Abstract
PINK1 and Parkin mutations cause recessive Parkinson's disease (PD). In Drosophila and SH-SY5Y cells, Parkin is recruited by PINK1 to damaged mitochondria, where it ubiquitinates Mitofusins and consequently promotes mitochondrial fission and mitophagy.
Here, we investigated the impact of mutations in endogenous PINK1 and Parkin on the ubiquitination of mitochondrial fusion and fission factors and the mitochondrial network structure. Treating control fibroblasts with mitochondrial membrane potential (Δψ) inhibitors or H2O2 resulted in ubiquitination of Mfn1/2 but not of OPA1 or Fis1. Ubiquitination of Mitofusins through the PINK1/Parkin pathway was observed within 1 h of treatment. Upon combined inhibition of Δψ and the ubiquitin proteasome system (UPS), no ubiquitination of Mitofusins was detected. Regarding morphological changes, we observed a trend towards increased mitochondrial branching in PD patient cells upon mitochondrial stress.
For the first time in PD patient-derived cells, we demonstrate that mutations in PINK1 and Parkin impair ubiquitination of Mitofusins. In the presence of UPS inhibitors, ubiquitinated Mitofusin is deubiquitinated by the UPS but not degraded, suggesting that the UPS is involved in Mitofusin degradation.
Introduction
Parkinson's disease (PD) is a progressive neurodegenerative disorder, clinically characterized by bradykinesia, tremor, and rigidity, with a monogenic cause in about 2–3% of the cases [1]. Studying the consequences of mutations in recessively inherited PD-associated genes, such as PTEN-induced putative kinase 1 (PINK1) or the E3 ubiquitin ligase Parkin, may help to understand the mechanisms underlying the disease also in sporadic, idiopathic PD patients.
Although the exact cause of sporadic PD is still elusive, mitochondrial dysfunction has long been connected with the disease. Impaired respiratory chain function has been found in sporadic PD patients and different PINK1 or Parkin knockdown models [2], [3], [4], [5]. Furthermore, Drosophila pink1 and parkin loss-of-function mutants showed defects in mitochondrial morphology [6], [7], [8], [9], [10]. Transgenic expression of parkin markedly ameliorated all pink1 loss-of-function phenotypes, but not vice versa, suggesting that parkin functions downstream of pink1 [6], [7], [8].
A series of experiments in Drosophila, SH-SY5Y cells, and primary mouse neurons provided evidence that the PINK1/Parkin pathway promotes mitochondrial fission and that loss of activity of either protein results in decreased fission and impaired tissue integrity [11], [12]. Inactivation of the dynamin-related protein 1 (drp1), a key factor of mitochondrial fission, enhances the pink1 and parkin-mutant phenotypes in Drosophila [11], [12], [13]. By contrast, increased drp1 gene dosage or inactivation of the mitochondrial fusion-promoting components optic atrophy 1 (opa1) and mitofusin (mfn) suppress the mitochondrial phenotype in Drosophila pink1 and parkin mutants [11], [12], [13]. Recently, these observations have been linked to mitophagy. Under stress conditions, PINK1 recruits Parkin to dysfunctional mitochondria [14], [15], [16], [17]. The subsequent ubiquitination of Mitofusins by Parkin inhibits mitochondrial fusion and thus promotes mitochondrial fragmentation as an initial step of mitophagy [18], [19], [20].
In PD patient fibroblasts, only the morphological effects of mutations in Parkin have been studied so far revealing that the degree of mitochondrial branching was higher than in controls [21].
In our present work, we used fibroblast cultures from PD patients carrying two mutated Parkin or PINK1 alleles to investigate the consequences of mutations in endogenous PINK1 and Parkin on the ubiquitination of mitochondrial fusion and fission factors. Furthermore, we evaluated the influence of these mutations on the structure of the mitochondrial network in human cells.
Results
Two fibroblast cultures with homozygous PINK1 mutations, p.Q456X or p.V170G, two cultures with homozygous Parkin mutations, p.V324fsX434 or p.R245fsX253, and fibroblasts from two age-matched mutation-negative healthy controls were included in the study. The effects of these mutations on PINK1 and Parkin mRNA levels are described elsewhere [16], [22]. Clinical features of the mutation carriers were compatible with idiopathic PD, with the exception of an earlier age of onset of 42.3+/−13.5 years [23], [24], [25]. All experiments were performed at least in triplicate and representative blots are shown.
Decreased Mfn2 levels after valinomycin or CCCP treatment in control fibroblasts
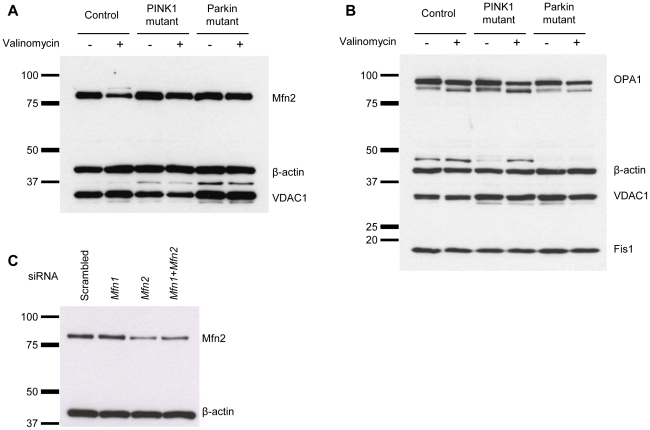
First, we determined the endogenous levels of Mfn2 in the PINK1 and Parkin mutants and controls under basal conditions and after exposure to 1 µM valinomycin for 12 h. This treatment caused a drop in the protein levels of Mfn2 in controls but not in either of the mutant cells (Figure 1A). Furthermore in controls, Mfn2 had an additional band on the Western blot, which was about 8 kDa larger in size than the non-modified form, consistent with monoubiquitination of the protein. By contrast, protein levels of OPA1 and Fis1 were unchanged in all cell cultures when incubated with valinomycin (Figure 1B) and modified forms of these proteins were not detectable. Protein levels of the mitochondrial marker voltage-dependent anion channel 1 (VDAC1) were comparable in all samples under basal and stress conditions (Figure 1A, B).
