed to an alternative rotamer configuration for Cys42, which is not disulfide-bonded.
Structure of crosslinked HIV-1 CA A42C/T54C/W184A/M185A (CC2)
Although we initially focused on a construct with very favorable properties, we also wondered whether one that crosslinked less efficiently still formed the correct hexamer structure. We therefore revisited the A42C/T54C mutant for further analysis: this construct displayed a ladder of 1 to 6-mers on SDS-PAGE (Fig. 2j, lane 9) and yet retained the ability to assemble into tubes (Fig. 2i). The crosslinked 6-mer band only represented ~30% of the total protein (Fig. 2j, lane 9), and maximal crosslinking was limited to ~50% even in the presence of stoichiometric amounts of βME (data not shown). On the basis of these observations, one might conclude that oxidation of the Cys42/Cys54 pair distorted the hexamer architecture or that the disulfide bond was not comfortably accommodated among the native contacts.
As for the CC1 construct, soluble A42C/T54C assemblies were obtained by the addition of the W184A and M185A mutations to the CTD; the A42C/T54C/W184A/M185A construct is hereinafter referred to as CC2. This construct also crystallized readily, but unlike CC1, all CC2 crystals had hexagonal morphology. Analysis of the diffraction data indicated that the CC2 crystals were isomorphous to the hexagonal crystal form of CC1. Accordingly, the CC2 diffraction data were reduced in space group P6 with one CA molecule in the asymmetric unit (see Materials and Methods for details). The structure was solved by molecular replacement, and refined at 1.9 Å resolution to Rfree = 25.5% (crystallographic statistics are presented in Table 3).
The refined model showed that the Cys42 and Cys54 sidechains were only partly disulfide-bonded. Consistent with the attenuated crosslinking efficiency for this construct, the difference density peak for the Cys54 sulfur atom indicated a single conformation, whereas peaks for the Cys42 sulfur atom indicated at least two alternative rotamer configurations (Fig. 4b). Nevertheless, the CC2 structure was practically identical to CC1, demonstrating that inefficient crosslinking of engineered disulfides does not necessarily imply distortion of the native protein structure.
The CC1 and CC2 hexamer structures superimposed with an average root mean squared deviation of 0.40 Å on equivalent backbone atoms (Fig. 5a). Sidechain conformations were also very similar across both structures, although certain residues appeared to have alternative rotamer configurations. At the positions occupied by the engineered cysteines, linear atomic displacements ranged from 0 to ~1 Å for backbone and Cβ atoms (Fig. 5b). These minor displacements appeared to be accommodated by torsion angle adjustments that were dispersed across the surrounding residues. The cysteine mutations also perturbed the configurations of ordered water molecules that mediate inter-subunit hydrogen bonds across the subunits in the NTD ring, although the perturbations were highly localized (not shown).
Figure 5.
