Abstract
Background
In response to environmental iron concentrations, many bacteria coordinately regulate transcription of genes involved in iron acquisition via the ferric uptake regulation (Fur) system. The genome of Nitrosomonas europaea, an ammonia-oxidizing bacterium, carries three genes (NE0616, NE0730 and NE1722) encoding proteins belonging to Fur family.
Results
Of the three N. europaea fur homologs, only the Fur homolog encoded by gene NE0616 complemented the Escherichia coli H1780 fur mutant. A N. europaea fur:kanP mutant strain was created by insertion of kanamycin-resistance cassette in the promoter region of NE0616 fur homolog. The total cellular iron contents of the fur:kanP mutant strain increased by 1.5-fold compared to wild type when grown in Fe-replete media. Relative to the wild type, the fur:kanP mutant exhibited increased sensitivity to iron at or above 500 μM concentrations. Unlike the wild type, the fur:kanP mutant was capable of utilizing iron-bound ferrioxamine without any lag phase and showed over expression of several outer membrane TonB-dependent receptor proteins irrespective of Fe availability.
Conclusions
Our studies have clearly indicated a role in Fe regulation by the Fur protein encoded by N. europaea NE0616 gene. Additional studies are required to fully delineate role of this fur homolog.
Background
The molecular basis for the coordinated regulation of iron acquisition systems by iron was first described for Escherichia coli [1]. Several bacteria are now known to regulate their iron acquisition systems via Fur (ferric uptake regulator) [2-5]. Fur is a sequence-specific DNA-binding protein that acts mainly as a negative regulator of transcription in vivo by complexing with ferrous (Fe2+) ion to repress the expression of iron-regulated genes [6]. Fur also activates the expression of many genes by either indirect or direct mechanisms [7]. Mutations in the fur gene resulted in constitutive expression of siderophores and outer membrane Fe3+-siderophore receptors potentially required for iron uptake [8].
Nitrosomonas europaea is an aerobic chemolithoautotroph that uses NH3 and CO2 for growth [9]. Mechanisms for iron transport are essential to this bacterium for maintaining the many cytochromes and other heme-binding proteins involved in ammonia metabolism [10,11]. The genome of N. europaea has 4% of its genes dedicated for iron acquisition, but no evidence for siderophore production was found [9]. N. europaea's inability to produce siderophores in Fe-replete or Fe-limited media was further confirmed by universal Chrome Azurol S assay [12]. N. europaea responds to iron limitation by elevating production of Fe3+-siderophore receptors normally repressed under iron-replete conditions [13,14]. Several N. europaea iron-repressible genes contain sequences similar to the E. coli Fur box (unpublished data) in their promoter regions; hence it is likely that a Fur-like repressor regulates iron uptake genes in N. europaea as well. Indeed, sequence annotation of N. europaea genome revealed three genes encoding fur homologs (NE0616, NE0730, NE1722) that contain characteristic Fur domains [9].
Multiple fur homologs have been described for several bacteria. Different species have a variable number of genes bearing the Fur domain. For example, E. coli [15] has two, Bacillus subtilis [16], Mycobacterium smegmatis have three, Staphylococcus aureus and some species of Brucella have four and Thermoanaerobacter tengcongensis has five fur homologs [17]. The apparent redundancy in fur homologs has been clarified by a considerable amount of experimental data obtained from genetic and biochemical analysis in bacteria such as E. coli and B. subtilis [15,16,18-20]. The experimental data suggests that the Fur protein family has several subclasses with different functions [19]. The major Fe-sensing Fur subclass is mainly involved in the control of iron homeostasis [21]. A second subclass controls the expression of genes involved in the response of bacteria to oxidative stress (i.e. PerR), but it does not appear to be involved in the cellular response to iron [16]. A third subclass called Zur (zinc uptake regulator) controls the uptake of zinc in E. coli [15,20] and B. subtilis [18].
The Fe-sensing Fur protein has been extensively studied and is shown to act as a global regulator in response to environmental iron concentration due to its involvement in the regulation of activities as varied as the acid tolerance response, the oxidative stress response, metabolic pathways, and virulence factors [6]. In this study, we aimed to characterize the regulatory role of a fur homolog from N. europaea. Using genetic complementation studies, we demonstrated that one fur homolog (NE0616) out of three in N. europaea encoded a functional Fur protein. Here we report the construction of the N. europaea fur promoter knockout mutant (fur:kanP) strain, its effect on the expression of Fe-regulated proteins and the physiology of N. europaea.
Results
Sequence analysis of N. europaea fur homologs
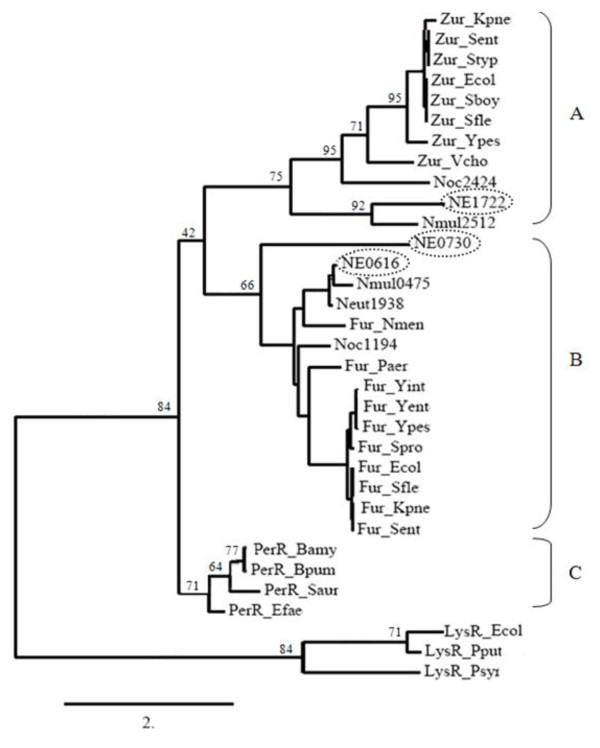
The three N. europaea Fur-like repressors encoded by NE0616, NE0730, NE1722 are only distantly related to each other with 25% to 35% amino acid identity. The Fur homolog encoded by NE0616 is most similar (~84% similar to E. coli Fur protein) in sequence to various Gram-negative Fe-sensing Fur proteins. The publication of the crystal structure of the Pseudomonas aeruginosa Fur protein provided considerable insight into its 2 metal binding sites. Binding Site 1 represents the putative iron binding regulatory site and is coordinated by amino acids H86, D88, E107, and H124 and Site 2 is coordinated by H32, E80, H89 and E100 [19]. All these residues are conserved only in the N. europaea NE0616 Fur homolog but not in Fur homologs encoded by NE0730 and NE1722 (Figure 1). Phylogenetic analysis of Fur homolog coding sequences from N. europaea with Fur proteins from other bacteria placed NE0616 in the group B comprised of Fe-sensing Fur proteins, NE1722 in the group A comprised of Zn-sensing Zur proteins. Surprisingly, NE0730 Fur homolog was also placed in group B. No Fur homologs of N. europaea grouped with peroxide sensing PerR proteins i.e., in group C (Figure 2).
Figure 1.
Alignment of N. europaea Fur homolog coding sequences with E. coli and P. aeruginosa Fur proteins using ClustalW [31]. Identical residues are shaded black, with similar residues shaded grey. Metal binding site 1 residues are indicated with circles, and site 2 residues are indicated with triangles, as identified from the crystal structure of P. aeruginosa Fur. Residues indicated by straight line highlight a motif thought to be involved in DNA binding.