Figure 1. Expression of mitochondrial fusion and fission proteins after valinomycin treatment.
Fibroblasts from a healthy control, a homozygous PINK1 mutant and a homozygous Parkin mutant were cultured under basal conditions or treated with 1 µM valinomycin for 12 h. The protein levels of (A) Mfn2, (B) OPA1 and Fis1 were investigated by means of Western blotting. Valinomycin exposure caused a decrease in Mfn2 levels in controls, but not in PINK1- or Parkin-mutant cells. The protein levels of OPA1 and Fis1 were not influenced by this treatment in any of the cell cultures. Under basal and stress conditions, expression of the mitochondrial marker VDAC1 was comparable in mutants and controls. (C) Mutant cells were incubated with scrambled siRNA, Mfn1 siRNA, Mfn2 siRNA or a combination of Mfn1 and Mfn2 siRNA for 40 h. Western blot analysis was performed with an antibody against Mfn2. The Mfn2 level decreased only when Mfn2 siRNA was employed, confirming the specificity of the anti-Mfn1 antibody used in our study. β-actin served as a loading control. Fis1 – fission 1; Mfn1 – mitofusin 1; Mfn2 – mitofusin 2; OPA1 – optic atrophy 1; VDAC1 – voltage-dependent anion channel 1.
To test whether the used Mfn2 antibody specifically binds Mfn2, a knock-down experiment with siRNA against Mfn1 or Mfn2 was performed (Figure 1C). This experiment showed a drop in Mfn2 level only when siRNA against Mfn2 was employed, confirming the specificity of the antibody.
In this experiment also the Mfn2 homolog mitofusin 1 (Mfn1) [26] was investigated and showed comparable effects (Figure S1). Since the available Mfn1 antibody was less sensitive than the Mfn2 antibody, out of the series of experiments for Mfn2 described in this article, only selected ones were repeated for Mfn1 (see supplementary material).
Reduced Mfn2 levels in controls but not in PINK1- and Parkin-mutant fibroblasts were also detected after incubation with the protonophore cyanide m-chlorophenylhydrazone (CCCP; 10 µM for 12 h) (Figure S2). Since valinomycin and CCCP had identical effects on Mfn2, only results from the experiments using valinomycin are shown.
In control fibroblasts, Mfn2 is ubiquitinated after valinomycin exposure and is detected in the mitochondrial fraction
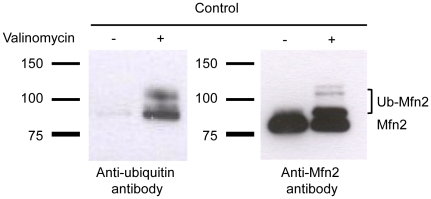
Next, we intended to verify whether the additional band on the Mfn2 blot, which is present only in controls after valinomycin- or CCCP-induced stress, is indeed explained by ubiquitination of the protein. For this, we performed immunoprecipitation using an antibody against Mfn2. Whole cell lysates from non-treated and valinomycin-treated (1 µM, for 6 h) controls were employed. The resulting immunoprecipitates were analyzed by Western blotting with an antibody against ubiquitin (Figure 2, left panel) or with an antibody against Mfn2 (Figure 2, right panel). On both blots, bands of the size of mono- and polyubiquitinated or multiple monoubiquitinated Mfn2 were only detected in valinomycin-treated but not in non-treated controls. Immunoprecipitation was also performed with cell lysates from valinomycin-treated PINK1- and Parkin-mutant fibroblasts. Western blot analysis with an Mfn2 antibody showed only the non-modified form of the protein (Figure S3). Taken together, these findings supported our previous results and underline that Mfn2 is ubiquitinated via the PINK1/Parkin pathway.
Figure 2. Ubiquitination of Mfn2 upon valinomycin treatment.
Fibroblasts from a healthy control were treated with 1 µM valinomycin for 6 h. Whole cell lysates from non-treated and valinomycin treated controls were immunoprecipitated using an antibody against Mfn2. Immunoprecipitates were analyzed by Western blotting using an antibody against ubiquitin (left panel). Subsequently, the membrane was washed and reprobed with an antibody against Mfn2 (right panel). Ubiquitinated forms of Mfn2 (mono- and polyubiquitinated) are present only in valinomycin treated controls. Mfn2 – mitofusin 2; Ub-Mfn2 – ubiquitinated mitofusin 2.
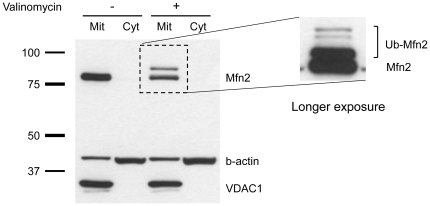
To determine the subcellular localization of ubiquitinated Mfn1 and 2 in control fibroblasts, cells were incubated with 1 µM valinomycin for 6 h and mitochondrial and cytosolic protein fractions separated. Western blot analysis revealed that Mfn1/2 and their ubiquitinated forms are exclusively localized in the mitochondrial fraction (Figure 3 and Figure S4). The same findings for Mfn2 were obtained when the fractionation experiment was repeated in SH-SY5Y cells (Figure S5).