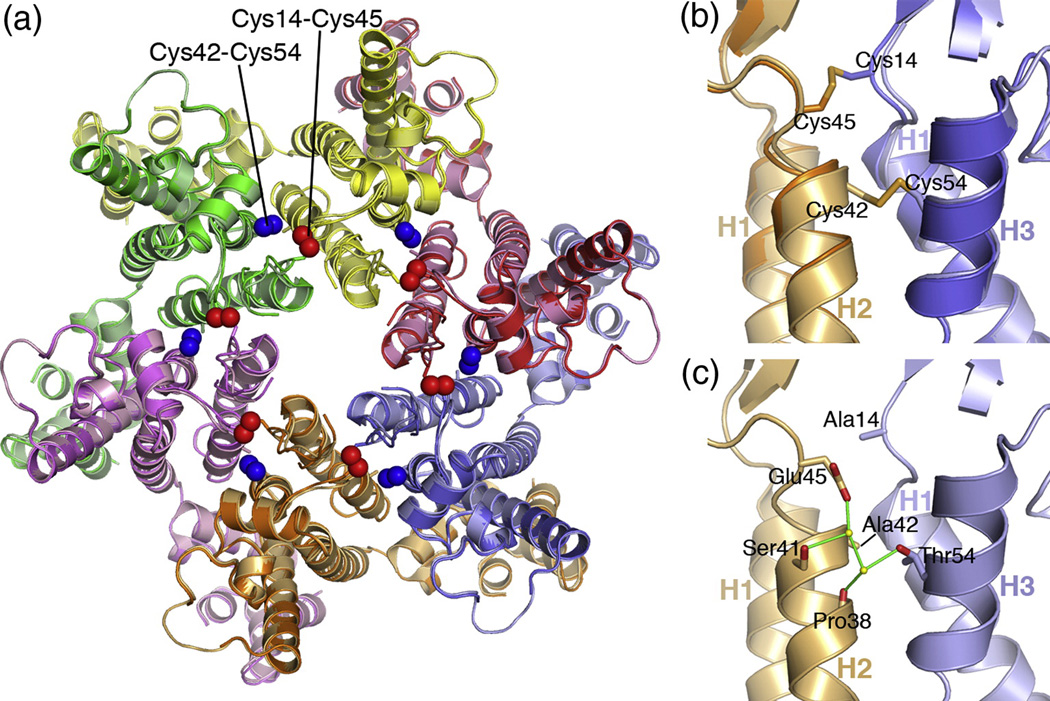
Composite structure of the HIV-1 CA hexamer. (A) Superposition of the CC1 and CC2 hexamer structures viewed as would be seen from the outside of the capsid. Each subunit is colored differently, with CC1 is brighter shades and CC2 in muted shades. The engineered Cys14/Cys45 and Cys42/Cys54 disulfide bonds are indicated by the red and blue spheres, respectively. (B) Close-up side view of the superposition in the vicinity of the engineered disulfides. (C) View of the composite hexamer interface model, in the same orientation as (B). Sidechains for Ala14, Ala42, Glu45, and Thr54 are modeled in their native conformations. A subset of ordered water molecules within the interface are shown as yellow spheres. Putative hydrogen bonds are shown by green lines.
Building a “native” CA hexamer structure
Since the CC1 and CC2 structures were very similar, we derived a composite hexamer model with a native set of NTD ring interactions in the following manner: (1) a single CC1 NTD subunit was superimposed on each of the six CC2 subunits on their backbone atoms; (2) the coordinates of the native Ala42 and Thr54 sidechains in CC1 were used to replace the Cys42 and Cys54 coordinates in CC2; (3) the water positions in the immediate vicinity of the mutated residues were individually inspected, and those that clashed with protein atoms were excluded from the model. Although this modeling approach was somewhat ad hoc, the resulting model had appropriate van der Waals distances, minimal steric clashes, and favorable hydrogen bonding distances between polar atoms, including water molecules. Interestingly, the composite model suggested that the Glu45 and Thr54 sidechains may interact across the hexamerization interface, via water-mediated hydrogen bonds (Fig. 5c). This feature was not observable in either of the CC1 or CC2 structures, and is consistent with studies indicating that both residues are required for optimal capsid assembly and virus infectivity.13,14,35
Significance for β-hairpin formation
In addition to forming the mature capsid, CA also forms essential lattice contacts within the immature capsid, as a domain of the Gag polyprotein. During virus maturation, CA is released as a mature protein through proteolysis of Gag by the viral protease. The processed Pro1 imino group folds back into the globular domain of mature CA, where it forms a buried salt bridge with the sidechain carboxyl group of Asp51.10,29 In the immature form, the Pro1-Asp51 salt bridge does not exist, and the β-hairpin is unfolded into an extended conformation.36,37 Biochemical and mutagenesis studies have shown that formation of the β-hairpin is a structural switch that facilitates disassembly of Gag and/or assembly of CA into the mature capsid.29,38
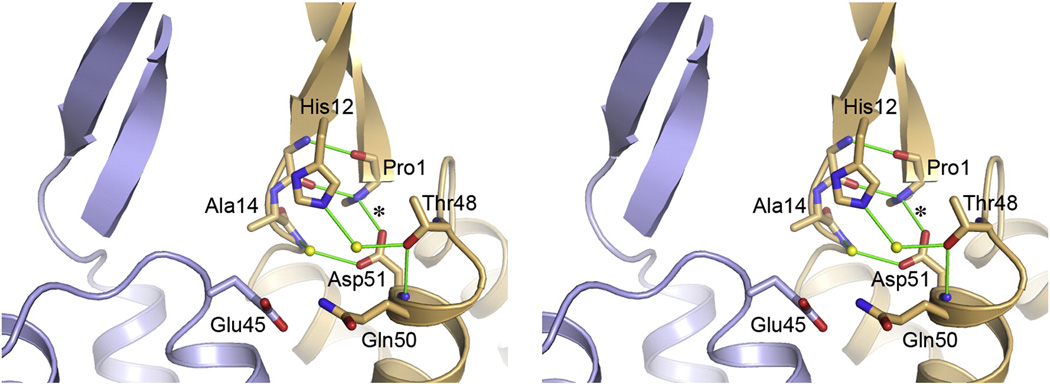