Figure 2.
Maximum-Likelihood tree of the Fur homologs. Phylogenetic tree of Fur encoding sequences generated by Phyml analysis. The numbers beside nodes are the percentages of bootstrap values calculated for 200 replicates: The three groups - A, B and C - mentioned in the text are indicated on the right side of the tree. Bamy, Bacillus amyloliquefaciens; Bpum, Bacillus pumilus; Ecol, Escherichia coli; Efae, Enterococcus faecalis; Kpne, Klebsiella pneumoniae; Nmen, Neisseria meningitidis; Paer, Pseudomonas aeruginosa; Pput, Pseudomonas putida; Psyr, Pseudomonas syringae; Saur, Staphylococcus aureus; Sboy, Shigella boydii; Sent, Salmonella enterica; Sfle, Shigella flexneri; Spro, Serratia proteamaculans;Styp, Salmonella typhimurium; Vcho, Vibrio cholerae; Yent, Yersinia enterocolitica; Yint, Yersinia intermedia; Ypes, Yersinia pestis; Ypse, Yersinia pseudotuberculosis; NE, Nitrosomonas europaea; Neut, Nitrosomonas eutropha; Nmul, Nitrosospira multiformis; Noc, Nitrosococcus oceanii.
Based on well-studied model systems, expression of the fur gene itself is iron regulated and there is strong evidence that this is through a mechanism of autoregulation [34,35]. Fur recognizes and binds specifically to a DNA sequence, known as the Fur box, that is typically located in proximity to the -10 and/or -35 promoter elements of target genes [6]. Analysis of several Fur-binding sites allowed the early definition of a 19-bp inverted repeat consensus Fur box in E. coli [6]. Since then, canonical Fur boxes have been described in several bacteria such as P. aeruginosa [36], Neisseria gonorrhoeae [37] and Vibrio cholerae [38]. The canonical Fur box identified by B. subtilis Fur revealed a different conserved 15-bp (7-1-7) inverted repeat present twice within this 19-bp consensus sequence [39]. We have used an in silico approach, fed with experimentally confirmed N. europaea Fur boxes (unpublished data), to identify candidate Fur-binding sites in promoter regions of all 3 N. europaea fur homologs. A potential Fur box (5'-TAATAATACGTATCTTTAT-3') in the promoter region of NE0616 gene, -121 bp upstream of the proposed initiation of translation of the fur gene was found. We were unable to find potential Fur boxes in the promoter region of the other N. europaea fur homologs, NE0730 and NE1722.
Complementation of an E. coli fur mutant by N. europaea fur homologs
In order to determine which fur homolog of N. europaea encodes the Fe-sensing Fur protein, pFur616, pFur730 and pFur1722 plasmids (Table 1) were used to transform the E. coli fur mutant H1780 [40]. E. coli H1780 strain was engineered to be fur deficient and to include the Fur-regulated gene fiu fused to a promoterless lacZ gene. This reporter gene, fiu-lacZ, cannot be repressed in this strain due to the fur mutation, and therefore the gene encoding the enzyme β-galactosidase is constitutively expressed and the strain always shows Lac+ phenotype [40]. The pFur616-kanC (Table 1) plasmid carrying kanamycin resistance cassette (Kmr) insertion in the C-terminal region of NE0616 gene was used to transform H1780 as a negative control.
Table 1.
Bacterial strains, plasmids and primers used in this study
| Strains or plasmid | Description | Reference |
|---|---|---|
| E. coli | ||
| DH5⟨ | F2ø80dlacZ⊗M15 endA1 recA1 gyrA96 thi-1 hsdR17(rK- mK+) supE44 relA1 deoR Δ(lacZYA-argF)U169 | [56] |
| H1717 | aroB fhuF::λplacMu | [40] |
| H1717 (pFur616) | E. coli H1717 carrying pFur616 | This study |
| H1717 (pFur616-kanP) | E. coli H1717 carrying pFur616-kanP | This study |
| H1717 (pFur616-kanC) | E. coli H1717 carrying pFur616-kanC | This study |
| H1780 | araD139∆aargF-lacU169rpsL150 relA1 flbB5301deoC1 ptsF25 rbsR fiu::lacZ fusion lacking Fur | [40] |
| H1780 (pFur616) | E. coli H1780 carrying pFur616 | This study |
| H1780 (pFur616-kanP) | E. coli H1780 carrying pFur616-kanP | This study |
| H1780 (pFur616-kanC) | E. coli H1780 carrying pFur616-kanC | This study |
| H1780 (pFur730) | E. coli H1780 carrying pFur730 | This study |
| H1780 (pFur1722) | E. coli H1780 carrying pFur1722 | This study |
| N. europaea | ||
| ATCC 19178 | Wild type | ATCC |
| fur:kanP | Insertion of kan cassette in the furbox upstream of NE0616 gene | This study |
| Plasmids | ||
| pGEM-T Easy | Vector for cloning PCR products; Ampr | Promega |
| pFur616 | pGEM-T Easy vector containing NE0616 u&d* region | This study |
| pFur616-kanP | In vitro transposon mutagenesis of pFur616 with EZ-Tn5 <KAN-2> with kan cassette insertion in fur box located in promoter region of NE0616 | This study |
| pFur616-kanC | In vitro transposon mutagenesis of pFur616 with EZ-Tn5 <KAN-2> with kan cassette insertion in C-terminal region of NE0616 | This study |
| pFur730 | pGEM-T Easy vector containing NE0730 u&d region | This study |
| pFur1722 | pGEM-T Easy vector containing NE1722 u&d region | This study |
| Primers used for cloning, mutagenesis and mutant confirmation | ||
| NE0616u&d-1 | 5'-ATCCTGGAAGAAAACGGTCA-3' | This study |
| NE0616u&d-2 | 5'-TGCAGGTTTCAAACGAAAAA-3' | This study |
| NE0730u&d-1 | 5'-TTTCAGACGTTGCTGACAAAA-3' | This study |
| NE0730u&d-2 | 5'-TCATTTTGGCTGTTCATTTCA-3' | This study |
| NE1722u&d-1 | 5'-TATGGCTTACGGAAAACGGTA-3' | This study |
| NE1722u&d-2 | 5'-ACAAAAACAGACACGGAGGAA-3' | This study |
*- u&d denotes upstream & downstream region.
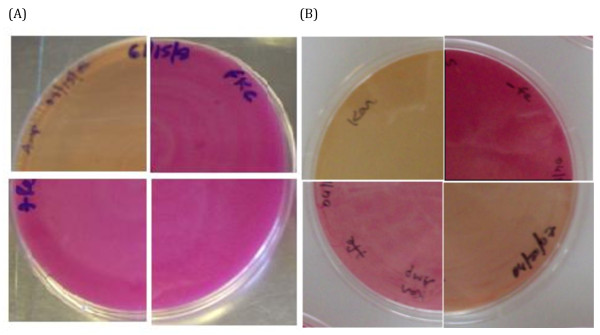
All strains evaluated for Lac phenotype were grown on McConkey Lactose plates with 30 μM iron supplement, since iron is required to ensure that Fur is functional as a repressor [6]. In these studies, E. coli H1780, H1780 (pFur616), H1780 (pFur616-kanC), H1780 (pFur730) and H1780 (pFur1722) strains were compared. Lac+ phenotype was observed for E. coli H1780 whether grown in the presence or absence of added Fe supplement as predicted since it is deficient in Fur protein (data not shown). Complementation of E. coli H1780 with pFur616 rescued the Fur defect of this strain and resulted in the repression of transcription of the fiu-lacZ reporter gene, as shown by the Lac- phenotype (Figure 3A; upper left quadrant). When pFur616-kanC plasmid containing the disrupted NE0616 gene, was transformed into the E. coli H1780 mutant, Lac+ phenotype was maintained (Figure 3A; upper right quadrant). When pFur730 and pFur1722 plasmids containing the N. europaea fur homologs NE0730 and NE1722 were transformed separately into E. coli H1780 strain, Lac+ phenotype was observed (Figure 3A; lower left and right quadrants). These results clearly demonstrate that the N. europaea NE0616 fur homolog is expressed in E. coli in a functional form and is capable of regulating the Fur-dependent fiu promoter in H1780. The other N. europaea fur homologs (NE0730 and NE1722) were not capable of regulating the fiu promoter in H1780. NE0616 is here after referred to as N. europaea fur.
