Abstract
We describe a new type of synthetic amphiphile that is intended to support biochemical characterization of intrinsic membrane proteins. Members of this new family displayed favorable behavior with four of five membrane proteins tested, and these amphiphiles formed relatively small micelles.
Membrane proteins (MPs) play crucial roles in biology, but these proteins are difficult to handle and analyze because of their physical properties.1 The native conformations of MPs display extensive nonpolar surfaces, which is necessary for residence in a lipid bilayer but leads to denaturation and/or aggregation in an aqueous medium. Detergents, such as dodecyl-β-d-maltoside (DDM), are typically employed to render MPs soluble by coating nonpolar protein surfaces.2 However, not all MPs can be maintained in native-like conformations when solubilized with conventional detergents.3 Moreover, even when a native conformation can be achieved, the MP-detergent complex may manifest unfavorable properties with regard to structural analysis (inability to crystallize and/or too large for NMR). Since our understanding of membrane protein structure and function remains poorly developed relative to soluble proteins, there is a persistent need for new amphiphilic "assistants" that can promote solubilization and manipulation of MPs. 4
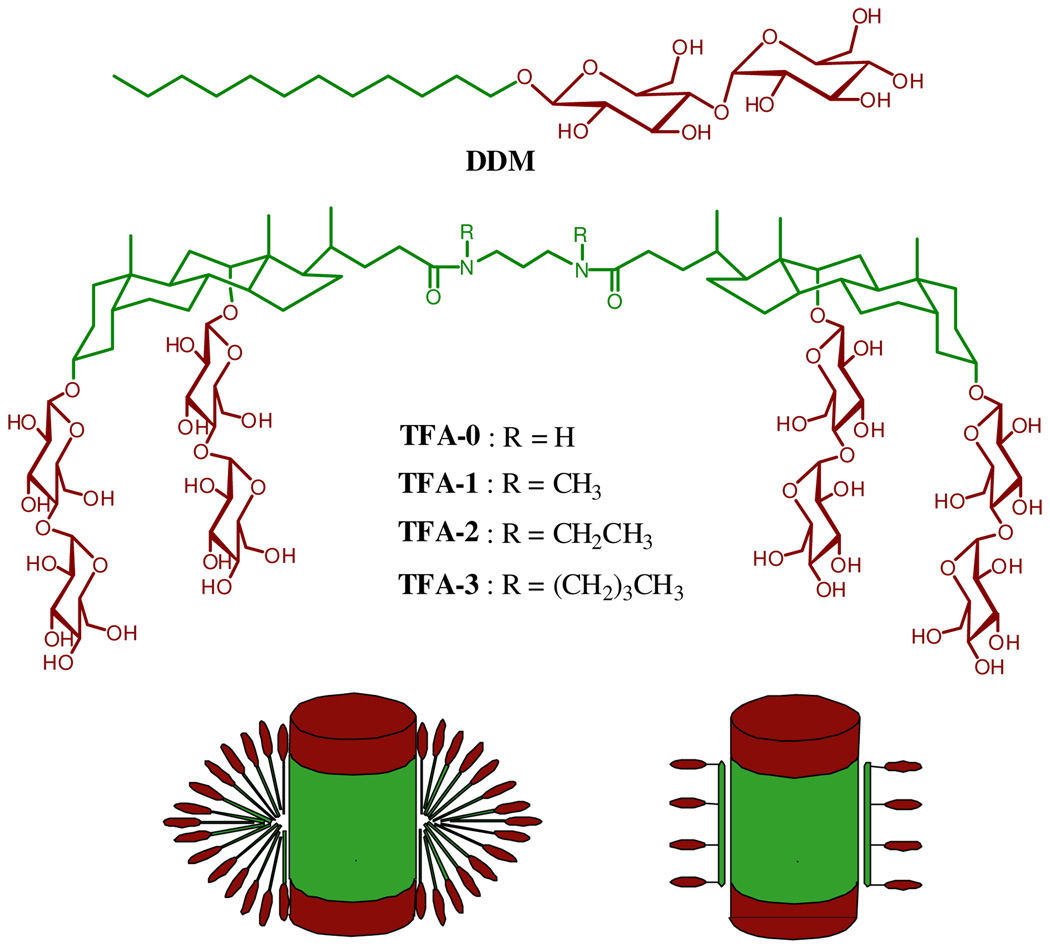
Several groups have reported creative implementations of the "facial amphiphile" concept for the design of novel amphiphiles that display favorable behavior with selected membrane proteins. 5 McGregor et al., for example, have reported lipopeptides that are intended to match the width of a lipid bilayer and to form a sheath around nonpolar surfaces of MPs.5c Zhang et al. have developed cholate-based amphiphiles in which hydrophilic maltose units project from one side of the rigid and hydrophobic steroidal skeleton.5d Here we disclose the design of "tandem facial amphiphiles" (TFAs), which contain a pair of maltose-functionalized deoxycholate units. Unlike previous cholate-based designs, the TFAs are long enough to match bilayer width,7 and unlike lipopeptides, the TFAs are readily synthesized in large quantities. We show that one TFA forms micelles containing only six molecules, and that simple TFAs can be used to maintain a variety of MPs in native-like states in aqueous solution.
