Abstract
Background
BtuB (B twelve uptake) is an outer membrane protein of Escherichia coli, it serves as a receptor for cobalamines uptake or bactericidal toxin entry. A decrease in the production of the BtuB protein would cause E. coli to become resistant to colicins. The production of BtuB has been shown to be regulated at the post-transcriptional level. The secondary structure switch of 5' untranslated region of butB and the intracellular concentration of adenosylcobalamin (Ado-Cbl) would affect the translation efficiency and RNA stability of btuB. The transcriptional regulation of btuB expression is still unclear.
Results
To determine whether the btuB gene is also transcriptionally controlled by trans-acting factors, a genomic library was screened for clones that enable E. coli to grow in the presence of colicin E7, and a plasmid carrying gadX and gadY genes was isolated. The lacZ reporter gene assay revealed that these two genes decreased the btuB promoter activity by approximately 50%, and the production of the BtuB protein was reduced by approximately 90% in the presence of a plasmid carrying both gadX and gadY genes in E. coli as determined by Western blotting. Results of electrophoretic mobility assay and DNase I footprinting indicated that the GadX protein binds to the 5' untranslated region of the btuB gene. Since gadX and gadY genes are more highly expressed under acidic conditions, the transcriptional level of btuB in cells cultured in pH 7.4 or pH 5.5 medium was examined by quantitative real-time PCR to investigate the effect of GadX. The results showed the transcription of gadX with 1.4-fold increase but the level of btuB was reduced to 57%.
Conclusions
Through biological and biochemical analysis, we have demonstrated the GadX can directly interact with btuB promoter and affect the expression of btuB. In conclusion, this study provides the first evidence that the expression of btuB gene is transcriptionally repressed by the acid responsive genes gadX and gadY.
Background
BtuB (B twelve uptake) is a 614 amino acid outer membrane protein of Escherichia coli. It is responsible for the uptake of cobalamins [1], such as vitamin B12 including cyanocobalamin, hydroxocobalamin, methylcobalamin, and adenosylcobalamin[2]. It also serves as the receptor for bacteriophage BF23 [3]. The synthesis of the BtuB protein in E. coli is regulated at the translational level by adenosylcobalamin (Ado-Cbl) which is produced by the BtuR protein (CobA in Salmonella typhimurium and CobO in Pseudomonas denitrificans) [4-6]. BtuR is an ATP:corrinoid adenosyltransferase and converts cobalamins to Ado-Cbl [4]. In the presence of Ado-Cbl, the stability of the btuB mRNA is reduced with a half-life of only 2 - 4 minutes [7]. In addition, Ado-Cbl binds to the leader region (5' untranslated region, 5' UTR) of the btuB mRNA and suppresses its translation [8,9]. A 25-nucleotide sequence designated as the B12-box located +138 - +162 nucleotides downstream from the transcription initiation site of btuB in E. coli has been suggested to be the binding site of Ado-Cbl [10]. A B12-box is also present in the 5' UTR of both btuB and cbiA genes of S. typhimurium [11]. The btuB gene of S. typhimurium is highly homologous to that of E. coli. The CbiA protein is a cobyrinic acid a, c-diamide synthase using cobyrinic acid as substrate [10,12]. Binding of Ado-Cbl to the 5' UTR of the mRNAs of these genes may interfere with ribosome binding and thus decrease their translation [7-9,13].
It is unknown whether BtuB synthesis is also controlled by regulatory proteins at the transcriptional level. Results of this study suggest that GadX (Glutamic acid decarboxylation) is a transcriptional regulator of btuB. GadX has been shown to suppress the expression of perA encoded by a plasmid of enteropathogenic E. coli [14], but activate gadX, gadA, gadB, and gadC in response to acid stress [15-19]. GadA and GadB are isozymes of glutamate decarboxylases that convert glutamate to γ-aminobutyric acid (GABA) which is then exported by the antiporter protein GadC [20,21]. An intracellular proton is consumed during GABA production [22], but the released GABA, which is less acidic than glutamate, provides local buffering of the extracellular environment.
The expression of gadX is activated by the alternative sigma factor RpoS during the stationary phase of growth [15,19,21], but is repressed during the exponential phase by the nucleoid protein H-NS due to its binding to the gadX promoter or its destabilizing effect on RpoS [23-25]. However, the acid stress increases the RpoS level and thus induces gadX expression even during the exponential phase of growth [26]. GadW, like GadX, belongs to the family of AraC-like regulators. It represses the expression of gadX and inhibits the activation of gadA and gadBC by GadX [15,18,27]. In addition to these trans-acting proteins, the production of GadX is also controlled by gadY that is located between gadX and gadW in an opposite orientation to gadX [28,29]. The gadY gene has no known protein products. It produces three RNA species of 105, 90, and 59 nucleotides with a common 3' end [28]. The 3' ends of gadX and gadY RNAs overlap by at least 30 nucleotides and are complementary to each other. Annealing of gadY RNA to the 3' end of gadX mRNA stabilizes gadX mRNA, resulting in an increased production of the GadX protein [28].
BtuB is also involved in the import of colicins such as colicin E7 (ColE7) [30-34]. ColE7 is composed of three domains responsible for the translocation of ColE7 through the OmpF porin, binding of ColE7 to BtuB, and cleavage of DNA [35,36], respectively. The import of ColE7 is dependent on the Tol (tolerance to colicin) system that is composed of TolQ, TolR, TolA, and TolB proteins [35,36]. Deletion or mutation of BtuB, OmpF, or any of the Tol proteins renders E. coli resistant to ColE7 [33,37,38]. Based on this information, we used a ColE7 resistance assay in this study to search for transcriptional regulators of btuB from a genomic library of E. coli strain DH5α and found that gadX and gadY genes down regulate the expression of btuB.
Results
Screening of genes conferring E. coli resistance to ColE7
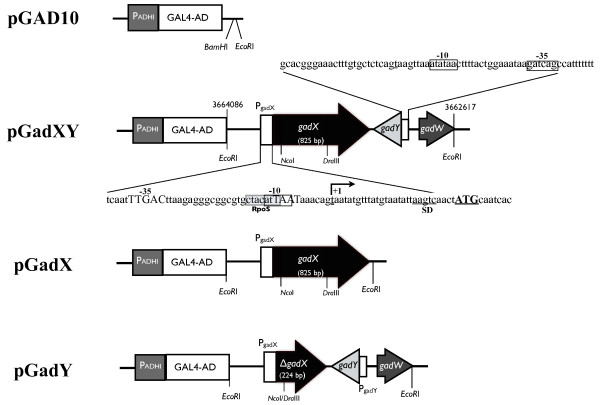
To search for genes that can confer E. coli resistance to ColE7, plasmids in the genomic library were transformed into the ColE7-sensitive E. coli strain DH5α, and the transformants were plated on LB agar plates containing 50 μg/ml of ampicillin and 5.0 ng/ml of His6-tagged ColE7/ImE7. Two colonies were seen after incubation at 37°C overnight. The plasmids of each colony were isolated after culturing in 3 ml LB medium containing 50 μg/ml of ampicillin then retransformed into DH5α. The new transformants were plated on agar plates containing 0, 1.3, 2.6, 3.9, or 5.2 ng/ml of His6-tagged ColE7/ImE7 to confirm their resistance to ColE7. The insert in the plasmid that conferred DH5α resistance to 5.2 ng/ml His6-tagged ColE7/ImE7 was sequenced. A 1,470-bp DNA region on the chromosome at position 3662617 to 3664086 was analyzed that contains both complete gadX and gadY genes. The plasmid was thus named pGadXY (Figure 1).