The Pro1-Asp51 salt bridge was observed in each of the 14 crystallographically independent subunits within the CC1 and CC2 hexamers, and this is consistent with the observation that a folded β-hairpin is an essential feature of mature CA.10,29 Both residues form part of a network of intramolecular hydrophilic interactions that hold together the bottom of the β-hairpin and the tops of helices 1 and 3 (Fig. 6). Specifically, the imino group of Pro1 makes an additional hydrogen bond with the backbone carbonyl of Glu13 from the opposite strand of the hairpin, and Asp51 forms a water-mediated hydrogen bond with the backbone amide of Ile15 (Fig. 6). His12 from the second strand also forms a water-mediated hydrogen bond with Thr48 (helix 3). However, there are no significant intermolecular interactions in this region, although it is conceivable that Glu45 from the adjacent subunit may participate in the hydrogen bond network if its sidechain adopts an alternative rotamer configuration. Likewise, residues in the strand regions do not form intermolecular β-barrel-type interactions. This lack of significant long-range interactions involving the β-hairpin has also been observed in the structure of the isolated CA NTD hexamer structure of murine leukemia virus,5 and is consistent with the biochemical finding that deletion of the first 13 residues of HIV-1 CA does not abolish assembly in vitro (although assembly is less efficient compared to full-length protein).38 Taken together, these observations support the model wherein formation of the β-hairpin indirectly promotes assembly of the mature capsid by inducing a CA tertiary conformation that is no longer capable of packing into the immature lattice.5,37
Figure 6.
Stereoview of the β-hairpin region between two adjacent subunits. Details of the hydrophilic interactions surrounding the Pro1-Asp51 salt bridge (asterisk) are shown for one subunit, as seen in the composite hexamer model. Similar arrangements were observed each of the crystallographically independent CC1 and CC2 hexamer structures. Note that there are essentially no intermolecular interactions involving either the salt bridge or strand regions of the β-hairpin.
Generality of the structure determination approach
There is increasing literature on the use of engineered disulfide bonds to stabilize discrete complexes to facilitate crystallization and obtain structures at atomic resolution. One notable example is the elongation complex of the HIV-1 reverse transcriptase enzyme, which was successfully crystallized only after a disulfide bond was used to tether the enzyme to its DNA:RNA hybrid substrate.39,40 Our studies demonstrate that a similar approach may also be applied to very large supramolecular complexes (e.g., the HIV-1 capsid). By combining hexamer-stabilizing disulfide bonds with mutations that disrupt formation of hexamer-to-hexamer linkages in the capsid lattice, we solubilized and crystallized a sub-complex composed of CA hexamers. Structures of different sub-complexes may be solved iteratively in this manner, and then combined into a model for the entire supramolecular complex. In the case of the HIV-1 capsid, structures of the hexameric building block [(21) and this study] and of the CTD-CTD dimer connectors are now available,9,18,20 but deriving a complete atomic model for the capsid would require the structure of the CA pentamer.