Figure 3. Mitochondrial localization of ubiquitinated Mfn2 after valinomycin treatment.
Control fibroblasts were cultured under basal conditions or treated with 1 µM valinomycin for 6 h. Cells were harvested and mitochondrial and cytosolic fractions were analyzed by Western blotting. The subcellular localization of Mfn2 was determined. Quality of cellular fractionation was confirmed using antibodies against VDAC1 and β-actin. The ubiquitinated forms of Mfn2, which were observed only after valinomycin stress, are exclusively found in the mitochondrial fraction. A longer exposure of the blot showed several Mfn2 bands with higher molecular weight, indicative of Mfn2 polyubiquitination (enlarged cutout). Cyt – cytosolic fraction; Mit – mitochondrial fraction; Mfn2 – mitofusin 2; VDAC1 – voltage-dependent anion channel 1.
Rescue of Mfn2 ubiquitination in mutant fibroblasts
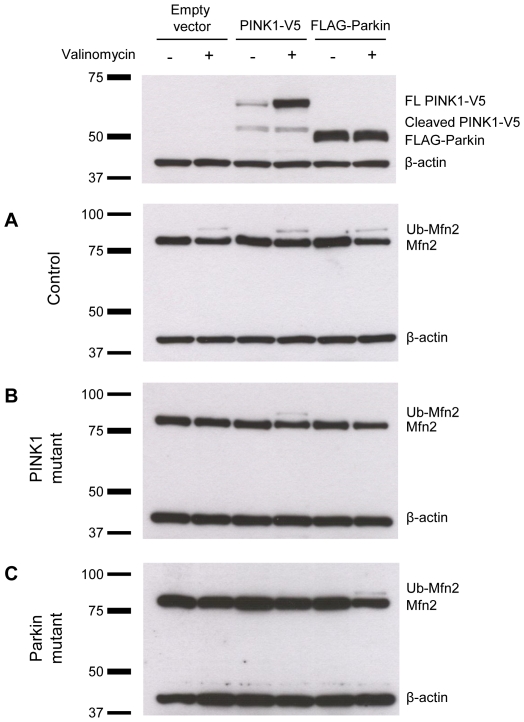
To test whether lack of ubiquitination of Mfn2 in the mutants can be rescued, we transfected control, PINK1- and Parkin-mutant fibroblasts with an empty vector, a vector containing PINK1-V5 or a vector containing FLAG-Parkin. Twenty-four hours after transfection, these cells were cultured under basal conditions or treated with 1 µM valinomycin for an additional 12 h. Whole cell lysates were analyzed by Western blotting. Using antibodies against V5 and FLAG, bands of the size of tagged full-length and cleaved PINK1 or Parkin were detected, confirming successful transfection (Figure 4, upper panel). Furthermore, using an antibody against Mfn2, ubiquitinated Mfn2 was detected in control cells under stress conditions (Figure 4A). In PINK1-mutant cells, ubiquitination of Mfn2 under stress was rescued through expression of PINK1-V5 but not through expression of FLAG-Parkin (Figure 4B). Similarly, in Parkin-mutant fibroblasts, ubiquitinated Mfn2 was only detected after transfection with FLAG-Parkin (Figure 4C).
Figure 4. Rescue of Mfn2 ubiquitination.
Fibroblasts of a control, a PINK1 and a Parkin mutant were transfected with an empty vector, a vector containing PINK1-V5 or a vector containing FLAG-Parkin. Twenty hours after transfection, cells were cultured under basal conditions or treated with 1 µM valinomycin for an additional 6 h. Western blot analysis using antibodies against V5 and FLAG showed bands of the size of full-length/cleaved PINK1 and Parkin, respectively, in treated and non-treated control cells after transfection, confirming the success of the experiment (upper panel). (A) In controls, ubiquitinated Mfn2 was detected in all three experiments after valinomycin treatment. (B) In PINK1-mutant cells, ubiquitination of Mfn2 under stress was rescued through overexpression of PINK1-V5 but not through overexpression of FLAG-Parkin. (C) In Parkin-mutant fibroblasts ubiquitinated Mfn2 was only detected after transfection with FLAG-Parkin. FL – full-length; Mfn2 – mitofusin 2; Ub-Mfn2 – ubiquitinated mitofusin 2.
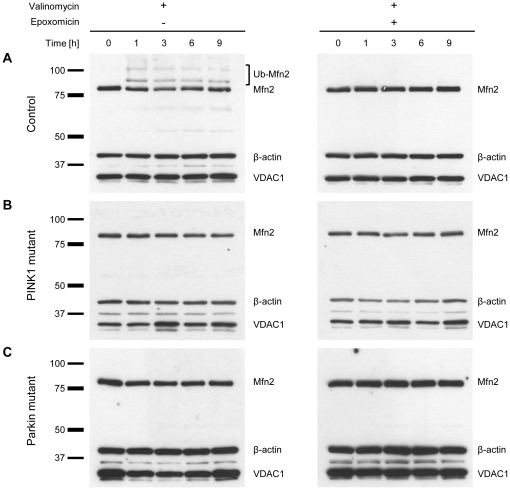
Ubiquitination of Mfn2 occurs within 1 h of valinomycin treatment and is prevented in the presence of epoxomicin
To explore whether the ubiquitinated forms of Mfn2 are degraded by the UPS, we treated control (Figure 5A), PINK1- (Figure 5B) and Parkin-mutant (Figure 5C) fibroblasts with 1 µM valinomycin alone (Figure 5, left panel) or in combination with 10 µM epoxomicin (Figure 5, right panel) and extracted proteins at different time points for Western blot analysis. In control cells, valinomycin treatment initiated the ubiquitination of Mfn2 within 1 h of incubation (Figure 5A, left panel). Mfn2 ubiquitination was prevented by simultaneous exposure to epoxomicin (Figure 5A, right panel). The same effect was observed with MG132 (Figure S6). By contrast, simultaneous treatment with valinomycin and the lysosomal inhibitor bafilomycin did not prevent Mfn2 ubiquitination in control cells (Figure 6). To exclude that proteasomal inhibition influences the effect of the potassium ionophore valinomycin, the mitochondrial membrane potential in control fibroblasts was monitored during 9 h of treatment with valinomycin alone or in combination with epoxomicin. Both culturing conditions caused a similar drop in membrane potential (Figure S7). The protein levels of non-modified Mfn2 remained unchanged in all samples over time when treated with valinomycin plus epoxomicin (Figure 5, right panel). To further test whether epoxomicin alone has an impact on the protein levels of Mfn2, we treated control cells with 10 µM epoxomicin but observed no change in Mfn2 levels during 9 h of incubation (Figure S8).