A set of four TFAs was generated from a deoxycholate-bis-maltoside building block via linkage with a diaminopropane unit (Figure 1). Molecular mechanics calculations suggest that an extended conformation of the TFA backbone has a length that is comparable to the width of a typical lipid bilayer (~30 Å).7 These TFAs vary in the appendage on the amide nitrogen atoms. Each amphiphile could be obtained in excellent purity (>98%) and good overall yield (~65%) in five straightforward synthetic steps with two chromatographic purifications.7 Multi-gram quantities are readily available.
Figure 1.
Chemical structures of DDM (top) tandem facial amphiphiles (TFAs, middle) and schematic representation of membrane proteins interacting with DDM (bottom left) and TFAs (bottom right).
The TFAs displayed interesting behavior in water. TFA-0 forms a hydrogel at concentrations > 0.4 wt %, and this compound was not studied further. The other three TFAs are soluble to 5–10 wt % in aqueous media. Critical micelle concentrations (CMC) were determined by monitoring solubilization of a hydrophobic fluorescent dye, dicyclohexatriene,6 and the hydrodynamic radii (Rh) of the micelles were determined via dynamic light scattering (DLS).7 Table 1 compares the data for TFAs with those for DDM, a conventional detergent that is very widely used for MP applications; DDM and our TFAs share maltose as their hydrophilic moieties. CMC values of the three TFAs are smaller than that of DDM, whether CMC is measured in units of mM or wt %. The micelles formed by TFA-1 and TFA-2 (Rh ~ 2.0 nm) are somewhat smaller than those formed by DDM, while micelles formed by TFA-3 are comparable to those of DDM (Rh ~ 3.4 nm).
Table 1.
Critical micelle concentration (CMC) of TFAs and hydrodynamic radii (Rh) of their micelles (mean ± SD, n = 3).
| MW a | CMC (µM) | CMC (wt %) | Rh (nm) b | |
|---|---|---|---|---|
| TFA-1 | 2148.4 | 13 ± 1.4 | 0.0028 ± 0.00030 | 1.9 ± 0.08 |
| TFA-2 | 2176.5 | 13 ± 1.8 | 0.0028 ± 0.00039 | 2.0 ± 0.03 |
| TFA-3 | 2232.6 | 7 ± 2.3 | 0.0016 ± 0.00051 | 3.3 ± 0.12 |
| DDM | 510.1 | 170 | 0.0087 | 3.4 ± 0.03 |
Molecular weight of detergents.
Hydrodynamic radius of micelles measured by dynamic light scattering.