Figure 1.
Structures of pGAD10, pGadXY, pGadX, and pGadY. pGAD10 was the vector used to clone gadXY, gadX, and gadY. pGadXY has a 1,470-bp fragment containing gadX, gadY, and a portion of gadW of E. coli K-12 genomic DNA inserted into the EcoRI site of pGAD10. pGadX contains a DNA fragment carrying the 825-bp gadX also inserted into the EcoRI site of pGAD10. pGadY is derived from pGadXY by deleting the 601-bp NcoI-DraIII fragment and thus contains a truncated gadX, the entire gadY, and a portion of gadW. Nucleotide sequences of the promoter regions of gadX and gadY are shown. The orientation of gadX is opposite to that of gadY. The sigma factor S (RpoS) recognition site and the Shine-Dalgarno (SD) sequence are shown in the 5' end region of gadX. PADH is the promoter of GAL4-AD and is not functional in E. coli.
To determine whether gadX or gadY was responsible for ColE7 resistance, pGadX, pGadY, and pGadXY that contain gadX, gadY, and gadXY, respectively, were separately introduced into E. coli strain DH5α and then assayed for their ability to confer ColE7 resistance. 1 × 105 cells containing pGadX, pGadY, or pGadXY were plated on LB agar containing 1.3, 2.6, 3.9, or 5.2 ng/ml of His6-tagged ColE7/ImE7. Cells containing the vector pGAD10 were also plated to serve as controls. The percent survival of cells containing pGAD10, pGadXY, pGadX, and pGadY in the presence of 1.3 ng/ml of His6-tagged ColE7/ImE7 were 41.7, 95.5, 71.4, and 73.5%, respectively, and 1.5, 63.9, 3.6, and 9.1%, respectively, in the presence of 2.6 ng/ml of His6-tagged ColE7/ImE7. Only pGadXY conferred ColE7 resistance to 3.9 and 5.2 ng/ml of His6-tagged ColE7/ImE7 with 29.1 and 17.1% survival rates, respectively (Table 1).
Table 1.
Effects of gadXY, gadX, and gadY on ColE7 resistance
| ColE7 conc./Bacteria | pGAD10/DH5α | pGadXY/DH5α | pGadX/DH5α | pGadY/DH5α |
|---|---|---|---|---|
| 1.3 ng/ml | 41.7% | 95.5% | 71.4% | 73.5% |
| 2.6 ng/ml | 1.5% | 63.9% | 3.6% | 9.1% |
| 3.9 ng/ml | 0 | 29.1% | 0 | 0 |
| 5.2 ng/ml | 0 | 17.1% | 0 | 0 |
Detection of protein whose expression is affected by gadXY
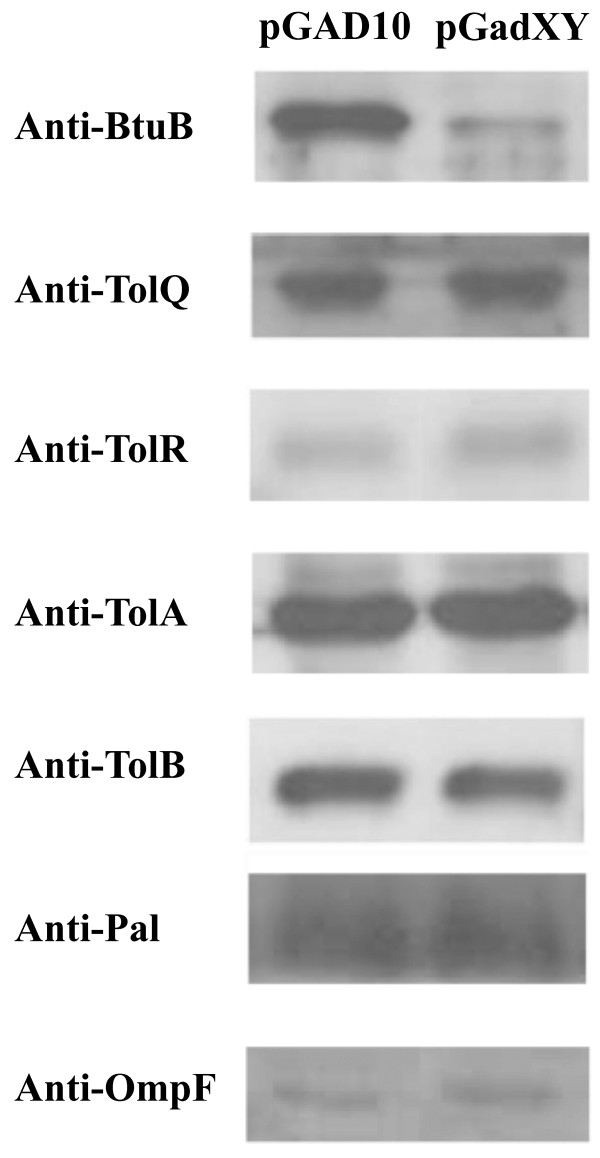
To investigate the mechanism by which gadXY affects ColE7 resistance, the expression levels of BtuB, TolQ, TolR, TolA, TolB, Pal, and OmpF that are involved in ColE7 import were determined by Western blotting, and BtuB was the only protein found to be affected. Its expression level was reduced by 93% in the presence of gadXY (Figure 2) as determined by densitometry.
Figure 2.
Effects of gadXY on the production of envelope proteins involved in ColE7 uptake. Cell lysates of E. coli DH5α cells harboring pGAD10 or pGadXY were examined by Western blotting using antibodies against envelope proteins BtuB, TolQ, TolR, TolA, TolB, Pal, and OmpF.