Figure 5. Ubiquitination of Mfn2 is prevented by exposure to epoxomicin.
Fibroblasts from (A) a healthy control, (B) a homozygous PINK1 mutant and (C) a homozygous Parkin mutant were treated with 1 µM valinomycin alone (left panel) or with 1 µM valinomycin plus 10 µM epoxomicin (right panel). Proteins were extracted at different time points and analyzed by Western blotting. In control cells, valinomycin treatment initiated the ubiquitination of Mfn2 after 1 h of incubation. This effect was prevented by simultaneous exposure to epoxomicin. The mitochondrial marker VDAC1 and the cytosolic marker β-actin served as loading controls. Mfn2 – mitofusin 2; Ub-Mfn2 – ubiquitinated mitofusin 2; VDAC1 – voltage-dependent anion channel 1.
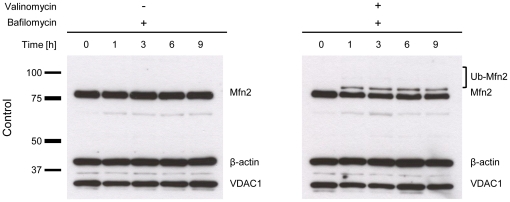
Figure 6. Exposure to bafilomycin has no impact on Mfn2 ubiquitination.
Control fibroblasts were treated with 10 nM bafilomycin alone (left panel) or with 10 nM bafilomycin plus 1 µM valinomycin (right panel). Proteins were extracted at different time points and analyzed by Western blotting. Bafilomycin had no effect on the Mfn2 levels and no ubiquitination was detected. When bafilomycin and valinomycin were combined, Mfn2 ubiquitination was initiated by valinomycin and not influenced by bafilomycin. In both experiments, the mitochondrial marker VDAC1 was unchanged. β-actin served as a loading control. Mfn2 – mitofusin 2; Ub-Mfn2 – ubiquitinated mitofusin 2; VDAC1 – voltage-dependent anion channel 1.
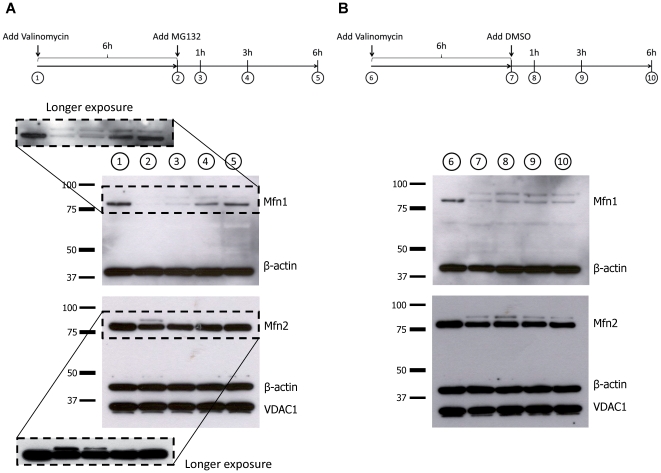
Next, we wanted to show that inhibition of the UPS is not only preventing ubiquitination of Mfn1 and Mfn2 (as shown above) but is actually causing deubiquitination of already ubiquitinated Mitofusins. For that we treated control fibroblasts with valinomycin to induce ubiquitination. After 6 h we added MG132 or DMSO (dissolvent for MG132) and harvested cells at different time points. Western blot analysis revealed that upon 6 h of UPS inhibition, levels of non-modified Mitofusins were almost at the same level as before treatment (Figure 7). This additionally confirms that the UPS is involved in the processing of ubiquitinated Mfn1 and Mfn2.
Figure 7. MG132 promotes deubiquitination of Mfn1 and Mfn2.
Control fibroblasts were treated with 1 µM valinomycin. After 6 h, MG132 (final concentration 10 µM) was added to the cells. Proteins were extracted at different time points and analyzed by Western blotting. Exposure to (A) an inhibitor of the UPS, i.e.MG132 but not to (B) DMSO alone, induced deubiquitination of both Mfn1 and Mfn2. Mfn1 – mitofusin 1; Mfn2 – mitofusin 2; VDAC1 – voltage-dependent anion channel 1.