Micelles formed by DDM or by TFA-1 in pH 7.0 buffer (20 mM HEPES, 150 mM NaCl) were further characterized by gel filtration using a triple-detector system8 (light scattering, refractive index, and differential pressure) (Table 2). In both cases the micelles are globular and monodisperse. TFA-1 micelles contain only 6 molecules, which contrasts with the ~175 molecules in a DDM micelle. TFA-2 (R = ethyl) seems to behave similarly to TFA-1 (R = methyl), given the similarity in Rh, but TFA-3 (R = butyl) forms larger micelles. A related trend was observed among lipopeptides, with increasing length of the alkyl appendages leading to increasing micelle size.5c
Table 2.
Detailed characterization of TFA-1 (mean ± SD, n = 6) and DDM micelles (mean ± SD, n = 8).
| MMW a | N b | Rh (nm) | IV c | Mw/Mn d | dn/dc e | |
|---|---|---|---|---|---|---|
| DDM | 89982 ± 663 | 176.4 ± 1.3 | 3.42 ± 0.04 | 0.028 ± 0.009 | 1.01 ± 0.00 | 0.130 |
| TFA-1 | 13279 ± 157 | 6.2 ± 0.07 | 1.96 ± 0.01 | 0.036 ± 0.005 | 1.00 ± 0.00 | 0.173 |
Molecular weight of micelles.
Aggregation number of micelles.
Intrinsic viscosity.
Weight-averaged molecular weight divided by number-averaged molecular weight.
Specific refractive index increment.
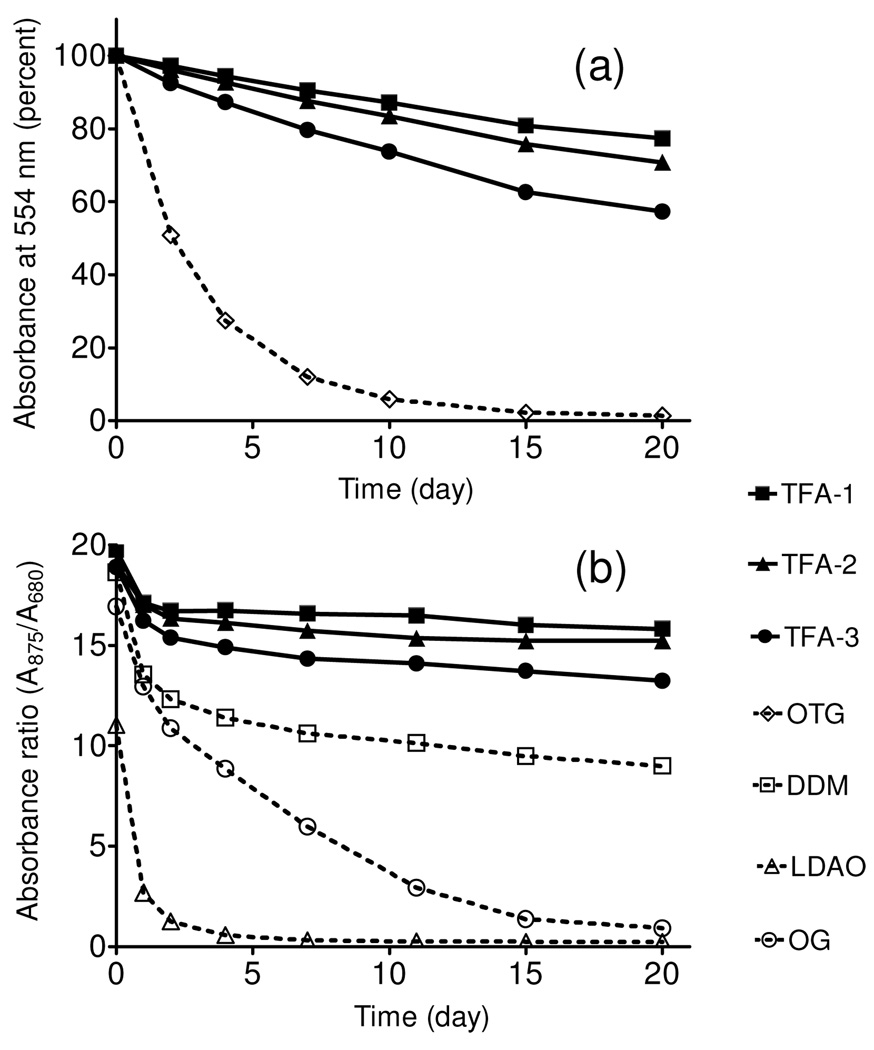
Bacteriorhodopsin (bR) has been widely employed for assessment of new amphiphiles because this membrane protein is readily available, and stability can be assessed via spectrophotometry (absorbance at 554 nm). Following standard protocols9, we used 2.0 wt % octyl-β-d-thioglucoside (OTG) to extract bR from the native purple membrane. After removal of insoluble debris via ultracentrifugation, the bR solution was diluted with amphiphile-containing solutions to generate samples containing 0.2 wt % OTG + 0.8 wt % TFA. A control sample had OTG added to give a total of 1.0 wt %. Figure 2a shows that all three TFA-containing samples were much more effective at maintaining native bR absorbance over 20 days relative to the sample containing only OTG. The bR was almost completely denatured by day 10 in the OTG-only sample, but ~80% intact at day 20 when solubilized with TFA-1.10, 11