Two important observations imply that the above technique may be broadly applied. Firstly, we found that engineered disulfides need not oxidize with high efficiency to stabilize functional complexes. Although we believe this does not obviate the need for careful selection of sites for cysteine mutagenesis, it implies that the predicted geometry of the engineered disulfide is not a critical criterion for site selection. Secondly, the correctness of disulfide-stabilized structures may be assessed by comparing independent structures of different crosslinked constructs that have the engineered cysteine pairs at different positions. We argue that if they converge on a single conformation, then the disulfide-stabilized structures very likely recapitulate the native conformation. This type of validation is particularly crucial in cases wherein reliable low-resolution structures or other supporting biochemical data are scarce or unavailable. An additional advantage of this approach is that each of the independent structures “fill-in” the information missing from the others, and one can then combine the non-mutated portions of the structures to obtain a “native” model.
Materials and Methods
Protein purification
Plasmid constructs were made as described previously.21 HIV-1 CA proteins were expressed by IPTG induction in E. coli BL21(DE3) cells for 6–12 hrs at 25 °C. All purification steps were performed at 4 °C. Cell pellets were resuspended in 50 mM Tris, pH 8, 50 mM NaCl, 200 mM βME, 0.2% (w/v) deoxycholate, supplemented with protease inhibitors, and lysed using a microfluidizer. After centrifugation at 45,000 g for 45 min, ammonium sulfate was added to the clarified supernatant to 25% saturation. The precipitate was pelleted by centrifugation at 8,000 g for 20 min, resuspended in Buffer A (25 mM MOPS, pH 6.8, 20–200 mM βME), and applied to a SP-Sepharose column (GE Healthcare). The column was developed with a linear gradient from 0 to 1 M NaCl in Buffer A. Peak fractions were pooled, dialyzed against 20 mM Tris, pH 8, 40 mM NaCl, 60 mM βME, and applied to a Q-Sepharose column (GE Healthcare). Homogeneously pure CA eluted in the flow-through, was concentrated to 30 mg/mL, flash-frozen in liquid nitrogen, and stored at −80 °C.
In vitro assembly and crosslinking
In initial experiments, CA constructs were assembled by overnight dialysis at 4 °C of 10 mg/mL protein into assembly buffer (1 M NaCl, 50 mM Tris, pH 8),27 containing 20 mM βME. In subsequent experiments, we found that varying the βME concentration from 0–200 mM had no significant effect on the assembly efficiency of the various constructs (data not shown). Assembly was also performed by direct dilution of protein into assembly buffer,14,29 which generally resulted in more highly curved particles (data not shown). To visualize crosslinked proteins, assembly reactions were diluted into buffer containing 2–40 mM methyl methanethiolsulfonate, as appropriate to the protein and βME concentrations, and separated by using non-reducing SDS-PAGE with Coommassie staining. For EM, assembly reactions were diluted 10-fold and a 5 µL aliquot was applied to a carbon-coated grid for 5 min. The grid was washed with 0.1 M KCl, and then stained with 2% uranyl acetate. Images were recorded on a Tecnai F20 electron microscope operating at 120 keV.