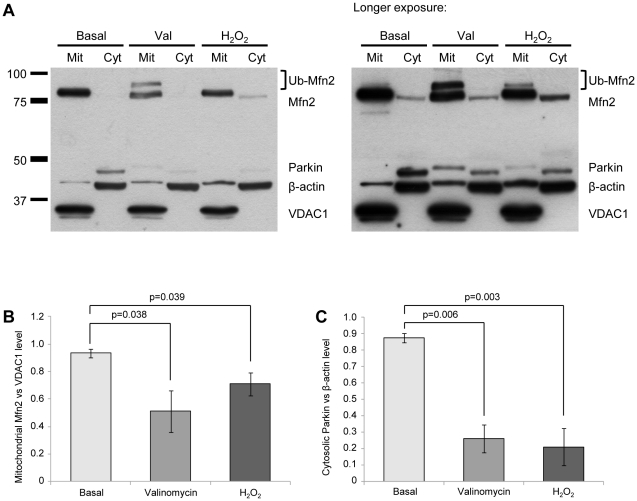
Exposure to H2O2 causes ubiquitination of Mfn2
Next, we investigated whether exposure of control fibroblasts to the superoxide generator H2O2 also results in ubiquitination of Mfn2. Therefore, cells were incubated with 100 µM H2O2 for 12 h and compared to cells stressed with 1 µM valinomycin for 6 h and non-treated cells. Mitochondrial and cytosolic fractions of these cells were analyzed by Western blotting using antibodies against Mfn2, Parkin, VDAC1 and β-actin. The predominant presence of VDAC1 in the mitochondrial and of β-actin in the cytosolic fraction indicated good quality of the fractionation (Figure 8A). Densitometric analysis revealed a significant drop in protein levels of non-modified Mfn2 in the mitochondrial fraction after valinomycin but also after H2O2 treatment (Figure 8A and B). Under both stress conditions, high-molecular-weight bands of Mfn2 were detected, indicative of Mfn2 ubiquitination. As already demonstrated in our recently published study on PINK1- and Parkin-mutant fibroblasts [16], both treatments caused a significant drop in protein levels of Parkin in the cytosolic fraction (Figure 8A and C). Longer exposure of the Western blots revealed mitochondrial translocation of endogenous Parkin after both treatments (Figure 8A, right panel).
Figure 8. Exposure to H2O2 causes ubiquitination of Mfn2.
Control fibroblasts were cultured under basal conditions, treated with 1 µM valinomycin for 6 h or with 100 µM H2O2 for 12 h. Cells were harvested and mitochondrial and cytosolic fractions were analyzed by Western blotting. (A) The subcellular localization of Mfn2 and Parkin was determined. Quality of cellular fractionation was confirmed using antibodies against VDAC1 and β-actin. (B) Densitometric analysis of the Western blot results revealed a significant drop in protein levels of non-modified Mfn2 (normalized against VDAC1 expression level) in the mitochondrial fraction after valinomycin or H2O2 treatment. (C) Furthermore, both treatments caused a significant drop in protein levels of Parkin (normalized against β-actin expression) in the cytosolic fraction. (A, right panel) A longer exposure of the Western blot shows mitochondrial translocation of Parkin after both treatments. For quantification of protein levels, blots of three independent experiments were evaluated. In the graphs mean intensities +/− standard deviation are given. Cyt – cytosolic fraction; H2O2 – hydrogen peroxide; Mit – mitochondrial fraction; Mfn2 – mitofusin 2; Ub-Mfn2 – ubiquitinated mitofusin 2; Val – Valinomycin; VDAC1 – voltage-dependent anion channel 1.
Branching of the mitochondrial network
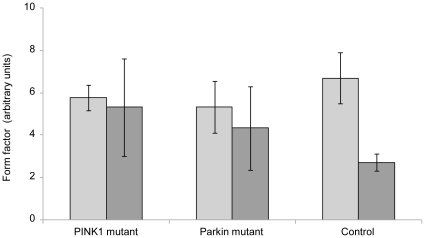
Finally, we determined the impact of the absence of Mfn2 ubiquitination in the mutants under stress conditions on the mitochondrial network. To evaluate the degree of mitochondrial branching, we measured the form factor [21] in cells from a PINK1 mutant, a Parkin mutant and a control. This showed a comparable degree of mitochondrial network branching in all investigated individuals under basal conditions. When we stressed the cells with 1 µM valinomycin for 12 h to initiate Mfn2 ubiquitination, the form factor decreased in all samples. Although there was a trend towards more fragmented mitochondria in control than in mutant cells, this difference did not reach significance (Figure 9).
Figure 9. Branching of mitochondrial network.
The mitochondrial form factor was determined in a control, a homozygous PINK1 and a homozygous Parkin mutant. The quantification revealed no differences in mitochondrial branching between mutant and control cells under basal conditions (light gray bars). After treatment with 1 µM valinomycin for 12 h, a decrease in branching occurred in all individuals, with the control being most severely affected (dark gray bars). In the graph, mean form factors +/− standard deviation are given.
Discussion
Mitochondrial dysfunction and changes in mitochondrial morphology have long been linked to the disease mechanisms underlying PD [2], [4], [5], [6], [7], [27]. However, only recently, several studies demonstrated that the various observed mitochondrial phenotypes can be ascribed to one common molecular cause: Apparently, a deficit in mitophagy leads to accumulation of dysfunctional mitochondria in the cell [14], [15], [16], [17]. The PD-associated proteins PINK1 and Parkin seem to play a central role in the initiation of mitophagy [18], [19], [20], [28]. In a recent study, we have established human fibroblasts with homozygous PINK1 and Parkin mutations as a suitable model system to investigate the PINK1/Parkin pathway [16]. Here, we expand our previous results using these PD patient cells to characterize effects of the PINK1/Parkin pathway on mitochondrial fusion and fission proteins on the endogenous level.