Figure 2.
Time course of the stability of bR (a) and R. capsulatus superassembly (b) at room tempearture. Detergents were tested at 0.2 wt % OTG + 0.8 wt % TFA and CMC + 0.04 wt % for bR and R. capuslutus superassembly, respectively
The promising results with bR stabilization led us to investigate a more challenging system, the photosynthetic superassembly formed by the light harvesting I (LHI) and reaction center (RC) complexes from Rhodobacter capsulatus.12 This superassembly contains 30–40 protein molecules (five different components), and maintenance of native quaternary structure can be assessed via spectrophotometry. The LHI-RC superassembly was extracted from native membranes with 1.0 wt % DDM and purified with DDM at its CMC (0.009 wt %). This preparation was diluted 20-fold with solutions containing TFA-1, TFA-2 or TFA-3, so that residual DDM was far below its CMC (0.0004 wt %). The final TFA concentrations were 0.043 wt % (well over the CMC in each case). A control sample had DDM added to a total concentration of 0.049 wt % (all samples were CMC + 0.04 wt %). Figure 2b shows that LHI-RC superassembly solubilized with any of the TFAs was more stable over 20 days than was superassembly solubilized by DDM. Controls involving other common biochemical detergents (lauryldimethylamine oxide or octyl-glucoside) showed rapid degradation of the superassembly.10
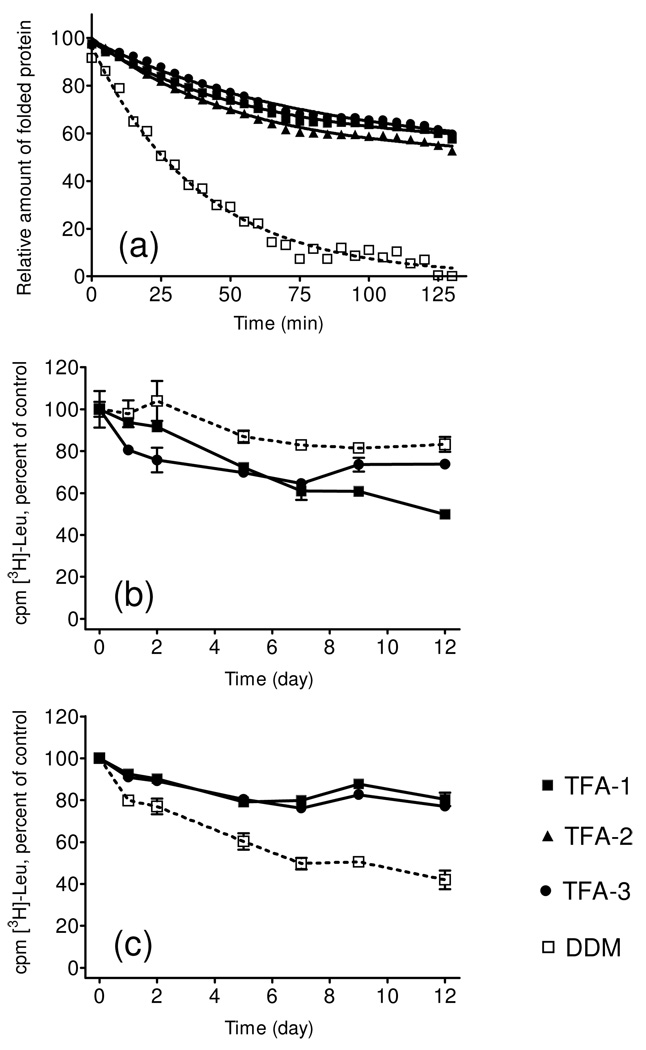
Each membrane protein (such as bR) or membrane protein assembly (such as LHI-RC) has unique requirements for maintenance in a native-like state in aqueous solution; therefore, it is important to assess the capabilities of new amphiphiles in multiple systems, in order to establish the breadth of their utility. We turned next to cytochrome bo3 ubiquinol oxidase (Cyt bo3), the structural stability of which was assessed at elevated temperature (40°C) with a reactive probe, (N-[4-(7-diethylamino-4-methyl-3-coumarinyl)phenyl]maleimide)(CPM).13 This maleimide derivative reacts with the thiol