Effect of gadXY on btuB promoter
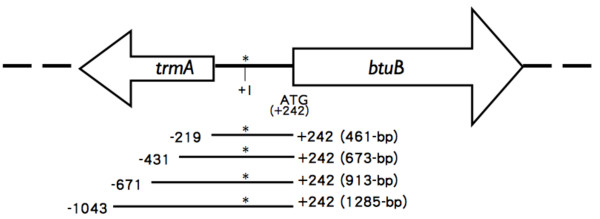
To determine whether gadXY affects the transcription of btuB, the β-galactosidase reporter assay was performed. The 461-, 673-, 913-, and 1285-bp DNA fragments (Figure 3) containing the promoter of btuB were fused with the lacZ coding sequence to generate pCB461lacZ, pCB673lacZ, pCB913lacZ, and pCB1285lacZ, respectively. Each of these single copy plasmid together with pGAD10 or pGadXY was transformed into E. coli strain DH5α. The transformed cells were grown in LB medium with 50 μg/ml of chloramphenicol and ampicilin to OD600~0.8 then assayed for β-galactosidase activity as described by Miller [39]. The β-galactosidase activity of cells containing pGadXY and a pCB derivative with the btuB promoter-lacZ fusion was divided by that of cells containing the control plasmid pGAD10 and the same pCB derivative to determine the percent decrease in btuB promoter activity in the presence of gadXY. The btuB promoter in the 461-, 673-, 913-, and 1285-bp DNA fragment was found to be decreased by 45.7, 47.1, 54.5, and 56.7%, respectively in the presence of gadXY, and was about 6 fold more active in the 1285-bp fragment than in other fragments (Table 2).
Figure 3.
DNA fragments containing the btuB promoter used for lacZ fusions. The btuB initiation codon ATG is located at nucleotide position +242. Asterisk indicates the first nucleotide of the btuB mRNA. The trmA (tRNA methyltransferase) gene is located upstream from btuB. It has no known effect on btuB expression.
Table 2.
Effect of gadXY on btuB promoter
| Plasmid | β-galactosidase activity a | % inhibition b |
|---|---|---|
| (A): pGAD10 + pC-lacZ | 0 | |
| (B): pGadXY + pC-lacZ | 0 | |
| (A): pGAD10 + pCB461lacZ | 6.4 ± 0.2 | 45.7 |
| (B): pGadXY + pCB461lacZ | 3.5 ± 0.2 | |
| (A): pGAD10 + pCB673lacZ | 7.2 ± 0.1 | 47.1 |
| (B): pGadXY + pCB673lacZ | 3.8 ± 0.1 | |
| (A): pGAD10 + pCB913lacZ | 4.8 ± 0.2 | 54.5 |
| (B): pGadXY + pCB913lacZ | 2.2 ± 0.5 | |
| (A): pGAD10 + pCB1285lacZ | 37.5 ± 0.7 | 56.7 |
| (B): pGadXY + pCB1285lacZ | 16.2 ± 0.5 | |
aMiller unit.
bCaculated according to the following equation: 1- [β-galactosidase activity of (B) ÷ β-galactosidase activity of (A)] × 100%.
To investigate the effect of gadX or gadY alone on the promoter activity of btuB, the same experiment was performed using DH5α cells containing pCB1285lacZ and pGAD10, pGadXY, pGadX, or pGadY. The β-galactosidase activity of cells containing pCB1285lacZ and pGadXY, pGadX, or pGadY was compared to those containing pGAD10 and pCB1285lacZ. The results indicated that btuB promoter activity was decreased 20.5% by gadX and 20.3% by gadY, but was decreased 54.4% by gadXY (Table 3).
Table 3.
Effect of gadX, gadY, and gadXY on btuB promoter
| Plasmids | β-galactosidase activity a | % inhibition b |
|---|---|---|
| (A): pGAD10/pC-lacZ | 0 | |
| (B): pGAD10/pCB1285lacZ | 48.8 ± 3.9 | |
| (C): pGadXY/pCB1285lacZ | 22.3 ± 0.7 | 54.4 |
| (D): pGadX/pCB1285lacZ | 38.9 ± 2.6 | 20.5 |
| (E): pGadY/pCB1285lacZ | 38.9 ± 2.0 | 20.3 |
aMiller unit
bCalculated according to the following equation: 1- [β-galactosidase activity of (C), (D), or (E) ÷ β-galactosidase activity of (A)] × 100%.
Binding of GadX to btuB promoter
GadX has been shown to be a DNA binding protein and can bind to the gadA or the gadB promoter. To determine whether GadX also binds to the btuB promoter, the DNA mobility shift assay was performed. Only GadX was assayed because gadY does not encode any proteins. The 461-bp DNA fragment containing the btuB promoter was labeled with 32P and incubated with 2, 4, or 6 pmoles of purified GadX protein (MalE-GadX) that was fused to the maltose binding protein. The DNA fragment containing the promoter of gadA or gadB was used as the positive control for GadX binding, and the DNA fragment containing the pal promoter was used as the negative control. As shown in Figure 4, DNA band shift was observed on gadA and gadB promoter fragments but not on the negative control. Band shift was also observed on the btuB promoter fragment in a dose-dependent manner, indicating that GadX binds to the btuB promoter.
Figure 4.
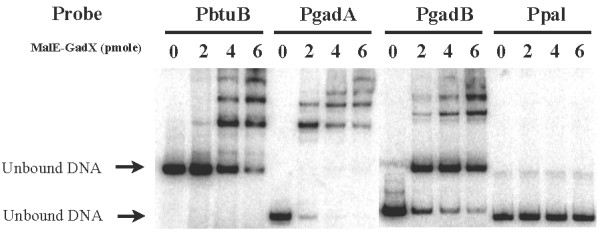
Binding of GadX to btuB promoter. 32P-labeled DNA fragments PbtuB, PgadA, PgadB, and Ppal containing the promoters of btuB, gadA, gadB, and pal, respectively, were incubated with GadX fused to the maltose binding protein (MalE-GadX) at 0, 2, 4, or 6 pmoles. The reaction mixtures were electrophoresed in a 5% native polyacrylamide gel. Band shift due to GadX binding was visualized by autoradiography. Arrows indicate bands of DNA probes not bound by GadX.
Identification of binding sequence of GadX on btuB promoter
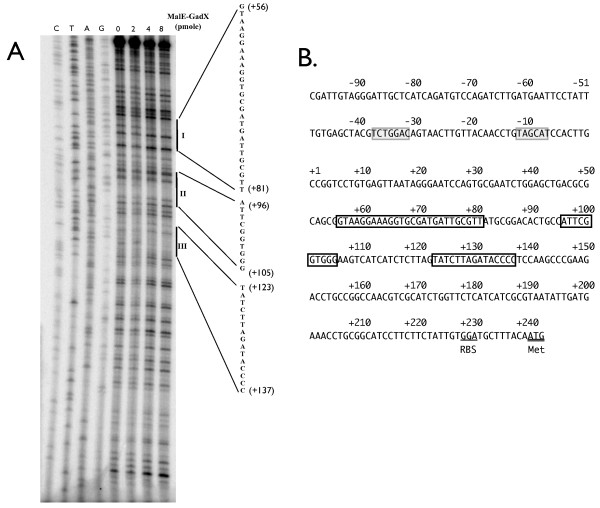
DNase I footprinting was then performed to determine the binding sequence of GadX on the btuB promoter. The 461-bp DNA fragment containing the btuB promoter was labeled with 32P and incubated with 0, 2, 4, or 8 pmoles of purified MalE-GadX protein and then digested with DNase I. Results shown in Figure 5 revealed three MalE-GadX protein binding sites that included nucleotide positions +56 - +81 (I), +96 - +105 (II) and +123 - +137 (III) on the 5' untranslated region of btuB.