Several studies reported that mitochondrial stress, such as exposure to membrane potential inhibitors, initiates the PINK1/Parkin mitophagy pathway [14], [15], [16], [17], [20], [28]. Therefore, we treated our fibroblast cell cultures with the mitochondrial uncoupling agents valinomycin and CCCP or the superoxide generator H2O2. All treatments resulted in decreased Mfn2 signal in the controls but not in the mutants. However, the effect after H2O2 incubation was the least pronounced. Moreover, high-molecular-weight Mfn2 bands were detected in the controls, indicative of Mfn2 poly- or multiple monoubiquitination. By contrast, the protein levels of OPA1 and Fis1 were not altered in mutants compared to controls under stress conditions. In Drosophila, the mitochondrial phenotype caused by pink1 and parkin loss-of-function mutations could at least partially be suppressed by opa1 knockdown [8], [11], [12], [13]. Conversely, in SH-SY5Y cells, overexpression of OPA1 prevented changes in mitochondrial morphology induced by PINK1 or Parkin knockdown. However, similar to our findings, no alterations in OPA1 processing were observed in these cells lacking PINK1 or Parkin [29]. These apparent discrepancies could be explained by differences in OPA1 function in arthropods compared to mammals [30].
By means of immunoprecipitation, the additional anti-Mfn2 reactive bands indeed proved to represent ubiquitinated forms of the protein. These findings are in line with recent publications reporting that in wild-type Drosophila and SH-SY5Y cells Mitofusins are ubiquitinated in response to mitochondrial stress. This modification was, however, impaired in treated Parkin or Pink1 knockdown cells [18], [19], [20], [28]. Furthermore, studies comparing wild-type flies with parkin or pink1 null mutants suggested that loss of parkin or pink1 increases the steady-state abundance of mfn [18], [19]. We did not detect any changes in protein levels when monitoring Mfn2 in PINK1- or Parkin-mutant human fibroblasts under stress conditions over time. Therefore, it is tempting to speculate that the “increased” mfn levels in pink1 or parkin knockdown flies may reflect an increase in mitochondrial biogenesis in these mutants [17].
Of note, all experiments performed for the Mfn2 interaction partner Mfn1 in this study indicate similar behavior of both Mitofusins in the PINK1/Parkin pathway.
Furthermore, we were able to rescue the ubiquitination of Mfn2 when PINK1 or Parkin was re-expressed in PINK1- or Parkin-mutant cells. In Drosophila, transgenic expression of parkin in pink1 loss-of-function mutants markedly ameliorated all mitochondrial phenotypes, but not vice versa, leading to the conclusion that parkin functions downstream of pink1 [6], [7], [8]. However, to our surprise, we were not able to detect ubiquitinated Mfn2 when PINK1 mutant cells were transfected with FLAG-Parkin. Given the weak signal of ubiquitinated Mfn2 after PINK1 transfection in the Parkin-mutant cells, a possible explanation for this discrepancy might be that the used antibody is not sensitive enough to detect the likely even lower levels of ubiquitinated Mfn2 in the PINK1-mutant fibroblasts.
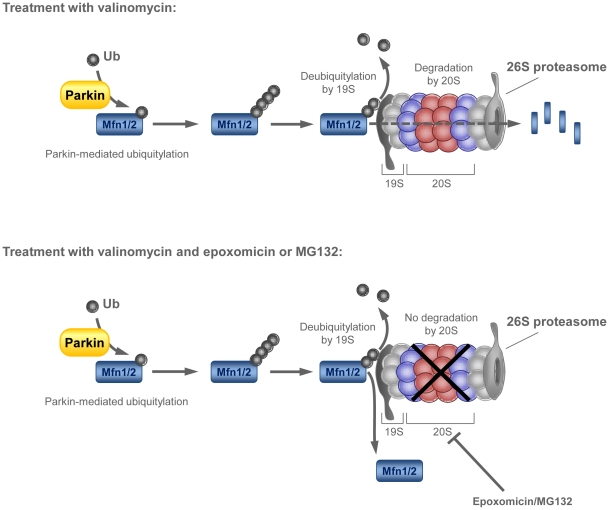
When we determined the subcellular localization of non-modified and ubiquitinated Mfn2 in control fibroblasts and SH-SY5Y cells, all Mfn2 forms were exclusively found in the mitochondrial fraction. Ubiquitination occurred already within one hour of treatment with valinomycin. Contrary to our expectations, inhibition of the UPS with epoxomicin or MG132 neither increased nor preserved the levels of ubiquitinated Mfn2 over time. Ubiquitination of Mfn2 in control cells was apparently absent using valinomycin in combination with a proteasomal inhibitor. However, this was not explained by interference of epoxomicin with the effect of the mitochondrial uncoupler valinomycin. According to the literature, proteins targeted for degradation can only enter the UPS after their ubiquitin chain has been removed. This deubiquitination is performed by the 19S regulatory complex of the UPS [31], [32]. Epoxomicin is a potent inhibitor of the 20S proteasome subunit, where protein degradation takes place, but does not influence the deubiquitinase activity of the 19S particle (Figure 10) [33]. Consequently, our data suggest that, in the presence of epoxomicin or MG132, ubiquitinated Mitofusins are solely deubiquitinated but not degraded by the UPS leading to constant levels of non-modified Mfn1/2 in the stressed control cells. Under stress conditions without UPS inhibition, however, the turnover of ubiquitinated Mfn1/2 in the cytosol is probably occurring too rapidly to be detected in a fractionation experiment. In line with our hypothesis, inhibition of lysosomal degradation did not prevent Mfn2 ubiquitination in control fibroblasts. At first sight, there appears to be a discrepancy between our data and a recently published report showing that the ubiquitinated forms of Mitofusins may be preserved upon treatment with the UPS inhibitor MG132 and the uncoupler of the mitochondrial membrane potential CCCP [28]. However, this can be explained by the fact that cells overexpressing Parkin were used in that study, whereas our experimental setup was based on endogenous levels of Parkin. It is conceivable that under conditions of overexpressed Parkin, the balance between Parkin-mediated ubiquitination and deubiquitination by the UPS is shifted towards ubiquitination, thus leading to accumulation of ubiquitinated Mitofusins in their system. Interestingly, the degradation of the yeast Mfn1/2 homologue Fzo1 is also dependent on the UPS [34].