groups of sterically accessible Cys side chains. The coumarin moiety of CPM is internally quenched by the maleimide unit, but thiol reaction causes the unit to become fluorescent. CPM can therefore be used to detect thermally-induced protein unfolding, via an increase in fluorescence, if the protein contains Cys residues that are buried in the native state but accessible upon unfolding. Cyt bo3 was initially extracted from the native membrane with DDM, and then diluted to generate solutions containing 0.043 wt % TFA-1, TFA-2 or TFA-3 (residual DDM = 0.0008 wt %). A control sample had DDM added to a total concentration of 0.049 wt % (CMC + 0.04 wt % for each amphiphile). Figure 3a shows that TFA-solubilized Cyt bo3 samples were more resistant to thermal denaturation than was the DDM-solubilized control.
Figure 3.
Experimental assessment of the time-course changes in stability of solubilized Cyt bo3 and activity of LeuT WT. (a) CPM assay for Cyt bo3 was performed at 40°C for 130 minutes using CMC + 0.04 wt % amphiphile. LeuT WT was kept at room termperature up to 12 days in the presence of CMC + 0.04 wt % (b) or CMC + 0.2 wt % (c) before determining binding activity by SPA.
The wild type of bacterial leucine transporter (LeuT WT) was examined because the functional state of this membrane protein is readily assessed by using a scintillation proximity assay (SPA)14 to monitor binding of radiolabelled leucine. LeuT was initially extracted with DDM and then diluted with amphiphile-containing solutions to generate final TFA concentrations of 0.04 wt % or 0.2 wt % (residual DDM = 0.005 wt %). Control samples had 0.05 wt % or 0.2 wt % DDM (overall, the final concentrations were CMC + 0.04 wt % or CMC + 0.2 wt %). At the lower amphiphile concentrations, DDM was slightly better than the TFAs at maintaining LeuT WT function over 12 days (Figure 3b), but the TFAs were clearly superior at the higher concentrations (Figure 3c). TFA-1 and TFA-3 at the higher concentration matched DDM at the lower concentration in maintaining LeuT WT activity over the time period.