Figure 5.
Binding sequence of GadX on btuB promoter. (A) The 461-bp DNA fragment containing btuB promoter was labeled at 5' end with 32P, incubated with 0, 16, 24, 32, or 40 pmoles of MalE-GadX, and then subjected to DNase I footprinting. A Sanger's DNA sequencing reaction was also done on the 461-bp fragment to reveal GadX binding sequences. All reactions were electrophoresed in a 6% urea-acrylamide gel, and the DNA bands were detected by autoradiography. The GadX bound regions are indicated with vertical lines, and the binding sequence of GadX are shown. (B) Sequence of the btuB promoter region. The boxed sequences are GadX binding sequences determined by the DNase I footprinting. The shaded sequences are -10 and -35 regions of the btuB promoter. The initiation codon of btuB is underlined.
Decreased btuB expression under acidic conditions
It is known that gadX and gadY are more highly expressed under acidic environments in stationary phase [15-19,28]. To determine whether the expression of btuB is also repressed in an acidic condition, wild type BW25113 cells were cultured in LB medium pH 7.4 or buffered with 100 mM MES pH5.5. Stationary phase cells grown in different culture media were collected and then assayed for the transcriptional level of btuB by quantitative real-time PCR. The cDNA amplification comparison results showed the transcription of gadX with 1.4-fold increase but the level of btuB was reduced to 57% (Table 4).
Table 4.
Fold changes of transcripts of gadX and btuB attribute to different pH medium (pH 5.5/pH 7.4) from early stationary phase.
| Gene | Fold increase a |
|---|---|
| gadX | 1.43 ± 0.07 |
| btuB | 0.57 ± 0.13 |
a Experiments were performed in triplicate and the data are presented as mean values ± SD.
Discussion
Although it has been suggested that the expression of btuB in E. coli is also regulated at the transcriptional level, the trans-acting regulators of btuB had not been identified [40,41]. In this study, we used the ColE7 resistance assay to search for such regulators and found that both gadX and gadY genes can repress the production of BtuB rendering E. coli DH5α cells resistant to ColE7. Introduction of pGadX, which contains a gadX gene, into DH5α cells caused 3.6% of the cells to become resistant to 2.6 ng/ml of ColE7. In a similar experiment, pGadY which contains the gadY gene enabled 9.1% of the cells to grow in the presence of the same concentration (2.6 ng/ml) of ColE7 (Table 1). Although gadY does not encode any proteins, it had a greater effect on making E. coli resistant to ColE7 than gadX. This is probably due to the binding of gadY RNA derived from pGadY to the gadX mRNA produced by the gadX gene in the chromosome. This binding stabilizes gadX mRNA so that more GadX protein is produced to suppress the production of BtuB, making the cells resistant to ColE7. The greatest effect (63.9% survival in 2.6 ng/ml ColE7) on ColE7 resistance was seen when pGadXY, which contains both gadX and gadY genes, was introduced into the cells. Since pGadXY is a high copy number plasmid, more gadX and gadY mRNAs would be produced and thus more GadX protein would be synthesized to suppress BtuB synthesis. However, excess GadX had adverse effects as over expression of GadX with a strong promoter, such as the T5-lacO promoter, was found to have toxic effect to E. coli [19]. Therefore, expression of gadX and gadY in this study was driven by their own promoters.
Since GadX is a known transcriptional regulator [14-16,18,19,42], the decrease in BtuB expression is due to its transcriptional repression by GadX. Our data showed that the btuB promoter activity was reduced by approximately 50% in the presence of gadXY (Table 2), most likely due to binding of GadX to the 5' untranslated region of btuB as DNase I footprinting experiment revealed binding of GadX to nucleotides positions +56 - +81 (I), +96 - +105 (II) and +123 - +137 (III) downstream from the btuB transcription initiation site (Figure 5). From the sequence alignment of GadX binding sites on btuB, gadA, and gadBC regulatory regions[42], we found that sequence in the region I (the 31 nucleotides) has 62.5% identity (+52-AGCGGTAAGGAAAGGTGCGATGATTGCGTTAT-+82, underlined nucleotides indicate the protected region) with gadBC and sequence in the region III (the 26 nucleotides) has 60.7% identity (+106-AAGTCATCATCTCTTAGTATCTTAGATA-+133, underlined nucleotides indicate the protected region) with gadA regulatory region. From the footprinting result, the GadX binding sites on 5' untranslated region of btuB share only partial homology with the 42 nucleotides consensus sequence which was reported by Tramonti et. al.[42]. The sequence analysis also revealed the btuB expression was regulated by the binding of GadX on its 5' untranslated region. Binding of transcriptional regulator to the 5' untranslated region to regulate gene expression is also seen in the glp regulon of E. coli, in which four repressor binding sites are located at -41 to -60, -9 to -28, +12 to -8, and +52 to +33 of the glpACB genes [43]. In addition, two IHF binding sites are present downstream from the glpT transcriptional start site at positions +15 to +51 and +193 to +227 [44].
In the btuB promoter assay experiment, different lengths of DNA fragments containing btuB promoter were fused to lacZ. The minimum length of DNA fragment with btuB promoter activity was 461 bp spanning -219 to + 242 nucleotides relative to the translation initiation site of btuB. No significant difference in promoter activity was observed when the 5' end of these fragments was extended to -671. However, a 6 fold (37.5 vs. 6.4 β-galactosidase units, Table 2) increase in promoter activity was detected when the DNA fragment was extended to -1043 with a total length of 1,285 bp as compared to that of the 461-bp fragment. It is very likely that a certain transcription regulator binds to the region between -1043 and -671 and enhances the expression of btuB. The β-galactosidase activity in these assays was not very high because the lacZ fusions were constructed using the single copy plasmid vector pCC1Bac™ (Epicentre). The purpose of using the single copy number plasmid in this experiment was to mimic the natural state of btuB expression in E. coli. In fact, the promoter activity of btuB is lower than other membrane protein, we have determined the ompC promoter activity, under the same test condition the Miller's Units of lacZ driven by ompC promoter is 8 folds higher than that of btuB (data not shown).
Although the results of footprinting and reporter assay revealed that the GadX binding sites on btuB 5' untranslated region share only partial homology with the GadX binding consensus sequence[42] and showing 50% down regulation in the reporter assay, the expression of btuB was indeed controlled by GadX.