Figure 3.
Fur Titration Assays (FURTA). (A) Complementation of an E. coli fur mutant H1780 by N. europaea Fur homologs.E. coli H1780 (pFur616)-upper left quadrant; H1780 (pFur616-kanC)-upper right quadrant; H1780 (pFur730)-lower left quadrant; H1780 (pFur1722)-lower right quadrant plated on McConkey medium with 30 μM Fe supplement and grown at 37°C for 24 hrs. (B) E. coli H1717 plated on McConkey medium with 30 μM Fe supplement-upper left quadrant, no Fe supplement-upper right quadrant; H1717 (pFur616)-lower left quadrant; H1717 (pFur616-kanP)-lower right quadrant plated on McConkey medium with 30 μM Fe supplement and grown at 37°C for 24 hrs.
The N. europaea fur promoter is repressed by Fur
Several studies have employed E. coli H1717 strain to allow the detection of iron-regulated promoters in bacteria such as E. coli and Salmonella typhimurium [41,42]. E. coli H1717 strain has a chromosomal iron-regulated fhuF promoter fused to lacZ. This fusion is exceptionally sensitive to small changes in iron concentration because of the weak affinity of the fhuF promoter for the Fur-Fe2+ repression complex. Introduction of a multi-copy plasmid carrying Fur-binding sites into the test strain depletes the intracellular Fur pool. This gives rise to the dissociation of the repressor from the fusion promoter, thereby allowing expression of enzyme β-galactosidase. We have screened plasmids pFur616 carrying intact Fur box and pFur616-kanP carrying disrupted Fur box using E. coli H1717 strain to determine NE0616 Fur box functionality. The pFur616-kanC plasmid (Table 1) carrying Kmr insertion in the C-terminal region of NE0616 gene was also used to transform E. coli H1717 as a positive control.
In these studies, E. coli H1717 in the presence and absence of Fe supplement, H1717 (pFur616), H1717 (pFur616-kanP) and H1717 (pFur616-kanC) strains were compared. Lac- phenotype was observed for E. coli H1717 when grown in the presence of 30 μM Fe supplement, since it does not carry any multi-copy plasmid with a functional Fur box on it (Figure 3B upper left quadrant). Lac+ phenotype was observed when H1717 was grown with no added Fe supplement, since there is not enough Fe to suppress fhuF-lacZ fusion (Figure 3B; upper right quadrant). When pFur616 carrying putative Fur box was transformed into E. coli H1717 and the resulting strain was grown in presence of 30 μM Fe supplement, it resulted in derepression of the fhuF-lacZ reporter gene, as shown by the Lac+ phenotype (Figure 3B; lower left quadrant). This result indicates that the predicted Fur box is functional and must have titrated the intracellular Fur-Fe pool. When a pFur616-kanP plasmid containing the disrupted NE0616 Fur box, was transformed into the E. coli H1717 strain, Lac- phenotype was restored (Figure 3B; lower right quadrant) indicating that the Kmr insertion led to disruption of Fur box functionality. When a pFur616-kanC plasmid containing Kmr insertion in the C-terminal region of NE0616 gene was transformed into E. coli H1717 strain, Lac+ phenotype was observed (data not shown) indicating that Kmr in C-terminal region of NE0616 did not affect its Fur box functionality. These results demonstrate that the promoter of N. europaea NE0616 fur homolog carries a Fur box and it is functional as recognized by E. coli Fur protein.
Isolation of the N. europaea fur:kanP mutant strain
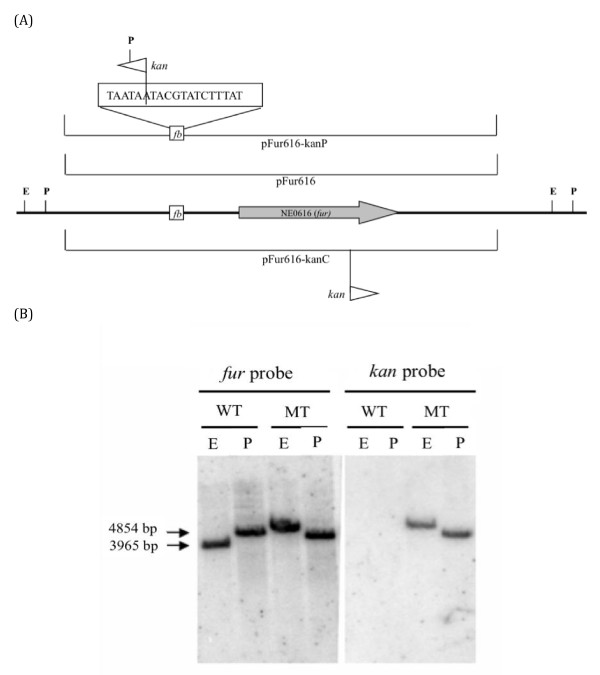
To address the physiological role fur plays in N. europaea, we attempted to generate an N. europaea fur null mutant but were unsuccessful. However, we were successful in isolating an N. europaea fur:kanP mutant strain with Kmr inserted in the Fur box located in the promoter region of NE0616 gene (Figure 4A). The pFur616-kanP plasmid was electroporated into N. europaea wild-type cells. The fur:kanP mutant was obtained through homologous recombination and confirmed by PCR (data not shown) and Southern hybridization (Figure 4B). The fur probe detected a 3.96 Kb EcoR1 fragment and a 4.85 Kb Pst1 fragment in wild type and a ~ 5 Kb EcoR1 fragment and a ~ 4.3 Kb Pst1 fragment (calculated size based on the DNA sequences) in fur:kanP mutant strain. The kanamycin-cassette probe detected the same ~ 5 Kb EcoR1 fragment and the ~ 4.3 Kb Pst1 fragment in fur:kanP mutant but not in the wild type. These results confirm that a single copy of Kmr was correctly inserted in the Fur box located in the promoter region of NE0616 gene of the N. europaea genome (Figure 4A). A fur transcript was not detected in the fur:kanP mutant by either RT-PCR or qRT-PCR analysis (up to 28 cycles) indicating the inactivation of fur gene due to Kmr insertion in its promoter region. Transcripts of ammonia monooxygenase C (amoC) component used as positive control both for the efficiency of the RT-PCR procedure and for RNA and cDNA recovery showed no significant difference in expression in wild type and the fur:kanP mutant (data not shown).
Figure 4.