As a final test, we examined the TFAs for the ability to stabilize a GPCR, the human β2 adrenergic receptor (β2AR).15 This assay employs a β2AR-T4-lysozyme fusion protein (β2AR-T4L) complexed to the inverse agonist carazolol; stability is assessed by following the fluorescence emission maximum of carazolol, which shifts from 341 nm in the bound state to 356 nm in aqueous solution (i.e., after release upon β2AR denaturation). Monitoring the 341:356 nm peak intensity ratio upon heating yields cooperative denaturation data. In this assay, the TFAs proved to be inferior to DDM.
We have introduced a new class of molecules, "tandem facial amphiphiles", that contain two deoxycholate-derived subunits and that are sufficiently long to span a lipid bilayer.7 These molecules can be easily prepared on a scale that would support biochemical research. One of the new amphiphiles, TFA-1, was shown to form small, discrete micelles in water (MW ~ 13 kD). In contrast, DDM, a popular biochemical detergent, forms much larger micelles (MW ~ 90 kD). Three TFAs have been evaluated for the ability to maintain intrinsic membrane proteins or protein assemblies in native-like forms in aqueous solution. In four of the five cases we examined, the TFAs proved to be comparable or superior to DDM for stabilizing the membrane protein. Given the great variation in structure and physical properties among membrane proteins, no single amphiphile or amphiphile family will be maximally effective for every case. Because the TFAs manifest favorable solubilization/stabilization behavior with several diverse membrane protein systems, relative to widely used conventional detergents (DDM or OTG), and because this new amphiphile class can form small assemblies, it seems likely that TFAs will be valuable tools for characterization of membrane proteins, perhaps including high-resolution structural analysis.3b
Supplementary Material
Acknowledgements
This work was supported by NIH grant P01 GM75913 (S.H.G), membrane protein expression center grant 2P50 GM073210 (R.S.), the European Community's Seventh Framework Programme FP7/2007-2013 under grant agreement n° HEALTH-F4-2007-201924, EDICT Consortium (K.G., B.B., U.G.), the Lundbeck Foundation (S.G.F.R., C.J.L., U.G.), and the Danish National Research Council (C.J.L., U.G.). R.R. was funded by the Defence, Science and Technology Laboratories (DSTL), Porton Down, UK. We thank Dr. P. Laible and N. Abbott for providing materials and allowing us to use DLS instrument, respectively.
Footnotes
Supporting Information Available: Experimental procedures for characterizations of new compounds, micelle characterization, and membrane protein stability assays. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.(a) Caffrey M. J. Struct. Biol. 2003;142:108–132. doi: 10.1016/s1047-8477(03)00043-1. [DOI] [PubMed] [Google Scholar]; (b) Lacapere JJ, Pebay-Peyroula E, Neumann JM, Etchebest C. Trends Biochem. Sci. 2007;32:259–270. doi: 10.1016/j.tibs.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 2.(a) White SH, Wimley WC. Annu. Rev. Biophys. Biomol. Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]; (b) Moller JV, le Maire J. J. Biol. Chem. 1993;268:18659–18672. [PubMed] [Google Scholar]
- 3.(a) Garavito RM, Ferguson-Miller S. J. Biol. Chem. 2001;276:32403–32406. doi: 10.1074/jbc.R100031200. [DOI] [PubMed] [Google Scholar]; (b) Privé GG. Methods. 2007;41:388–397. doi: 10.1016/j.ymeth.2007.01.007. [DOI] [PubMed] [Google Scholar]; (c) Bowie JU. Curr. Opin. Struct. Biol. 2001;11:397–402. doi: 10.1016/s0959-440x(00)00223-2. [DOI] [PubMed] [Google Scholar]; (d) Sanders CR, Sonnichsen F. Magn. Reson. Chem. 2006;44:S24–S40. doi: 10.1002/mrc.1816. [DOI] [PubMed] [Google Scholar]; (e) Serrano-Vega MJ, Magnani F, Shibata Y, Tage CG. Proc. Natl. Acad. Sci. USA. 2008;105:877–882. doi: 10.1073/pnas.0711253105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Loll PJ. J. Struct. Biol. 2003;142:144–153. doi: 10.1016/s1047-8477(03)00045-5. White SH. Protein Sci. 2004;13:1948–1949. doi: 10.1110/ps.04712004. (c) For a continuously updated database of MP structures, see: http://blanco.biomol.uci.edu/Membrane_Proteins_xtal.html.