Both gadX and gadY genes belong to a group of genes that are induced by acid stress under stationary growth phase [44]. Our data showed that the expression of btuB was indeed reduced when E. coli cells were grown to stationary phase in an acidic medium as compared to the same cells grown in neutral medium (Table 4). The reduction in the production of btuB in response to acid stress probably represents a physiological regulatory mechanism of bacteria facing environmental challenges such as low pH. Under stress environment, bacteria need to alter their metabolism to adapt to the environmental change. The transportation of cobalamin by BtuB receptor is driven by proton motive force (PMF)[45]. Since the PMF of bacteria is increased at low pH[46], the cobalamin transportation may be enhanced by increased PMF. The higher concentration of cobalamin in cytoplasm will initiate riboswitch mechanism to repress the translation of BtuB receptor, which is in good accord with the repression of btuB transcription by the acid-induced GadX for bacteria to decrease the production of BtuB in response to this acidic stress.
Conclusions
Through biological and biochemical analysis, we have demonstrated the GadX can directly interact with btuB promoter and affect the expression of btuB. When bacteria were grown to stationary phase in an acidic medium, the increased gadX expression would repress the btuB transcription to help bacteria to adapt to acidic shock. In conclusion, this study provides the first evidence that the expression of btuB gene is transcriptionally repressed by the acid responsive genes gadX and gadY.
Methods
Plasmid constructions
To produce the His6-tagged ColE7/Im7 protein complex for the ColE7 resistance assay, pQE30ColE7-Im7 was constructed. The cea7-cei7 genes encoding the colicin E7 and immunity proteins, that form an active ColE7 complex, were amplified from plasmid K317 [47] by PCR using primers F/cea7-BamHI and R/cei-PstI (Table 5). The 1,996-bp PCR product thus generated was inserted between BamHI and PstI sites of pQE30 (Qiagen), fusing the His6-tag to the N terminus of ColE7. For searching transcriptional regulators of btuB, a genomic library of E. coli K-12 strain constructed with the pGAD10 vector (Figure 1) was purchased from Clontech (catalog number XL4001AB) and transformed into E. coli strain DH5α. The plasmid pGadXY (Figure 1) was isolated from the library in this study. To investigate the effect of GadX on btuB expression, pGadX was constructed as follows. A 1,077-bp DNA fragment containing gadX was generated by PCR using pGadXY (Figure 1) as the template and the MATCHMAKER 5' insert screening sequence 5'-TACCACTACAATGGATG-3' (Clontech) and R/gadX-PstI (Table 5) as primers. This 1.1-kb PCR fragment was inserted into pGEM-TEasy (Promega) by TA cloning, generating pGEMgadX. The 1.1-kb fragment was then isolated from pGEMgadX by EcoRI digestion and inserted into the EcoRI site of pGAD10, resulting in pGadX (Figure 1). To investigate the effect of gadY on btuB expression, pGadY (Figure 1) was constructed by deleting the NcoI-DraIII fragment containing gadX from pGadXY.
Table 5.
Oligonucleotide primers used in this study
| Primer | Sequence 5 '- 3' |
|---|---|
| F/cea7-BamHI | GGATCCATGAGCGGTGGAGATGGACG |
| R/cei7-PstI | CTGCAGTCAGCCCTGTTTAAATCC |
| F/btuB-219-XbaI | GGCTCTAGAAAACGGTGCCATCATACTTTG |
| R/btuB+242-HindIII | GGCAAGCTTATCATTGTAAAGCATCCACAATAG |
| F/btuB-767 | GTTCACCGTTGCTCGATACC |
| R/btuB-1087 | TCAGATAGATGCCGGTATTACG |
| F/btuB-431-XbaI | GCTCTAGAACGGGATTATTACGC |
| F/btuB-671-XbaI | GCTCTAGATCATCTCTTTCCC |
| F/btuB-1043-XbaI | GCTCTAGACCGCTGCGCGGA |
| R/lacZ | TTATTTTTGACACCAGACC |
| F/gadA-176 | GATCGCCCGAACAGCAA |
| R/gadA+77 | CGTGAATCGAGTAGTTC |
| F/gadB-173 | AATAACAGCATAAAACA |
| R/gadB+77 | CGTGAATCGAGTAGTTCC |
| F/pal-XbaI | TCTAGAGAGGCGTACAAGTTCTG |
| R/pal-HindIII | AAGCTTATCATTTCAATGATTCCTTTAC |
| F/gadX-BamHI | GGATCCATGCAACCACTACATGG |
| R/gadX-PstI | CTGCAGCTATAATCTTATTCCTT |
| F/gadX-393 | TATACCGCTGCTTCTGAACG |
| R/gadX-726 | TCGCTCCTGATACTCTGTGG |
| F/rrsA-483 | CGTTACCCGCAGAAGAAGC |
| R/rrsA-808 | GTGGACTACCAGGGTATCTAATCC |
The underlined letters indicate restriction sites.
To assay btuB promoter activity, DNA fragments (461, 673, 913, and 1,285 bp) containing different portions (Figure 3) of the btuB promoter was fused to lacZ. These fragments were generated by PCR using primers F/btuB-219-XbaI, F/btuB-431-XbaI, F/btuB-671-XbaI, and F/btuB-1043-XbaI paired with the 3' primer R/btuB +242-HindIII (Table 5). The resulting PCR products were digested with XbaI and HindIII and then inserted into corresponding sites on pKM005 that carries a promoterless lacZ gene [48], generating pKMbtuB461-lacZ, pKMbtuB673-lacZ, pKMbtuB913-lacZ, and pKMbtuB1285-lacZ. To mimic native expression of btuB, these btuB-lacZ fusions were transferred to the single copy plasmid vector pCC1 (Epicentre). The fragments containing btuB promoter and lacZ on pKM005 derivatives were amplified with primers F/btuB-219-XbaI, F/btuB-431-XbaI, F/btuB-671-XbaI, and F/btuB-1043-XbaI paired with the 3' primer R/lacZ (Table 5), and the resulting 3.3, 3.5, 3.74, and 4.1-kb DNA fragments were separately inserted into pGEM-TEasy (Promega) by TA cloning. The 3.3, 3.5, 3.74, and 4.1-kb fragments were then isolated from these pGEM-TEasy derivatives by NotI digestion and inserted into the NotI site of pCC1 vector, generating pCB461lacZ, pCB673lacZ, pCB913lacZ, and pCB1285lacZ. The plasmid pC-lacZ that contains a promoterless lacZ gene inserted into pCC1 vector was used as a negative control. To produce GadX for DNA binding assay, pMalE-GadX that contains maltose-binding protein fused to GadX (MalE-GadX) was constructed. The 825-bp DNA fragment containing gadX was generated by PCR using pGadXY as the template and F/gadX-BamHI and R/gadX-PstI (Table 5) as primers and then ligated between the BamHI and PstI sites of pMAL-C2X (New England Biolab), resulting in pMalE-GadX.