Crystallization and structure determination
Soluble crosslinked hexamers were prepared as described previously.21 Crystals were obtained in sitting drops, by mixing protein and precipitant at a 2:1 ratio. As described previously, CC1 formed orthorhombic crystals at 20 °C in 10–12% PEG 8,000, 100 mM Tris (pH 7–9) and hexagonal crystals at 4 °C in 10–12% PEG 8,000, 100 mM sodium malonate (pH 6.5–7.5).21 Subsequent screening identified conditions that more reproducibly induced crystallization of the hexagonal form (10–14% PEG 8,000, 2% Tacsimate, 100 mM Tris, pH 7.6–8.6). Surprisingly, CC2 formed only hexagonal crystals under all three conditions.
Structure determination of CC1 crystals have been described in detail.21 For CC2 crystals, diffraction data were recorded at beamline 5.0.1 at the Advanced Light Source and processed with HKL2000.41 The crystals belonged to space group P3 (a = b = 156.4 Å, c = 56.5 Å), with one hexamer in the asymmetric unit, and were perfectly merohedrally twinned (Supplementary Table 1). Furthermore, the native Patterson map showed a strong off-origin peak at fractional unit cell coordinates (1/3, 2/3, 0) (70% of origin), and the diffraction data showed a characteristic pattern of strong and weak reflections, which indicated the presence of translational pseudosymmetry. The strong reflections obeyed the selection rule (h, h ± 3n, l) and on average had ~8-fold higher intensities than the weak reflections (Supplementary Fig. 1). The off-origin Patternson peak had equal magnitude to the origin peak when calculated with only the strong subset. As described previously for the isomorphous CC1 crystals, we therefore ignored the weak reflections and indexed the data in the smaller pseudo-cell (a = b = 90.3 Å, c = 56.5 Å, Rsym = 7.5% to 1.9 Å; Table 3), which belonged to space group P6 with one CA molecule in the asymmetric unit.21
Prior to refinement, the CC2 test set was matched to the published test set for the CC1 structure (PDB code 3h47). The CC1 structure (minus water and sidechains for Ala14, Cys42, Glu45, and Cys54) was used as the starting point for the refinement. The model-phased map after rigid body refinement clearly showed positive difference density for the Cys42 and Cys54 sulfur atoms and omitted water molecules. The entire model was rebuilt with COOT,42 and multiple rounds of refinement with simulated annealing were performed with PHENIX.43 The current model has Rwork and Rfree values of 23.0% and 25.5%, respectively, and the Ramachandran plot shows no regions in disallowed regions (Table 3). The CC2 structure factors and coordinates are available for download at the PDB database (http://www.rcsb.org) as 3mge.
The structure in the pseudo-cell setting is effectively an “average” of both the pseudotranslationally related molecules and the two twin domains. Further refinement in the true cell setting of P3 was not attempted, because the combination of translational pseudosymmetry and twinning produced severe intensity correlations in the data. The expectation, however, is that the atomic model would change minimally if we were to do so, based on published examples.
PDB accession numbers
The atomic coordinates and observed structure factors of CC2 have been deposited in the Research Collaboratory for Structural Bioinformatics PDB with code 3MGE
Supplementary Material
Table 2.
Crystallographic statistics for CC2
| Data collection | |
| Beam line | ALS 5.0.1 |
| Space group | P6 |
| Cell dimensions | a = b = 90.3 Å, c = 56.5 Å |
| α = β = 90°, γ = 120° | |
| Resolution range, Å | 50-1.90 (1.97-1.90) |
| Rsym, % | 7.5 (53.3) |
| Mean <I>/σ<I> | 27.9 (3.6) |
| Completeness, % | 99.9 (100) |
| Average redundancy | 11.2 (11.2) |
| Average mosaicity, ° | 0.46 |
| Wilson B-factor, Å2 | 23.6 |
| Refinement | |
| Resolution range | 35-1.9 (2.0-1.9) |
| No. of unique reflections | 20,802 (2,774) |
| Reflections in free set | 1,022 (143) |
| Rwork, % | 23.0 (23.6) |
| Rfree, % | 25.5 (27.9) |
| No. of nonhydrogen atoms | |
| protein | 1,606 |
| solvent | 141 |
| Average B-factor, Å2 | |
| protein | 29.5 |
| solvent | 37.6 |
| Coordinate deviations | |
| bond lengths, Å | 0.908 |
| bond angles, ° | 0.006 |
| Ramachandran plot | |
| favorable | 0.99 |
| allowed | 0.01 |
| disallowed | 0 |
| PDB ID | 3mge |
Values in parenthesis are for the highest resolution shell.