In vitro transposon mutagenesis scheme and mutant confirmation. (A) The physical structure of a 5,810-bp fragment of the N. europaea chromosome is shown in the center (heavy black line), with positions of NE0616 (fur) gene shown as grey arrow, the fur box (fb) located in NE0616 promoter region shown as white rectangle. The regions covered by the plasmids pFur616, pFur616-kanP, pFur616-kanC whose DNA sequences were determined are shown as thin black lines with the names of the respective plasmids shown below each line. The position and relative orientation of each in vitro-constructed Tn5-Kan2 cassette insertion mutation are indicated by a flag on the lines. The restriction endonuclease sites P (Pst1) and E (EcoR1) used for Southern blot confirmation are indicated. (B) Verification of mutagenesis of fur:kanP in N. europaea by Southern hybridization. Genomic DNA from the wild type (WT), fur:kanP mutant (MT) were digested with E (EcoRI) and P (Pst1), and probed with (left) fur ORF sequence and (right) kan sequence.
Effect of fur:kanP mutation on growth of N. europaea
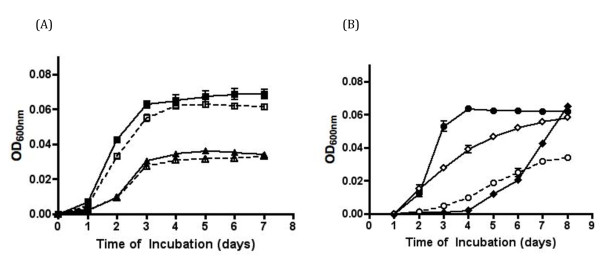
Growth of the N. europaea fur:kanP strain was compared to that of the wild-type strain in both Fe-replete (10 μM Fe) and Fe-limited (0.2 μM Fe) media. Surprisingly, there was no significant difference in growth of fur:kanP in both Fe-replete and Fe-limited media compared to the wild-type strain (Figure 5A). The fur:kanP mutant did not exhibit a growth advantage over the wild type when iron was limiting or show increased sensitivity to iron-induced redox stress when grown in the presence of Fe (up to 250 μM Fe; data not shown). However, growth of fur:kanP mutant was affected when grown in medium containing 500 μM Fe (Figure 5B). The mutant was unable to grow in media containing more than 500 μM Fe (data not shown). Growth of wild type was inhibited only when concentrations of Fe exceeded 1 mM [14].
Figure 5.
Growth curves of the N. europaea wild type (solid lines, filled symbols) and fur:kanP mutant (dotted lines, open symbols) as measured by OD. (A) Fe-replete (squares) and Fe-limited (triangles) medium. (B) 500 μM Fe medium (circles) and in Fe-limited medium with 10 μM ferrioxamine (diamonds). Data shown are means of triplicates, with variation less than 10%. The experiment was repeated several times and produced similar results. Error bars represent the standard error of the mean.
N. europaea can use the siderophore ferrioxamine for its iron uptake after a 3 to 4 day lag period suggesting that the ferrioxamine uptake system in N. europaea requires induction [13,14]. When N. europaea fur:kanP mutant was grown in Fe-limiting media containing ferrioxamine, there was no lag phase (Figure 5B) indicating that the ferrioxamine uptake system was already induced in the fur:kanP mutant.
Effect of fur:kanP mutation on induction of Fe-regulated outer membrane proteins in N. europaea
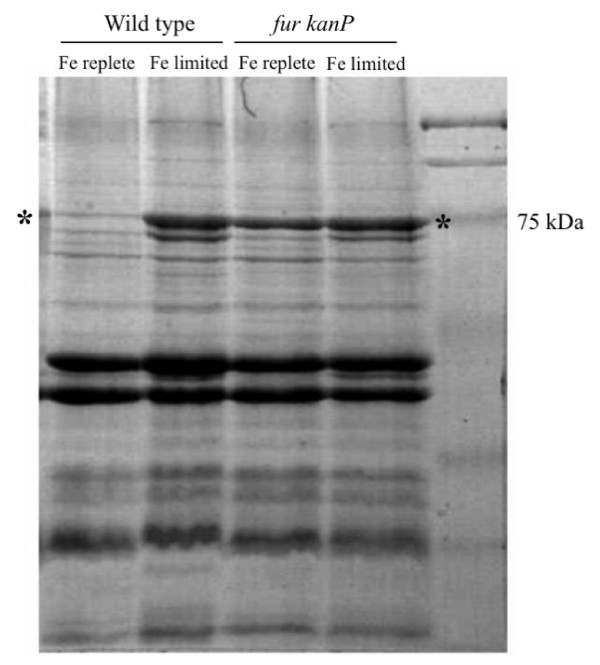
Previous studies have shown that N. europaea grown in Fe-limited medium stimulated expression of several Fe-regulated outer membrane proteins (TonB-dependent receptors) with molecular masses of ~ 80 kDa [13,14]. To determine whether the expression of these proteins was regulated by fur, the N. europaea wild type and the fur:kanP mutant strains were cultured in Fe-replete and Fe-limited media and their total outer membrane proteins were isolated. SDS-PAGE analysis of the outer membrane protein profiles demonstrated that fur:kanP mutant shared a major protein band (Figure 6) with wild type cells grown in Fe-limited media irrespective of the concentration of iron in the medium. This band contained several TonB-dependent OM Fe3+-siderophore receptors [13,14]. This result is consistent with the model in which the TonB-dependent receptors with putative roles in iron uptake are regulated by fur [6].
Figure 6.
SDS-PAGE Analysis of total membrane proteins. N. europaea wild type and fur:kanP mutant in Fe-replete (10 μM) (lanes 1, 3) and Fe-limited (0.2 μM) media (lanes 2, 4). Over-expression of proteins with molecular weights similar to outer membrane Fe-siderophore receptors indicated by * was observed in fur:kanP mutant in both Fe-replete and Fe-limited media.
Effect of fur:kanP mutation on Fe and heme c contents of N. europaea
Fur deficient mutants generally express iron transport systems constitutively (with respect to iron), and have increased free cellular iron levels (although total cellular iron levels are actually reduced, due to low levels of iron-storage and iron-containing proteins) [43,44]. To determine the effect of fur:kanP mutation on iron contents of N. europaea, wild type and fur:kanP mutant cells were cultured in Fe-replete and Fe-limited media and their total cellular iron contents were measured by ICP-OES analysis. N. europaea Fe-limited cells showed significantly (P-value <0.0001) lower total cellular iron contents compared to Fe-replete cells irrespective of the fur mutation as observed previously (Table 2) [14]. The fur:kanP mutant had 1.5-fold significantly (P-value <0.001) more total cellular iron than the wild-type cells when grown in Fe-replete media (Table 2). The total iron contents of wild type and the fur:kanP mutant did not show significant (P-value = 0.47) variation when grown in Fe-limited medium (Table 2). The fur:kanP mutation also influenced both the amount of soluble cytochromes produced and the proportion of iron distributed to cytochromes (Table 2). These data suggest that in N. europaea, Fur regulates the concentration of intracellular iron through modulation of iron acquisition and iron consumption, and that, in the absence of Fur, N. europaea is unable to regulate its iron acquisition.
Table 2.