Production of ColE7
To produce the His6-tagged ColE7/ImE7 complex, E. coli strain XL1-Blue containing plasmid pQE30ColE7-Im7 was cultured in LB medium containing ampicillin (50 μg/ml) and tetracycline (20 μg/ml). When the bacterial growth reached OD600 ~1.0, IPTG was added to a final concentration of 1 mM. After a 2-hr induction, bacteria were harvested by centrifugation at 6,500 × g for 20 min and then resuspended in HB buffer (20 mM Tris, 150 mM NaCl, 30 mM imidazole, pH8.0). The resuspended bacteria were lysed with a French Pressure Cell (SLM Instruments, Inc. Urbane, IL), and the cell lysate was centrifuged at 48,000 × g for 1 hour. The clarified supernatant was passed through a ProBond™ nickel-nitrilotriacetic acid resin affinity column (Invitrogen, Carlsbad, CA) to purify the His6-tagged ColE7/ImE7 according to manufacture's protocol (Invitrogen, Carlsbad, CA).
Antibody preparation for detection of protein whose expression is affected by gadXY
To prepare antibodies against envelope proteins BtuB, TolQ, TolR, TolA, TolB, Pal, and OmpF, the coding region of each protein was fused inframe with His6-tag in the plasmid pQE30 (Qiagen), respectively. The His6-tagged proteins were then expressed and purified using the same method as described for His6-tagged ColE7/ImE7 and sent to the company to prepare polyclonal antibodies. The specificities of these antibodies were confirmed by Western blotting using these antibodies as reported by Pan et. al[49].
DNA binding assay
The electrophoretic mobility shift assay was performed to investigate binding of GadX to the btuB promoter. To obtain purified MalE-GadX protein, E. coli strain XL-1 Blue containing pMalE-GadX was grown in 100 ml of LB containing ampicillin (50 μg/ml) and 0.2% glucose to OD600 ~1.0. IPTG was then added to a final concentration of 1 mM. After 2 hr of incubation, the cells were pelleted, resuspended in maltose binding buffer (20 mM Tris-HCl pH 8.0, 200 mM NaCl), and lysed with a French Pressure Cell. The cell lysate was centrifuged at 48,000 × g for 1 hr, and the supernatant was subjected to an amylose resin affinity chromatography (New England BioLabs) to purify the MalE-GadX protein.
To make probes for the DNA binding assay, a 461-bp (Figure 3, -219 - +242) DNA fragment containing the btuB promoter was amplified with primers F/btuB-219-XbaI and R/btuB+242-HindIII (Table 5) by PCR. The DNA fragment containing the promoter of gadA (-176 - +77, 253 bp) or gadB (-173 - +77, 250 bp) was used as the positive control, which were amplified with primer pairs F/gadA-176 and R/gadA+77 and F/gadB-173 and R/gadB+77 (Table 5), respectively, as described by Tramonti et al. [19]. The DNA fragment containing the upstream noncoding region of pal was used as the negative control, which was amplified with primers F/pal-XbaI and R/pal-HindIII (Table 5). These DNA fragments were end-labeled with [γ-32P] ATP by T4 polynucleotide kinase (New England BioLabs). The labeled DNA fragments (6 fmol) were incubated with the MalE-GadX protein at room temperature for 20 min in 10 μl of binding buffer [19]. Samples were then loaded on a 5% nondenaturing polyacrylamide gel in 0.5X TBE buffer and electrophoresed for 35 min at room temperature. The gels were then dried and autoradiographed.
DNase I footprinting
DNase I footprinting was performed to determine the binding sequence of MalE-GadX on btuB promoter as described by Tramonti et al [19]. Thirty μl of reaction mixture that contains 5 ng of 32P-labeled 461-bp btuB promoter fragment, various amounts of the MalE-GadX protein, and reaction buffer (40 mM HEPES pH 8.0, 100 mM potassium chloride, and 10 mM magnesium acetate) was incubated at room temperature for 20 min. At the end of the incubation, 0.5 U DNase I (Roche Biochemicals, Indianapolis, IN) was added to each reaction mixture and then incubated at 37°C for 1 min followed by addition of 3 μl of quench solution (0.1% xylene cyanol, 4% SDS, and 50% glycerol) to stop the DNase I digestion. The partially digested product was passed through a Sephadex G25 spin column (GE Healthcare), and the eluate was subjected to 30 cycles of asymmetric PCR (SequiTherm Excel™II, Epicentre) using 5'-end 32P-labeled primer R/btuB+242-HindIII (Table 5). The PCR-generated products were electrophoresed on a 6% sequencing gel. The gel was then dried and autoradiographed. To determine the binding sequence of GadX, the 461-bp btuB DNA probe was sequenced by the Sanger's sequencing method using the 5'-end 32P-labeled primer R/btuB+242-HindIII (Table 5).
Quantitative Real-Time Polymerase Chain Reaction
Total RNA of wild type Escherichia coli strain BW25113 grown under LB (pH 7.4) or LB/MES (LB buffered with 100 mM MES, pH 5.5) to early stationary phase were isolated using a modified hot-phenol extraction method[21]. This was followed by further purification using RNAspin Mini RNA purification kit (GE) to remove contaminating genomic DNA and enhance the quality of RNA. Each cDNA sample was synthesized from 0.1 µg total RNA with specific primers of rrsA, gadX and btuB using RevertAid™ First strand cDNA synthesis kit (Fermentas). Following reverse transcription, specific gene transcription levels were determined by quantitative real-time PCR using the ABI PRISM 7700 Sequence Detection System (Applied Biosystem). Real-time PCR was performed with each specific primer pair using SYBR Green PCR Master mix (MBI). For rrsA, primer pair rrsA F and rrsA R was used; for gadX, primer pair gadX F and gadX R was used; and for btuB, primer pair btub F and btub R was used (Table 5). The rrsA of 16S rRNA was chosen as the normalizing gene. The expression levels of gadX and btuB of cells grown in medium with different pH and different growth were compared.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
GSL designed and performed most of the experiments, analyzed data and wrote the manuscript. STH designed, supervised all the experiments, analyzed data and wrote the manuscript. KFC provided ColE7 for colicin assay and gave suggestions. PHL provided the antibodies against BtuB, TolQ, TolR, TolA, TolB, Pal, and OmpF for this research. WJS and WSH gave suggestions and analyzed data for this research.
All the authors have read and approved the final manuscript.
Contributor Information
Guang-Sheng Lei, Email: d87108@ym.edu.tw.
Wan-Jr Syu, Email: wjsyu@ym.edu.tw.
Po-Huang Liang, Email: phliang@gate.sinica.edu.tw.
Kin-Fu Chak, Email: kfchak@ym.edu.tw.
Wensi S Hu, Email: huws@ym.edu.tw.
Shiau-Ting Hu, Email: tingnahu@ym.edu.tw.
Acknowledgements
We thank Dr. Chao-Hung Lee for discussion and critical editing of this manuscript. This work was supported by grants from Ministry of Education, Aim for the Top University Plan (96A-D-T130, 97A-C-T130, 98A-C-T131, and 99A-C-T130) to S.-T. H, and the National Science Council, Taiwan R. O. C. (NSC92-2321-B-010-007, NSC93-2321-B-010-008, and NSC94-2321-B-010-002) to S.-T. H.