- 5.(a) Cheng Y, Ho DM, Gottlieb CR, Kahne D, Bruck MA. J. Am. Chem. Soc. 1992;114:7319–7320. [Google Scholar]; (b) Schafmeister CE, Meircke LJW, Stroud RM. Science. 1993;262:743–738. doi: 10.1126/science.8235592. [DOI] [PubMed] [Google Scholar]; (c) McGregor C-L, Chen L, Pomroy NC, Hwang P, Go S, Chakrabartty A, Prive GG. Nat. Biotech. 2003;21:171–176. doi: 10.1038/nbt776. [DOI] [PubMed] [Google Scholar]; (d) Zhang Q, Ma X, Ward A, Hong W-X, Jaakola V-P, Stevens RC, Finn MG, Chang G. Angew. Chem. Int. Ed. 2007;119:7153–7155. doi: 10.1002/anie.200701556. [DOI] [PubMed] [Google Scholar]
- 6.Chattopadhyay A, London E. Anal. Biochem. 1984;139:408–412. doi: 10.1016/0003-2697(84)90026-5. [DOI] [PubMed] [Google Scholar]
- 7.See the Supporting Information
- 8.Gatta AL, Rosa MD, Marzaioli L, Busico T, Schiraldi C. Anal. Biochem. 2010;404:21–29. doi: 10.1016/j.ab.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 9.Bazzacco P, Sharma KS, Durand G, Giusti F, Ebel C, Popot J-L, Pucci B. Biomacromolecules. 2009;10:3317–3326. doi: 10.1021/bm900938w. [DOI] [PubMed] [Google Scholar]
- 10.See the Supporting Information for parallel studies at a different amphiphile concentration.
- 11.It is known that DDM is not efficient at bR extraction from the native membrane, but extracted bR is stable in the presence of DDM; Milder SJ, Thorgeirsson TE, Miercke LJ, Stroud RM, Kliger DS. Biochemistry. 1991;30:1751–1761. doi: 10.1021/bi00221a004..
- 12.(a) Laible PD, Kirmaier C, Udawatte CS, Hofman SJ, Holten D, Hanson DK. Biochemistry. 2003;42:1718–1730. doi: 10.1021/bi026959b. [DOI] [PubMed] [Google Scholar]; (b) Youvan DC, Ismail S, Bylina EJ. Gene. 1985;33:19–30. doi: 10.1016/0378-1119(85)90199-4. [DOI] [PubMed] [Google Scholar]
- 13.(a) Alexandrov A, Mileni M, Chien EY, Hanson MA, Stevens RC. Structure. 2008;16:351–359. doi: 10.1016/j.str.2008.02.004. [DOI] [PubMed] [Google Scholar]; (b) Hanson MA, Cherezov V, Griffith MT, Roth CB, Jaakola V-P, Chien EYT, Velasquez J, Kuhn P, Stevens RC. Structure. 2008;16:897–905. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quick M, Javitch JA. Proc. Natl. Acad. Sci. USA. 2007;104:3603–3608. doi: 10.1073/pnas.0609573104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SGF, Thian FS, Kobilka TS, Choi H-J, Yao X-J, Weis WI, Stevens RC, Kobilka BK. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.