Physiological characteristics of N. europae a wild type and fur:kanP mutant grown under Fe-replete (10 μM) and Fe-limited (0.2 μM) conditions*
| Physiological Characteristic | Wild type | fur:kanP mutant | ||
|---|---|---|---|---|
| Fe-replete | Fe-limited | Fe-replete | Fe-limited | |
|
Heme c content in cell's soluble fraction |
||||
| Heme c (nmol/ml culture) | 0.85 ± 0.02 | 0.38 ± 0.05 | 0.48 ± 0.02 | 0.21 ± 0.04 |
| Heme c (nmol/mg protein) | 7.77 ± 0.23 | 4.04 ± 0.53 | 5.67 ± 0.31 | 5.04 ± 0.91 |
| Whole Cell Fe content | ||||
| Fe (nmol/ml culture) | 1.36 ± 0.15 | 0.15 ± 0.01 | 2.04 ± 0.09 | 0.11 ± 0.01 |
| Fe (nmol/mg protein) | 90.4 ± 6.0 | 26.4 ± 2.0 | 136.2 ± 14.0 | 24.9 ± 3.0 |
| Cellular Fe concentration (mM) | 8.27 ± 0.94 | 1.99 ± 0.13 | 12.4 ± 0.6 | 1.98 ± 0.18 |
| Whole cell enzyme-catalyzed activity | ||||
| NH4+-dependent O2 consumption (nmol/(min × OD600 nm) | 94.5 ± 4.1 | 38.1 ± 6.0 | 88.2 ± 2.5 | 21.7 ± 0.6 |
| NH4+-dependent O2 consumption (nmol/(min × mg protein) | 1500 ± 63 | 779 ± 17 | 1446 ± 40 | 680 ± 18 |
| NH2OH-dependent O2 consumption (nmol/(min × OD600 nm) | 25.9 ± 0.2 | 10.9 ± 2.4 | 25.7 ± 4.8 | 4.6 ± 0.2 |
| NH2OH-dependent O2 consumption (nmol/(min × mg protein) | 412 ± 3.0 | 222 ± 5.0 | 421 ± 2.0 | 146 ± 6.0 |
*Data are means of triplicates, with variation less than 10%. The experiment was repeated several times and produced similar results. Data are means ± S.D.
Effect of fur:kanP mutation on NH4+- and NH2OH-dependent O2 uptake activities of N. europaea
As indicators of the overall cell activity, NH4+- and NH2OH-dependent O2 uptake rates in wild type and fur:kanP mutant cells grown in Fe-replete and Fe-limited media were measured. N. europaea Fe-limited cells showed significantly (P-value <0.0001) lower activities compared to Fe-replete cells irrespective of the fur mutation as observed previously (Table 2) [14]. The activities of wild type and fur:kanP mutant strains did not show significant (P-value ≤ 0.4) variation when grown in Fe-replete media (Table 2). The NH4+-dependent O2 uptake activities, which require both ammonia monooxygenase and hydroxylamine oxidoreductase activity, when measured at per mg basis were not affected; however the NH2OH-dependent O2 uptake activity, which requires hydroxylamine oxidoreductase, but not ammonia monooxygenase activity, was significantly (P-value <0.0001) two-fold lower in fur:kanP Fe-limited cells compared to wild type Fe-limited cells (Table 2). This result is consistent with our observation of lower heme contents in fur:kanP mutant than wild type. Hydroxylamine oxidoreductse contains 24 hemes per enzyme [45] and the lower NH2OH-dependent O2 uptake activity in Fe-limited cells of fur:kanP mutant than wild type might be due to the low availability of heme under Fe-limited conditions. This data also suggests that the fur:kanP mutation led to an improper balance of iron allocation in N. europaea.
Discussion
We provide several lines of evidence that the Fur homolog encoded by N. europaea gene NE0616 is the Fe-sensing Fur protein. First, we have shown that NE0616 shares all eight of the metal binding amino acid residues of P. aeruginosa Fur (Figure 1) [19] and that the Fur homolog encoded by NE0616 is clustered with Fe-sensing Fur proteins from other bacteria (Figure 2). An E. coli Fur titration assay (FURTA) system for Fur analysis was utilized as a second method to confirm that the cloned NE0616 fur encodes a functional protein. The H1780 (pFur616) strain carrying NE0616 fur homolog on a plasmid was evaluated for its ability to utilize lactose as described by Hantke et al., [40]. Utilization of lactose by H1780 (pFur616) strain was detected by color change of colonies from white to red on McConkey lactose plates indicating the formation of lactic acid. Lactose utilization was not detected when H1780 strain carrying plasmids pFur616-kanC, pFur730, pFur1722 were plated on McConkey lactose plates (Figure 3A).
One of the major limitations in our research on the role of Fur has been the inability to make a fur null mutant. Null mutations have been successfully isolated for E. coli [46,47], V. cholerae [48], Shigella flexneri [49], Neisseria meningitidis [34]. Unsuccessful attempts to isolate insertional null mutants were reported for P. aeruginosa [50], Pseudomonas putida [51], and N. gonorrhoeae [52]. To date, multiple attempts to generate a N. europaea fur mutant have been unsuccessful. Loss of the fur gene may be a lethal mutation in N. europaea, as occurs in some other gram-negative bacteria [50]. However, we were successful in generating an N. europaea fur promoter knockout mutant (fur:kanP) (Figure 4A). Southern analysis with probes internal to fur or the Kmr corroborated insertion of Kmr in the promoter region of the fur gene (Figure 4B) and hence fur:kanP mutant strain was selected for further analysis. Although we were unable to detect the NE0616 transcript in fur:kanP mutant strain by RT-PCR or qRT-PCR, it is possible that there is some leaky transcription of fur in our mutant strain, since it is a promoter knockout mutant. This could be the reason why we were able to generate a promoter knockout mutant but not a fur null mutant.
The effects of fur:kanP mutation on N. europaea were broad. Inactivation of the fur gene (resulting in deregulation of iron metabolism) increases sensitivity to redox stress when grown under iron-rich conditions in some bacteria such as E. coli [53]. The N. europaea, wild-type and the fur:kanP mutant strain showed similar growth patterns when grown in Fe-replete (10 μM Fe) and Fe-limited (0.2 μM Fe) media (Figure 5A). However, the fur:kanP mutant did not grow well when cultured in media containing 500 μM iron (Figure 5B). The fur:kanP mutant was unable to grow beyond 500 μM Fe concentrations while the wild-type strain was able to withstand iron concentrations up to 1 mM (data not shown). These results indicate that N. europaea Fur plays a role in regulating uptake of iron when present in excess and also probably helps to overcome oxidative stress.
Increased intracellular free iron is likely to result from deregulated iron uptake by the fur mutant [43]. The N. europaea fur:kanP mutant strain grown to mid exponential phase in Fe-replete media (10 μM Fe) contained 1.5-fold higher total cellular iron than that of the wild-type strain as measured by ICP-OES (Table 2). Our measurements of total acid-soluble non-heme iron cannot distinguish between free iron and iron bound to proteins. Hence we measured the heme contents of wild type and fur:kanP mutant strains and observed that the fur:kanP mutant had 1.4-fold lower heme contents compared to wild type (Table 2). In addition, the activity of iron-rich hydroxylamine oxidoreductase enzyme was lower in fur:kanP mutant strain (Table 2). These results indicated that the balance between acquiring enough iron and allocating it to various Fe-dependent proteins is lost in N. europaea fur:kanP mutant.
N. europaea protein profiles showed over expression of several outer membrane proteins upon Fe-limitation [13,14]. We have observed similar over expression of outer membrane proteins in N. europaea fur:kanP mutant (Figure 6 band indicated by *) irrespective of iron availability. These data are consistent with previous studies describing fur mutations in other bacterial species [54,55].