Figure 10. Possible scenario of Mfn1/2 deubiquitination and degradation at the UPS.
Under valinomycin stress, Mitofusins are ubiquitinated by Parkin. Poly-ubiquitinated Mitofusins are subsequently recognized by the intrinsic ubiquitin-binding activity of the 19S particle of the 26S proteasome. At the 19S regulatory complex the ubiquitin chain is disassembled, and the substrate is unfolded before it can enter the cavity of the 20S subunit where proteolysis takes place [31]. Simultaneous treatment with epoxomicin (or MG132) inhibits the degradation function of the 20S core particle but does not influence the deubiquitylase activity of the 19S subunit. Consequently, poly-ubiquitinated Mitofusins are deubiquitinated at the proteasome but cannot be degraded. Mfn1/2 – mitofusin 1/2; Ub – ubiquitin; UPS – ubiquitin proteasome system.
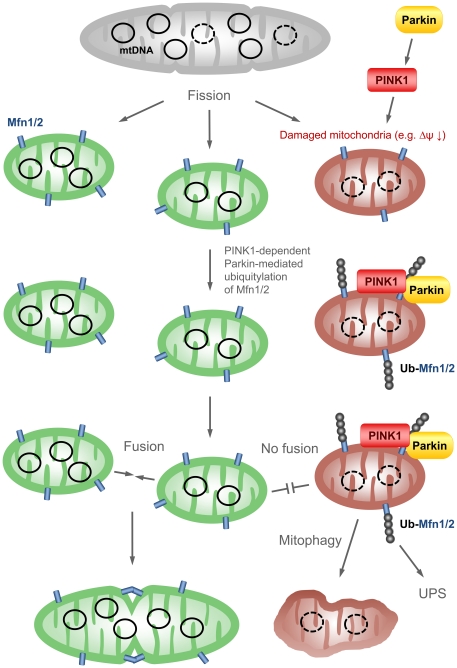
In a recent publication, the mitochondrial membrane potential was identified as an important cellular parameter to differentiate between functional and dysfunctional mitochondria. Following fission, the refusion of daughter mitochondria requires a membrane potential beyond a certain threshold [35]. We hypothesize that Parkin-mediated ubiquitination and subsequent degradation of Mfn1/2 prevents this refusion. Such isolated dysfunctional mitochondria likely undergo mitophagy and require both functional Parkin and PINK1. This notion is supported by colocalization of PINK1 and partially also of Parkin with microtubule-associated protein 1 light chain 3 (LC3), a marker of autophagosomes [36], and by abrogation of Parkin-induced mitophagy upon treatment with bafilomycin, a lysosomal inhibitor [37]. For a schematic representation of the putative Parkin/PINK1 mitophagy pathway, see Figure 11.
Figure 11. Scheme summarizing PINK1's and Parkin's putative function in mitophagy.
By means of fission, mitochondria are randomly divided. Damaged mitochondria can be distinguished from functional mitochondria, for instance, by a difference in membrane potential. Dysfunctional mitochondria with low membrane potential are detected by PINK1, which recruits Parkin. Next, Parkin ubiquitinates Mfn1/2 which localize to the outer mitochondrial membrane. Subsequently, ubiquitinated Mitofusins are degraded by the UPS, preventing fusion of dysfunctional with functional mitochondria. By this, dysfunctional mitochondria are selected out from the general pool of mitochondria and subsequently undergo mitophagy. Δψ – mitochondrial membrane potential; Mfn1/2 – mitofusin 1/2; PINK1 – PTEN-induced putative kinase 1; Ub-Mfn1/2 – ubiquitinated mitofusin 1/2; UPS – ubiquitin proteasome system.
According to our current knowledge, mitophagy is the only mechanism by which mitochondria are recycled [38]. Therefore, impairment of mitophagy due to PINK1 or Parkin mutations presumably leads to accumulation of dysfunctional mitochondria in the cell. This scenario may be an explanation for mitochondrial phenotypes, such as respiratory chain dysfunction [3], [4], [21] and elevated mitochondrial DNA mutational load [39], [40], [41] which have been observed in PINK1 or Parkin knockout models as well as PD patients with mutations in either gene (Figure 11). In accordance with our hypothesis, two studies in HeLa cells provided evidence that overexpression of Parkin leads to a significant loss of mitochondria [36], [37]. However, when we compared the expression of various mitochondrial markers in our PINK1- and Parkin-mutant as well as control fibroblasts, no changes indicative of differences in mitochondrial mass were identified. A possible explanation for this discrepancy is that mitophagy is highly selective in an endogenous model and thus does not result in a readily observable reduction in mitochondrial mass [38], [42].