Acknowledgements
We thank W.I. Sundquist (U. of Utah) for sharing unpublished data. Electron microscopy experiments were conducted at the National Resource for Automated Molecular Microscopy (NRAMM), which is supported by the National Institutes of Health (NIH) through the National Center for Research Resources’ P41 program (RR17573). Diffraction data were collected at the Advanced Light Source, which is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. This study was funded by NIH grants to M.Y. (R01-GM066087 and P50-GM082545).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ganser-Pornillos BK, Yeager M, Sundquist WI. The structural biology of HIV assembly. Curr Opin Struct Biol. 2008;18:203–217. doi: 10.1016/j.sbi.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jin Z, Jin L, Peterson DL, Lawson CL. Model for lentivirus capsid core assembly based on crystal dimers of EIAV p26. J Mol Biol. 1999;286:83–93. doi: 10.1006/jmbi.1998.2443. [DOI] [PubMed] [Google Scholar]
- 3.Li S, Hill CP, Sundquist WI, Finch JT. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature. 2000;407:409–413. doi: 10.1038/35030177. [DOI] [PubMed] [Google Scholar]
- 4.Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI. Assembly and analysis of conical models for the HIV-1 core. Science. 1999;283:80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- 5.Mortuza GB, Haire LF, Stevens A, Smerdon SJ, Stoye JP, Taylor IA. High-resolution structure of a retroviral capsid hexameric amino-terminal domain. Nature. 2004;431:481–485. doi: 10.1038/nature02915. [DOI] [PubMed] [Google Scholar]
- 6.Cornilescu CC, Bouamr F, Yao X, Carter C, Tjandra N. Structural analysis of the N-terminal domain of the human T-cell leukemia virus capsid protein. J Mol Biol. 2001;306:783–797. doi: 10.1006/jmbi.2000.4395. [DOI] [PubMed] [Google Scholar]
- 7.Campos-Olivas R, Newman JL, Summers MF. Solution structure and dynamics of the Rous sarcoma virus capsid protein and comparison with capsid proteins of other retroviruses. J Mol Biol. 2000;296:633–649. doi: 10.1006/jmbi.1999.3475. [DOI] [PubMed] [Google Scholar]
- 8.Khorasanizadeh S, Campos-Olivas R, Summers MF. Solution structure of the capsid protein from the human T-cell leukemia virus type-I. J Mol Biol. 1999;291:491–505. doi: 10.1006/jmbi.1999.2986. [DOI] [PubMed] [Google Scholar]
- 9.Gamble TR, Yoo S, Vajdos FF, von Schwedler UK, Worthylake DK, Wang H, McCutcheon JP, Sundquist WI, Hill CP. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science. 1997;278:849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- 10.Gitti RK, Lee BM, Walker J, Summers MF, Yoo S, Sundquist WI. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science. 1996;273:231–235. doi: 10.1126/science.273.5272.231. [DOI] [PubMed] [Google Scholar]
- 11.Macek P, Chmelik J, Krizova I, Kaderavek P, Padrta P, Zidek L, Wildova M, Hadravova R, Chaloupkova R, Pichova I, Ruml T, Rumlova M, Sklenar V. NMR structure of the N-terminal domain of capsid protein from the Mason-Pfizer monkey virus. J Mol Biol. 2009;392:100–114. doi: 10.1016/j.jmb.2009.06.029. [DOI] [PubMed] [Google Scholar]
- 12.Lanman J, Lam TT, Barnes S, Sakalian M, Emmett MR, Marshall AG, Prevelige PE., Jr Identification of novel interactions in HIV-1 capsid protein assembly by high-resolution mass spectrometry. J Mol Biol. 2003;325:759–772. doi: 10.1016/s0022-2836(02)01245-7. [DOI] [PubMed] [Google Scholar]