Conclusions
In summary, we have identified and characterized through insertional inactivation one of the three N. europaea Fur homologs. The N. europaea Fur protein encoded by gene NE0616 has extensive homology to the E. coli Fur protein and was able to complement an E. coli fur mutant. The N. europaea fur:kanP mutant is unable to regulate its intracellular iron and heme concentrations and appears to induce its iron acquisition systems constitutively. Additional studies are required to fully delineate the role of this N. europaea fur homolog.
Methods
Bacterial cultures and siderophore feeding experiments
N. europaea (ATCC 19178) was cultured as described with minor modifications [22,23]. The standard (Fe-replete) medium contained 10 μM Fe3+ (FeCl3) complexed with EDTA to prevent Fe precipitation. Fe-limited medium was made from reagent-grade chemicals, without addition of any Fe salt, and contained 0.2 μM Fe [14]. All media, buffers and other reagents were made in double-deionized water. All glassware was soaked in 1% HNO3 overnight, and then rinsed thoroughly with double-deionized water. Fe-free Desferal (deferoxamine/DFX mesylate) was purchased from Sigma (St. Louis, MO). Desferal was dissolved in double deionized water, filter sterilized, and added to Fe-limited medium in the siderophore feeding experiments. In this study 10 μM Desferal was used to ensure the complete chelation of Fe (0.2 μM) in the Fe-limited medium. N. europaea cultures were grown at 30°C on a rotary shaker, and mid-exponential-phase cells were collected by centrifugation and thorough washes for the analyses. E. coli DH5α, E. coli H1780 strain lacking fur gene, and E. coli H1717 strain were cultured on Luria-Bertani (LB) agar plates or in liquid LB medium in the presence of the appropriate antibiotic (ampicillin [100 μg ml-1] and/or kanamycin [20 μg ml-1]) under the conditions described above.
DNA preparation, PCR, cloning, mutagenesis and mutant isolation
General DNA preparation, restriction digestions and agarose gel electrophoresis were done as described by [24]. The three N. europaea fur homologs (Figure 1) were amplified by PCR using Taq DNA polymerase (Promega, Madison, WI) on an iCycler Thermal Cycler (Bio-Rad, Hercules, CA), as described by the manufacturers (see Table 1 for primers). The resulting DNA fragments were cloned into the pGEM-T Easy vector (Promega), sequenced to confirm that no mutations have been introduced and named pFur616, pFur730 and pFur1722 respectively. E. coli DH5α was used for plasmid amplification. For insertion of kanamycin resistance cassette (Kmr) into plasmid pFur616, the EZ::TN <KAN2> kit from Epicentre (Madison, WI) was used to insert a transposon conferring Kmr into the promoter region (pFur-kanP) and C-terminal region (pFur-kanC) of fur following the directions of the manufacturer. The insertion of the Kmr gene was localized by nucleotide sequence determination at 117 nt upstream of the ATG start codon of fur (pFur-kanP) and 312 nt downstream of the ATG start codon of fur (pFur-kanC) in plasmid pFur616. The pFur616-kanP plasmid construct with the Kmr insertion was introduced back into the N. europaea wild type cells by electroporation on the ElectroPorator (Invitrogen, Carlsbad, CA) at 1300 V, with a capacitance at 50 μF, and a load resistance at 500 Ω. Successful transformants were selected in liquid medium using kanamcyin sulfate (20 μg ml-1). Aliquots from these cultures were streaked onto Nylon disk membranes, which were placed on semisolid plates, to isolate clonal mutant strains, as described [25]. The mutant was verified by Southern analysis (Figure 4B, and Results). Southern blotting, labeling of DNA probes, hybridization and imaging were done as described previously [26]. Attempts to generate fur null mutant by using pFur-kanC construct were unsuccessful.
Fur Titration Assays (FURTA)
Plasmids (listed in Table 1) were introduced into E. coli H1717 and H1780 (fur inactivated) strains and lacZ expression was assessed by visualization of a change in colony color from white to red on MacConkey lactose plates (Difco) supplemented with 30 μM ferrous ammonium sulfate. Plates were examined after 24 h of growth at 37°C. The assays were performed in triplicate for each sample.
Determination of Fe and heme c contents and O2-uptake activities
Total Fe contents in thoroughly washed N. europaea cells were determined by the ferrozine assay following HNO3 (5%) digestion of cells at 100°C [27]. Measurements of Fe concentrations below 10 μM were made using a Teledyne Leeman Prodigy ICP-OES (Hudson, NH) at the W.M. Keck Collaboratory for Plasma Spectrometry, Oregon State University. Preparations of a cell-soluble fraction, and determination of heme contents following extraction with pyridine, were done as described [14,28]. Whole cell NH3-dependent and hydroxylamine dependent O2 uptake activities were measured as described [14,29]. The significance (P-values) for the physiological changes of the strains due to the treatments (Table 2) was assessed using Student's t-test. The P-values below 0.01 were considered statistically significant.
Cell fractionation, protein quantification and SDS-PAGE analyses
Total cell membranes were prepared as previously described [14]. Briefly, cells were broken by ultrasonication, the sonicated material was centrifuged at 1500 g for 1 min to pellet unlysed cells, and the top phase (cell lysate) was transferred to ultracentrifuge tubes. Crude total membranes were collected by ultracentrifugation of the cell lysates, and washed thoroughly by homogenization in Tris buffer (0.1 M, pH 7.8) containing 1 M KCl. Total membranes were collected again by ultracentrifugation, and resuspended in Tris buffer (50 mM, pH 7.8). Protein contents in whole cells and cell fractions were estimated by using the Micro BCA Protein Assay kit (Pierce), and BSA was used as a protein standard. The peptide composition of cell membranes was analyzed using SDS-PAGE [with 12% (w/v) acrylamide in the resolving gels], as described [14,30].
Phylogenetic tree construction
ClustalW was used for sequence alignment applying default parameters (altered gap penalties were not applied) [31]. Gaps in the alignment were not omitted. The phylogenetic tree was built by Phyml 3.0 with the distance matrix generated by ClustalW and was represented with the program TreeDyn 198.3 available at http://www.phylogeny.fr/[32]. The reliability of each node was established by bootstrap methods.
Hidden Markov Model-based Fur binding site prediction
A group of experimentally validated Fur boxes from E. coli, S. typhimurium, P. aeruginosa and S. aureus used by Quatrini et al., [33] along with 3 experimentally confirmed N. europaea Fur boxes were used to build HMM profiles and to search for fur binding sites in the promoter regions (600 nucleotides upstream of the proposed initiation of translation) of the potential target genes. Alignment of these promoters with the ClustalW multiple-sequence alignment program yielded a putative Nitrosomonas Fur box consensus sequence that has 80% homology with the E. coli Fur box consensus binding sequence.
N. europaea sequence data was obtained from DOE Joint Genome Institute (JGI) website http://genome.ornl.gov/microbial/neur/. Sequence similarity searches of the available nucleotide and protein databases were performed with the BLAST program, available at the National Center for Biotechnology Information website http://www.ncbi.nlm.nih.gov/blast/.
Authors' contributions
NV, LS, PB and DA conceived the study and participated in its design and coordination. NV collected and analyzed the data and wrote the manuscript. LS, PB and DA assisted in the drafting and provided substantial editorial advice and a critical revision of the manuscript. All authors have read and approved the manuscript.
Contributor Information
Neeraja Vajrala, Email: vajralan@science.oregonstate.edu.
Luis A Sayavedra-Soto, Email: sayavedl@science.oregonstate.edu.