References
- Heller K, Mann BJ, Kadner RJ. Cloning and expression of the gene for the vitamin B12 receptor protein in the outer membrane of Escherichia coli. J Bacteriol. 1985;11(3):896–903. doi: 10.1128/jb.161.3.896-903.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth JR, Lawrence JG, Bobik TA. Cobalamin (coenzyme B12): synthesis and biological significance. Annu Rev Microbiol. 1996;11:137–181. doi: 10.1146/annurev.micro.50.1.137. [DOI] [PubMed] [Google Scholar]
- Bradbeer C, Woodrow ML, Khalifah LI. Transport of vitamin B12 in Escherichia coli: common receptor system for vitamin B12 and bacteriophage BF23 on the outer membrane of the cell envelope. J Bacteriol. 1976;11(3):1032–1039. doi: 10.1128/jb.125.3.1032-1039.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundrigan MD, Kadner RJ. Altered cobalamin metabolism in Escherichia coli btuR mutants affects btuB gene regulation. J Bacteriol. 1989;11(1):154–161. doi: 10.1128/jb.171.1.154-161.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante-Semerena JC, Suh SJ, Roth JR. cobA function is required for both de novo cobalamin biosynthesis and assimilation of exogenous corrinoids in Salmonella typhimurium. J Bacteriol. 1990;11(1):273–280. doi: 10.1128/jb.172.1.273-280.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouzet J, Levy-Schil S, Cameron B, Cauchois L, Rigault S, Rouyez MC, Blanche F, Debussche L, Thibaut D. Nucleotide sequence and genetic analysis of a 13.1-kilobase-pair Pseudomonas denitrificans DNA fragment containing five cob genes and identification of structural genes encoding Cob(I)alamin adenosyltransferase, cobyric acid synthase, and bifunctional cobinamide kinase-cobinamide phosphate guanylyltransferase. J Bacteriol. 1991;11(19):6074–6087. doi: 10.1128/jb.173.19.6074-6087.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nou X, Kadner RJ. Coupled changes in translation and transcription during cobalamin-dependent regulation of btuB expression in Escherichia coli. J Bacteriol. 1998;11(24):6719–6728. doi: 10.1128/jb.180.24.6719-6728.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nou X, Kadner RJ. Adenosylcobalamin inhibits ribosome binding to btuB RNA. Proc Natl Acad Sci USA. 2000;11(13):7190–7195. doi: 10.1073/pnas.130013897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, Breaker RR. Genetic control by a metabolite binding mRNA. Chem Biol. 2002;11(9):1043–1049. doi: 10.1016/S1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- Richter-Dahlfors AA, Andersson DI. Cobalamin (vitamin B12) repression of the Cob operon in Salmonella typhimurium requires sequences within the leader and the first translated open reading frame. Mol Microbiol. 1992;11(6):743–749. doi: 10.1111/j.1365-2958.1992.tb01524.x. [DOI] [PubMed] [Google Scholar]
- Ravnum S, Andersson DI. Vitamin B12 repression of the btuB gene in Salmonella typhimurium is mediated via a translational control which requires leader and coding sequences. Mol Microbiol. 1997;11(1):35–42. doi: 10.1046/j.1365-2958.1997.1761543.x. [DOI] [PubMed] [Google Scholar]
- Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J Biol Chem. 2003;11(42):41148–41159. doi: 10.1074/jbc.M305837200. [DOI] [PubMed] [Google Scholar]
- Nahvi A, Barrick JE, Breaker RR. Coenzyme B12 riboswitches are widespread genetic control elements in prokaryotes. Nucleic Acids Res. 2004;11(1):143–150. doi: 10.1093/nar/gkh167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S, Castanie-Cornet MP, Foster JW, Crawford JA, Brinkley C, Kaper JB. An activator of glutamate decarboxylase genes regulates the expression of enteropathogenic Escherichia coli virulence genes through control of the plasmid-encoded regulator, Per. Mol Microbiol. 2001;11(5):1133–1150. doi: 10.1046/j.1365-2958.2001.02570.x. [DOI] [PubMed] [Google Scholar]
- Ma Z, Richard H, Tucker DL, Conway T, Foster JW. Collaborative regulation of Escherichia coli glutamate-dependent acid resistance by two AraC-like regulators, GadX and GadW (YhiW) J Bacteriol. 2002;11(24):7001–7012. doi: 10.1128/JB.184.24.7001-7012.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanie-Cornet MP, Penfound TA, Smith D, Elliott JF, Foster JW. Control of acid resistance in Escherichia coli. J Bacteriol. 1999;11(11):3525–3535. doi: 10.1128/jb.181.11.3525-3535.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hommais F, Krin E, Laurent-Winter C, Soutourina O, Malpertuy A, Le Caer JP, Danchin A, Bertin P. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol Microbiol. 2001;11(1):20–36. doi: 10.1046/j.1365-2958.2001.02358.x. [DOI] [PubMed] [Google Scholar]
- Ma Z, Richard H, Foster JW. pH-Dependent modulation of cyclic AMP levels and GadW-dependent repression of RpoS affect synthesis of the GadX regulator and Escherichia coli acid resistance. J Bacteriol. 2003;11(23):6852–6859. doi: 10.1128/JB.185.23.6852-6859.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tramonti A, Visca P, De Canio M, Falconi M, De Biase D. Functional characterization and regulation of gadX, a gene encoding an AraC/XylS-like transcriptional activator of the Escherichia coli glutamic acid decarboxylase system. J Bacteriol. 2002;11(10):2603–2613. doi: 10.1128/JB.184.10.2603-2613.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman SR, Small PL. Transcriptional expression of Escherichia coli glutamate-dependent acid resistance genes gadA and gadBC in an hns rpoS mutant. J Bacteriol. 2003;11(15):4644–4647. doi: 10.1128/JB.185.15.4644-4647.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Biase D, Tramonti A, Bossa F, Visca P. The response to stationary-phase stress conditions in Escherichia coli: role and regulation of the glutamic acid decarboxylase system. Mol Microbiol. 1999;11(6):1198–1211. doi: 10.1046/j.1365-2958.1999.01430.x. [DOI] [PubMed] [Google Scholar]
- Homola AD, Dekker EE. Decarboxylation of gamma-hydroxyglutamate by glutamate decarboxylase of Escherichia coli (ATCC 11246) Biochemistry. 1967;11(8):2626–2634. doi: 10.1021/bi00860a046. [DOI] [PubMed] [Google Scholar]
- Giangrossi M, Zattoni S, Tramonti A, De Biase D, Falconi M. Antagonistic role of H-NS and GadX in the regulation of the glutamate decarboxylase-dependent acid resistance system in Escherichia coli. J Biol Chem. 2005;11(22):21498–21505. doi: 10.1074/jbc.M413255200. [DOI] [PubMed] [Google Scholar]