- 13.von Schwedler UK, Stray KM, Garrus JE, Sundquist WI. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J Virol. 2003;77:5439–5450. doi: 10.1128/JVI.77.9.5439-5450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ganser-Pornillos BK, von Schwedler UK, Stray KM, Aiken C, Sundquist WI. Assembly properties of the human immunodeficiency virus type 1 CA protein. J Virol. 2004;78:2545–2552. doi: 10.1128/JVI.78.5.2545-2552.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ganser-Pornillos BK, Cheng A, Yeager M. Structure of full-length HIV-1 CA: a model for the mature capsid lattice. Cell. 2007;131:70–79. doi: 10.1016/j.cell.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 16.Mayo K, Huseby D, McDermott J, Arvidson B, Finlay L, Barklis E. Retrovirus capsid protein assembly arrangements. J Mol Biol. 2003;325:225–237. doi: 10.1016/s0022-2836(02)01176-2. [DOI] [PubMed] [Google Scholar]
- 17.Mayo K, Vana ML, McDermott J, Huseby D, Leis J, Barklis E. Analysis of Rous sarcoma virus capsid protein variants assembled on lipid monolayers. J Mol Biol. 2002;316:667–678. doi: 10.1006/jmbi.2001.5354. [DOI] [PubMed] [Google Scholar]
- 18.Worthylake DK, Wang H, Yoo S, Sundquist WI, Hill CP. Structures of the HIV-1 capsid protein dimerization domain at 2.6 Å resolution. Acta Crystallogr D Biol Crystallogr. 1999;55:85–92. doi: 10.1107/S0907444998007689. [DOI] [PubMed] [Google Scholar]
- 19.Bailey GD, Hyun JK, Mitra AK, Kingston RL. Proton-linked dimerization of a retroviral capsid protein initiates capsid assembly. Structure. 2009;17:737–748. doi: 10.1016/j.str.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 20.Byeon IJ, Meng X, Jung J, Zhao G, Yang R, Ahn J, Shi J, Concel J, Aiken C, Zhang P, Gronenborn AM. Structural convergence between Cryo-EM and NMR reveals intersubunit interactions critical for HIV-1 capsid function. Cell. 2009;139:780–790. doi: 10.1016/j.cell.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pornillos O, Ganser-Pornillos BK, Kelly BN, Hua Y, Whitby FG, Stout CD, Sundquist WI, Hill CP, Yeager M. X-ray structures of the hexameric building block of the HIV capsid. Cell. 2009;137:1282–1292. doi: 10.1016/j.cell.2009.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cardone G, Purdy JG, Cheng N, Craven RC, Steven AC. Visualization of a missing link in retrovirus capsid assembly. Nature. 2009;457:694–698. doi: 10.1038/nature07724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petersen MT, Jonson PH, Petersen SB. Amino acid neighbours and detailed conformational analysis of cysteines in proteins. Protein Eng. 1999;12:535–548. doi: 10.1093/protein/12.7.535. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt B, Ho L, Hogg PJ. Allosteric disulfide bonds. Biochemistry. 2006;45:7429–7433. doi: 10.1021/bi0603064. [DOI] [PubMed] [Google Scholar]
- 25.Ehrlich LS, Agresta BE, Carter CA. Assembly of recombinant human immunodeficiency virus type 1 capsid protein in vitro. J Virol. 1992;66:4874–4883. doi: 10.1128/jvi.66.8.4874-4883.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campbell S, Vogt VM. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J Virol. 1995;69:6487–6497. doi: 10.1128/jvi.69.10.6487-6497.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gross I, Hohenberg H, Kräusslich HG. In vitro assembly properties of purified bacterially expressed capsid proteins of human immunodeficiency virus. Eur J Biochem. 1997;249:592–600. doi: 10.1111/j.1432-1033.1997.t01-1-00592.x. [DOI] [PubMed] [Google Scholar]