Peter J Bottomley, Email: peter.bottomley@oregonstate.edu.
Daniel J Arp, Email: Dan.J.Arp@oregonstate.edu.
Acknowledgements
We thank Andy Ungerer (College of Oceanic and Atmospheric Sciences, OSU) for help with Fe determination by ICP-OES. This research was supported by grant DE-FG03-01ER63149 to D. J. A. and the Oregon Agricultural Experiment Station.
References
- Hantke K. Cloning of the repressor protein gene of iron-regulated systems in Escherichia coli K12. Mol Gen Genet. 1984;197(2):337–341. doi: 10.1007/BF00330982. [DOI] [PubMed] [Google Scholar]
- Ernst FD, Bereswill S, Waidner B, Stoof J, Mader U, Kusters JG, Kuipers EJ, Kist M, van Vliet AH, Homuth G. Transcriptional profiling of Helicobacter pylori Fur- and iron-regulated gene expression. Microbiology. 2005;151(Pt 2):533–546. doi: 10.1099/mic.0.27404-0. [DOI] [PubMed] [Google Scholar]
- Holmes K, Mulholland F, Pearson BM, Pin C, McNicholl-Kennedy J, Ketley JM, Wells JM. Campylobacter jejuni gene expression in response to iron limitation and the role of Fur. Microbiology. 2005;151(Pt 1):243–257. doi: 10.1099/mic.0.27412-0. [DOI] [PubMed] [Google Scholar]
- McHugh JP, Rodriguez-Quinones F, Abdul-Tehrani H, Svistunenko DA, Poole RK, Cooper CE, Andrews SC. Global iron-dependent gene regulation in Escherichia coli. A new mechanism for iron homeostasis. J Biol Chem. 2003;278(32):29478–29486. doi: 10.1074/jbc.M303381200. [DOI] [PubMed] [Google Scholar]
- Mey AR, Wyckoff EE, Kanukurthy V, Fisher CR, Payne SM. Iron and fur regulation in Vibrio cholerae and the role of fur in virulence. Infect Immun. 2005;73(12):8167–8178. doi: 10.1128/IAI.73.12.8167-8178.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escolar L, Perez-Martin J, de Lorenzo V. Opening the iron box: transcriptional metalloregulation by the Fur protein. J Bacteriol. 1999;181(20):6223–6229. doi: 10.1128/jb.181.20.6223-6229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Helmann JD. Functional specialization within the Fur family of metalloregulators. Biometals. 2007;20(3-4):485–499. doi: 10.1007/s10534-006-9070-7. [DOI] [PubMed] [Google Scholar]
- Crosa JH. Genetics and molecular biology of siderophore-mediated iron transport in bacteria. Microbiol Rev. 1989;53(4):517–530. doi: 10.1128/mr.53.4.517-530.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chain P, Lamerdin J, Larimer F, Regala W, Lao V, Land M, Hauser L, Hooper A, Klotz M, Norton J. et al. Complete genome sequence of the ammonia-oxidizing bacterium and obligate chemolithoautotroph Nitrosomonas europaea. J Bacteriol. 2003;185(9):2759–2773. doi: 10.1128/JB.185.9.2759-2773.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker M, Bergmann D, Arciero D, Hooper AB. Electron transfer during the oxidation of ammonia by the chemolithotrophic bacterium Nitrosomonas europaea. Biochim Biophys Acta. 2000;1459(2-3):346–355. doi: 10.1016/S0005-2728(00)00171-7. [DOI] [PubMed] [Google Scholar]
- Upadhyay AK, Petasis DT, Arciero DM, Hooper AB, Hendrich MP. Spectroscopic characterization and assignment of reduction potentials in the tetraheme cytochrome C554 from Nitrosomonas europaea. J Am Chem Soc. 2003;125(7):1738–1747. doi: 10.1021/ja020922x. [DOI] [PubMed] [Google Scholar]
- Schwyn B, Neilands JB. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160(1):47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- Wei X, Sayavedra-Soto LA, Arp DJ. Characterization of the ferrioxamine uptake system of Nitrosomonas europaea. Microbiology. 2007;153(Pt 12):3963–3972. doi: 10.1099/mic.0.2007/010603-0. [DOI] [PubMed] [Google Scholar]
- Wei X, Vajrala N, Hauser L, Sayavedra-Soto LA, Arp DJ. Iron nutrition and physiological responses to iron stress in Nitrosomonas europaea. Arch Microbiol. 2006;186(2):107–118. doi: 10.1007/s00203-006-0126-4. [DOI] [PubMed] [Google Scholar]
- Patzer SI, Hantke K. The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol Microbiol. 1998;28(6):1199–1210. doi: 10.1046/j.1365-2958.1998.00883.x. [DOI] [PubMed] [Google Scholar]
- Bsat N, Herbig A, Casillas-Martinez L, Setlow P, Helmann JD. Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol Microbiol. 1998;29(1):189–198. doi: 10.1046/j.1365-2958.1998.00921.x. [DOI] [PubMed] [Google Scholar]
- Hernandez JA, Lopez-Gomollon S, Bes MT, Fillat MF, Peleato ML. Three fur homologues from Anabaena sp. PCC7120: exploring reciprocal protein-promoter recognition. FEMS Microbiol Lett. 2004;236(2):275–282. doi: 10.1111/j.1574-6968.2004.tb09658.x. [DOI] [PubMed] [Google Scholar]
- Gaballa A, Helmann JD. Identification of a zinc-specific metalloregulatory protein, Zur, controlling zinc transport operons in Bacillus subtilis. J Bacteriol. 1998;180(22):5815–5821. doi: 10.1128/jb.180.22.5815-5821.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl E, Haller JC, Mijovilovich A, Meyer-Klaucke W, Garman E, Vasil ML. Architecture of a protein central to iron homeostasis: crystal structure and spectroscopic analysis of the ferric uptake regulator. Mol Microbiol. 2003;47(4):903–915. doi: 10.1046/j.1365-2958.2003.03337.x. [DOI] [PubMed] [Google Scholar]
- Patzer SI, Hantke K. The zinc-responsive regulator Zur and its control of the znu gene cluster encoding the ZnuABC zinc uptake system in Escherichia coli. J Biol Chem. 2000;275(32):24321–24332. doi: 10.1074/jbc.M001775200. [DOI] [PubMed] [Google Scholar]
- Hall HK, Foster JW. The role of fur in the acid tolerance response of Salmonella typhimurium is physiologically and genetically separable from its role in iron acquisition. J Bacteriol. 1996;178(19):5683–5691. doi: 10.1128/jb.178.19.5683-5691.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensign SA, Hyman MR, Arp DJ. In vitro activation of ammonia monooxygenase from Nitrosomonas europaea by copper. J Bacteriol. 1993;175(7):1971–1980. doi: 10.1128/jb.175.7.1971-1980.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LY, Arp DJ. Loss of ammonia monooxygenase activity in Nitrosomonas europaea upon exposure to nitrite. Appl Environ Microbiol. 1998;64(10):4098–4102. doi: 10.1128/aem.64.10.4098-4102.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: a Laboratory Manual. 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Hommes NG, Sayavedra-Soto LA, Arp DJ. Mutagenesis of hydroxylamine oxidoreductase in Nitrosomonas europaea by transformation and recombination. J Bacteriol. 1996;178(13):3710–3714. doi: 10.1128/jb.178.13.3710-3714.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Sayavedra-Soto LA, Arp DJ. The transcription of the cbb operon in Nitrosomonas europaea. Microbiology. 2004;150(Pt 6):1869–1879. doi: 10.1099/mic.0.26785-0. [DOI] [PubMed] [Google Scholar]
- Carter P. Spectrophotometric determination of serum iron at the submicrogram level with a new reagent (ferrozine) Anal Biochem. 1971;40(2):450–458. doi: 10.1016/0003-2697(71)90405-2. [DOI] [PubMed] [Google Scholar]