- Yamashino T, Ueguchi C, Mizuno T. Quantitative control of the stationary phase-specific sigma factor, sigma S, in Escherichia coli: involvement of the nucleoid protein H-NS. Embo J. 1995;11(3):594–602. doi: 10.1002/j.1460-2075.1995.tb07035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth M, Marschall C, Muffler A, Fischer D, Hengge-Aronis R. Role for the histone-like protein H-NS in growth phase-dependent and osmotic regulation of sigma S and many sigma S-dependent genes in Escherichia coli. J Bacteriol. 1995;11(12):3455–3464. doi: 10.1128/jb.177.12.3455-3464.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge-Aronis R. Back to log phase: sigma S as a global regulator in the osmotic control of gene expression in Escherichia coli. Mol Microbiol. 1996;11(5):887–893. doi: 10.1046/j.1365-2958.1996.511405.x. [DOI] [PubMed] [Google Scholar]
- Ma Z, Gong S, Richard H, Tucker DL, Conway T, Foster JW. GadE (YhiE) activates glutamate decarboxylase-dependent acid resistance in Escherichia coli K-12. Mol Microbiol. 2003;11(5):1309–1320. doi: 10.1046/j.1365-2958.2003.03633.x. [DOI] [PubMed] [Google Scholar]
- Opdyke JA, Kang JG, Storz G. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J Bacteriol. 2004;11(20):6698–6705. doi: 10.1128/JB.186.20.6698-6705.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker DL, Tucker N, Ma Z, Foster JW, Miranda RL, Cohen PS, Conway T. Genes of the GadX-GadW regulon in Escherichia coli. J Bacteriol. 2003;11(10):3190–3201. doi: 10.1128/JB.185.10.3190-3201.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Masi DR, White JC, Schnaitman CA, Bradbeer C. Transport of vitamin B12 in Escherichia coli: common receptor sites for vitamin B12 and the E colicins on the outer membrane of the cell envelope. J Bacteriol. 1973;11(2):506–513. doi: 10.1128/jb.115.2.506-513.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley MA. Positive selection for colicin diversity in bacteria. Mol Biol Evol. 1993;11(5):1048–1059. doi: 10.1093/oxfordjournals.molbev.a040054. [DOI] [PubMed] [Google Scholar]
- James R, Kleanthous C, Moore GR. The biology of E colicins: paradigms and paradoxes. Microbiology. 1996;11(Pt 7):1569–1580. doi: 10.1099/13500872-142-7-1569. [DOI] [PubMed] [Google Scholar]
- Kadner RJ. Repression of synthesis of the vitamin B12 receptor in Escherichia coli. J Bacteriol. 1978;11(3):1050–1057. doi: 10.1128/jb.136.3.1050-1057.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurisu G, Zakharov SD, Zhalnina MV, Bano S, Eroukova VY, Rokitskaya TI, Antonenko YN, Wiener MC, Cramer WA. The structure of BtuB with bound colicin E3 R-domain implies a translocon. Nat Struct Biol. 2003;11(11):948–954. doi: 10.1038/nsb997. [DOI] [PubMed] [Google Scholar]
- Lazdunski C, Bouveret E, Rigal A, Journet L, Lloubes R, Benedetti H. Colicin import into Escherichia coli cells requires the proximity of the inner and outer membranes and other factors. Int J Med Microbiol. 2000;11(4-5):337–344. doi: 10.1016/S1438-4221(00)80037-5. [DOI] [PubMed] [Google Scholar]
- Lazdunski CJ, Bouveret E, Rigal A, Journet L, Lloubes R, Benedetti H. Colicin import into Escherichia coli cells. J Bacteriol. 1998;11(19):4993–5002. doi: 10.1128/jb.180.19.4993-5002.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isnard M, Rigal A, Lazzaroni JC, Lazdunski C, Lloubes R. Maturation and localization of the TolB protein required for colicin import. J Bacteriol. 1994;11(20):6392–6396. doi: 10.1128/jb.176.20.6392-6396.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanteur D, Schirmer T, Fourel D, Simonet V, Rummel G, Widmer C, Rosenbusch JP, Pattus F, Pages JM. Structural and functional alterations of a colicin-resistant mutant of OmpF porin from Escherichia coli. Proc Natl Acad Sci USA. 1994;11(22):10675–10679. doi: 10.1073/pnas.91.22.10675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY; 1972. [Google Scholar]
- Franklund CV, Kadner RJ. Multiple transcribed elements control expression of the Escherichia coli btuB gene. J Bacteriol. 1997;11(12):4039–4042. doi: 10.1128/jb.179.12.4039-4042.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundrigan MD, Koster W, Kadner RJ. Transcribed sequences of the Escherichia coli btuB gene control its expression and regulation by vitamin B12. Proc Natl Acad Sci USA. 1991;11(4):1479–1483. doi: 10.1073/pnas.88.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tramonti A, De Canio M, De Biase D. GadX/GadW-dependent regulation of the Escherichia coli acid fitness island: transcriptional control at the gadY-gadW divergent promoters and identification of four novel 42 bp GadX/GadW-specific binding sites. Mol Microbiol. 2008;11(4):965–982. doi: 10.1111/j.1365-2958.2008.06458.x. [DOI] [PubMed] [Google Scholar]
- Larson TJ, Cantwell JS, van Loo-Bhattacharya AT. Interaction at a distance between multiple operators controls the adjacent, divergently transcribed glpTQ-glpACB operons of Escherichia coli K-12. J Biol Chem. 1992;11(9):6114–6121. [PubMed] [Google Scholar]
- Yang B, Gerhardt SG, Larson TJ. Action at a distance for glp repressor control of glpTQ transcription in Escherichia coli K-12. Mol Microbiol. 1997;11(3):511–521. doi: 10.1046/j.1365-2958.1997.3651733.x. [DOI] [PubMed] [Google Scholar]
- Bradbeer C. The proton motive force drives the outer membrane transport of cobalamin in Escherichia coli. J Bacteriol. 1993;11(10):3146–3150. doi: 10.1128/jb.175.10.3146-3150.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashket ER. Proton motive force in growing Streptococcus lactis and Staphylococcus aureus cells under aerobic and anaerobic conditions. J Bacteriol. 1981;11(1):369–376. doi: 10.1128/jb.146.1.369-376.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu FM, Chak KF. Two overlapping SOS-boxes in ColE operons are responsible for the viability of cells harboring the Col plasmid. Mol Gen Genet. 1996;11(4):407–411. doi: 10.1007/BF02172368. [DOI] [PubMed] [Google Scholar]
- Masui Y, Coleman J, Inouye M. In: Multipurose expression cloning vehicles in E. coli In experimental manipulation of gene expression. Inouye M, editor. Academic press, Inc., NY; 1983. [Google Scholar]
- Pan YH, Liao CC, Kuo CC, Duan KJ, Liang PH, Yuan HS, Hu ST, Chak KF. The critical roles of polyamines in regulating ColE7 production and restricting ColE7 uptake of the colicin-producing Escherichia coli. J Biol Chem. 2006;11(19):13083–13091. doi: 10.1074/jbc.M511365200. [DOI] [PubMed] [Google Scholar]