- 28.Barklis E, Alfadhli A, McQuaw C, Yalamuri S, Still A, Barklis RL, Kukull B, Lopez CS. Characterization of the in vitro HIV-1 capsid assembly pathway. J Mol Biol. 2009;387:376–389. doi: 10.1016/j.jmb.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.von Schwedler UK, Stemmler TL, Klishko VY, Li S, Albertine KH, Davis DR, Sundquist WI. Proteolytic refolding of the HIV-1 capsid protein aminoterminus facilitates viral core assembly. EMBO J. 1998;17:1555–1568. doi: 10.1093/emboj/17.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Briggs JA, Wilk T, Welker R, Kräusslich HG, Fuller SD. Structural organization of authentic, mature HIV-1 virions and cores. EMBO J. 2003;22:1707–1715. doi: 10.1093/emboj/cdg143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lanman J, Lam TT, Emmett MR, Marshall AG, Sakalian M, Prevelige PE., Jr Key interactions in HIV-1 maturation identified by hydrogen-deuterium exchange. Nat Struct Mol Biol. 2004;11:676–677. doi: 10.1038/nsmb790. [DOI] [PubMed] [Google Scholar]
- 32.Yoo S, Myszka DG, Yeh C, McMurray M, Hill CP, Sundquist WI. Molecular recognition in the HIV-1 capsid/cyclophilin A complex. J Mol Biol. 1997;269:780–795. doi: 10.1006/jmbi.1997.1051. [DOI] [PubMed] [Google Scholar]
- 33.Stremlau M, Perron M, Lee M, Li Y, Song B, Javanbakht H, Diaz-Griffero F, Anderson DJ, Sundquist WI, Sodroski J. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5α restriction factor. Proc Natl Acad Sci U S A. 2006;103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kerfeld CA, Sawaya MR, Tanaka S, Nguyen CV, Phillips M, Beeby M, Yeates TO. Protein structures forming the shell of primitive bacterial organelles. Science. 2005;309:936–938. doi: 10.1126/science.1113397. [DOI] [PubMed] [Google Scholar]
- 35.Forshey BM, von Schwedler U, Sundquist WI, Aiken C. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J Virol. 2002;76:5667–5677. doi: 10.1128/JVI.76.11.5667-5677.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang C, Ndassa Y, Summers MF. Structure of the N-terminal 283-residue fragment of the immature HIV-1 Gag polyprotein. Nat Struct Biol. 2002;9:537–543. doi: 10.1038/nsb806. [DOI] [PubMed] [Google Scholar]
- 37.Kelly BN, Howard BR, Wang H, Robinson H, Sundquist WI, Hill CP. Implications for viral capsid assembly from crystal structures of HIV-1 Gag1–278 and CAN133–278. Biochemistry. 2006;45:11257–11266. doi: 10.1021/bi060927x. [DOI] [PubMed] [Google Scholar]
- 38.Gross I, Hohenberg H, Huckhagel C, Kräusslich HG. N-terminal extension of human immunodeficiency virus capsid protein converts the in vitro assembly phenotype from tubular to spherical particles. J Virol. 1998;72:4798–4810. doi: 10.1128/jvi.72.6.4798-4810.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang H, Chopra R, Verdine GL, Harrison SC. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 40.Huang H, Harrison SC, Verdine GL. Trapping of a catalytic HIV reverse transcriptase·template:primer complex through a disulfide bond. Chem Biol. 2000;7:355–364. doi: 10.1016/s1074-5521(00)00113-7. [DOI] [PubMed] [Google Scholar]
- 41.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 42.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 43.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.