- Berry EA, Trumpower BL. Simultaneous determination of hemes a, b, and c from pyridine hemochrome spectra. Anal Biochem. 1987;161(1):1–15. doi: 10.1016/0003-2697(87)90643-9. [DOI] [PubMed] [Google Scholar]
- Shiemke AK, Arp DJ, Sayavedra-Soto LA. Inhibition of membrane-bound methane monooxygenase and ammonia monooxygenase by diphenyliodonium: implications for electron transfer. J Bacteriol. 2004;186(4):928–937. doi: 10.1128/JB.186.4.928-937.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman MR, Arp DJ. An electrophoretic study of the thermal- and reductant-dependent aggregation of the 27 kDa component of ammonia monooxygenase from Nitrosomonas europaea. Electrophoresis. 1993;14(7):619–627. doi: 10.1002/elps.1150140197. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008. pp. W465–469. [DOI] [PMC free article] [PubMed]
- Quatrini R, Lefimil C, Veloso FA, Pedroso I, Holmes DS, Jedlicki E. Bioinformatic prediction and experimental verification of Fur-regulated genes in the extreme acidophile Acidithiobacillus ferrooxidans. Nucleic Acids Res. 2007;35(7):2153–2166. doi: 10.1093/nar/gkm068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delany I, Ieva R, Alaimo C, Rappuoli R, Scarlato V. The iron-responsive regulator fur is transcriptionally autoregulated and not essential in Neisseria meningitidis. J Bacteriol. 2003;185(20):6032–6041. doi: 10.1128/JB.185.20.6032-6041.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delany I, Spohn G, Pacheco AB, Ieva R, Alaimo C, Rappuoli R, Scarlato V. Autoregulation of Helicobacter pylori Fur revealed by functional analysis of the iron-binding site. Mol Microbiol. 2002;46(4):1107–1122. doi: 10.1046/j.1365-2958.2002.03227.x. [DOI] [PubMed] [Google Scholar]
- Ochsner UA, Vasil ML. Gene repression by the ferric uptake regulator in Pseudomonas aeruginosa: cycle selection of iron-regulated genes. Proc Natl Acad Sci USA. 1996;93(9):4409–4414. doi: 10.1073/pnas.93.9.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai PJ, Angerer A, Genco CA. Analysis of Fur binding to operator sequences within the Neisseria gonorrhoeae fbpA promoter. J Bacteriol. 1996;178(16):5020–5023. doi: 10.1128/jb.178.16.5020-5023.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watnick PI, Butterton JR, Calderwood SB. The interaction of the Vibrio cholerae transcription factors, Fur and IrgB, with the overlapping promoters of two virulence genes, irgA and irgB. Gene. 1998;209(1-2):65–70. doi: 10.1016/S0378-1119(98)00018-3. [DOI] [PubMed] [Google Scholar]
- Baichoo N, Helmann JD. Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence. J Bacteriol. 2002;184(21):5826–5832. doi: 10.1128/JB.184.21.5826-5832.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantke K. Selection procedure for deregulated iron transport mutants (fur) in Escherichia coli K 12: fur not only affects iron metabolism. Mol Gen Genet. 1987;210(1):135–139. doi: 10.1007/BF00337769. [DOI] [PubMed] [Google Scholar]
- Stojiljkovic I, Baumler AJ, Hantke K. Fur regulon in gram-negative bacteria. Identification and characterization of new iron-regulated Escherichia coli genes by a fur titration assay. J Mol Biol. 1994;236(2):531–545. doi: 10.1006/jmbi.1994.1163. [DOI] [PubMed] [Google Scholar]
- Tsolis RM, Baumler AJ, Stojiljkovic I, Heffron F. Fur regulon of Salmonella typhimurium: identification of new iron-regulated genes. J Bacteriol. 1995;177(16):4628–4637. doi: 10.1128/jb.177.16.4628-4637.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdul-Tehrani H, Hudson AJ, Chang YS, Timms AR, Hawkins C, Williams JM, Harrison PM, Guest JR, Andrews SC. Ferritin mutants of Escherichia coli are iron deficient and growth impaired, and fur mutants are iron deficient. J Bacteriol. 1999;181(5):1415–1428. doi: 10.1128/jb.181.5.1415-1428.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyer K, Imlay JA. Superoxide accelerates DNA damage by elevating free-iron levels. Proc Natl Acad Sci USA. 1996;93(24):13635–13640. doi: 10.1073/pnas.93.24.13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arciero DM, Hooper AB. Hydroxylamine oxidoreductase from Nitrosomonas europaea is a multimer of an octa-heme subunit. J Biol Chem. 1993;268(20):14645–14654. [PubMed] [Google Scholar]
- Bagg A, Neilands JB. Mapping of a mutation affecting regulation of iron uptake systems in Escherichia coli K-12. J Bacteriol. 1985;161(1):450–453. doi: 10.1128/jb.161.1.450-453.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantke K. Regulation of ferric iron transport in Escherichia coli K12: isolation of a constitutive mutant. Mol Gen Genet. 1981;182(2):288–292. doi: 10.1007/BF00269672. [DOI] [PubMed] [Google Scholar]
- Litwin CM, Calderwood SB. Analysis of the complexity of gene regulation by fur in Vibrio cholerae. J Bacteriol. 1994;176(1):240–248. doi: 10.1128/jb.176.1.240-248.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt MP, Payne SM. Genetics and regulation of enterobactin genes in Shigella flexneri. J Bacteriol. 1988;170(12):5579–5587. doi: 10.1128/jb.170.12.5579-5587.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince RW, Cox CD, Vasil ML. Coordinate regulation of siderophore and exotoxin A production: molecular cloning and sequencing of the Pseudomonas aeruginosa fur gene. J Bacteriol. 1993;175(9):2589–2598. doi: 10.1128/jb.175.9.2589-2598.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturi V, Ottevanger C, Bracke M, Weisbeek P. Iron regulation of siderophore biosynthesis and transport in Pseudomonas putida WCS358: involvement of a transcriptional activator and of the Fur protein. Mol Microbiol. 1995;15(6):1081–1093. doi: 10.1111/j.1365-2958.1995.tb02283.x. [DOI] [PubMed] [Google Scholar]
- Thomas CE, Sparling PF. Isolation and analysis of a fur mutant of Neisseria gonorrhoeae. J Bacteriol. 1996;178(14):4224–4232. doi: 10.1128/jb.178.14.4224-4232.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews SC, Robinson AK, Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27(2-3):215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- Horsburgh MJ, Ingham E, Foster SJ. In Staphylococcus aureus, fur is an interactive regulator with PerR, contributes to virulence, and Is necessary for oxidative stress resistance through positive regulation of catalase and iron homeostasis. J Bacteriol. 2001;183(2):468–475. doi: 10.1128/JB.183.2.468-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staggs TM, Fetherston JD, Perry RD. Pleiotropic effects of a Yersinia pestis fur mutation. J Bacteriol. 1994;176(24):7614–7624. doi: 10.1128/jb.176.24.7614-7624.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166(4):557–580. doi: 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]