To obtain insight in the consequences of altered Mfn1/2 ubiquitination on mitochondrial morphology, we determined the degree of branching of the mitochondrial network by measuring the form factor [21] in PINK1-, Parkin-mutant and control cells. In line with our qualitative observations, this quantitative assessment showed no differences between mutant and control fibroblasts under basal conditions. By contrast, mitochondrial branching was found to be significantly increased in non-treated Parkin-mutants in an earlier study on fibroblasts [21]. When we stressed the mutant and control cells with valinomycin, the form factor decreased in all three samples. However, we detected a trend towards more fragmented mitochondria in control cells, supporting our hypothesis. It will be interesting to see whether this trend holds up in a larger sample of control, PINK1- and Parkin-mutant fibroblast cultures. In the above-mentioned study [21], fibroblasts were exposed to rotenone, an inhibitor of the respiratory chain complex I. This treatment caused mitochondrial fragmentation in Parkin-mutant and control cells similar to the effect of valinomycin in our cells. In this published study, however, no differences in branching were detected between the investigated groups after exposure to mitochondrial stress [21]. Since mitochondrial fusion and fission are transient events, dynamic quantification methods would be useful to determine the impact of PINK1 and Parkin mutations on mitochondrial morphology.
Confirming the results from arthropod studies and expanding on our previous findings, we showed that endogenous mutations in PINK1 and Parkin impair ubiquitination of mitofusins in human fibroblasts. In addition, our results imply that the UPS is involved in the degradation of Mitofusins under mitochondrial stress conditions.
Materials and Methods
Ethics statement
Written informed consent was obtained from all study participants and the study was approved by the local ethics committee of the University of Lübeck.
Tissue culture
Human dermal primary fibroblasts used in the present study were described before [16], [22], [43]. Fibroblasts and commercially available SH-SY5Y were cultured in high glucose Dulbecco's Modified Eagle's Medium supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin (all PAA) at 37°C, 5% CO2. In all assays, fibroblast passage numbers were matched (<10).
To induce mitochondrial stress, fibroblasts and SH-SY5Y cells were treated with the potassium ionophore valinomycin (1 µM, Sigma), the protonophore CCCP (10 µM, Sigma) or with the superoxide generator H2O2 (100 µM, Sigma). For inhibition of the proteasome system epoxomicin (10 µM, Sigma) or MG132 (10 µM, Sigma) were used. To inhibit the acidification of lysosomes bafilomycin was employed (10 nM, Sigma).
Mitochondrial preparation
Mitochondria were isolated from fibroblasts as previously described [43]. In brief, cells were harvested and homogenized in buffer containing 250 mM sucrose, 10 mM Tris and 1 mM EDTA, pH 7.4. After that, nuclei and unbroken cells were removed by centrifugation at 1,500×g for 20 min. The supernatant containing intact mitochondria was transferred into a new tube and centrifuged at 12,000×g for 10 min. Supernatant (“cytosolic fraction”) was transferred into another new tube and the mitochondria-enriched pellet (“mitochondrial fraction”) was dissolved in radioimmunoprecipitation assay (RIPA) buffer containing a cocktail of protease and phosphatase inhibitors (Roche Diagnostics).
Cytoplasmic fractions were concentrated by using centricon YM-10 devices (Millipore) according to the manufacturer's instructions. Proteins of the mitochondrial and cytoplasmic fractions were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS PAGE) and detected by Western blot analysis using appropriate antibodies.
Protein extraction
Proteins were extracted using RIPA buffer containing 0.1% SDS (50 mM Tris-HCl pH 7.6, 150 mM NaCl, 1% DOC, 1% NP-40 and 0.1% SDS). Cells or mitochondria-enriched pellets were dissolved in the appropriate amount of buffer and incubated on ice for 30 min. After that, the lysates were centrifuged at 16,000×g for 20 min at 4°C. The supernatant was transferred into a new tube and used for Western blotting.
Immunoprecipitation
Fibroblasts plated in 15 cm Petri dishes were treated with 1 µM valinomycin. Next, cells were harvested and resuspended in 1 ml of lysis buffer (150 mM NaCl, 50 mM Tris-HCl pH 7.6, 1% NP-40, 0.1% SDS, protease inhibitor cocktail (Roche Diagnostics)). Lysates were incubated on ice for 30 min and cleared by centrifugation at 13,000×g for 10 min. Samples were equalized for the protein concentration and incubated with 5 µl of anti-Mfn2 antibody overnight on a rotator. Fifty µl of washed protein G agarose beads (Roche Diagnostics) were added to the samples. This was followed by incubation for 2 h on a rotator. Next, the beads were pelleted by centrifugation and the supernatant was discarded. The beads were washed three times with lysis buffer followed by resuspension into 2× loading buffer (Invitrogen) and incubation at 95°C for 5 min. After centrifugation, the supernatant was analyzed by Western blotting.
Western blot analysis
SDS PAGE was performed using NuPAGE 4–12% Bis-Tris gels (Invitrogen). After electrophoresis, proteins were transferred to the nitrocellulose membrane (Protran) and probed with antibodies raised against Mfn1 (Abcam, # ab60939), Mfn2 (Abcam, # ab56889), β-actin (Sigma, # A 5316), VDAC1 (Abcam, # ab14734), OPA1 (Abcam, # ab42364), Fis1 (Alexis Biochemicals, # ALX-210-907), FLAG M2 (Sigma, # F 1804), V5 (Invitrogen, # R960-25) and ubiquitin (Boston Biochem, # AB-001). All Western blot analyses were performed in triplicates for all available mutants (PINK1: p.Q456X and p.V170G; Parkin: p.V324fsX434 and p.R245fsX253) and two controls, and representative blots are shown in the figures. For densitometric analyses TotalLab software (Nonlinear Dynamics) was used.
Transient transfection
Fibroblasts were transiently transfected with pcDNA3.1 V5/His (Invitrogen) containing full-length wild-type PINK1 cDNA (FL PINK1). For overexpression of Parkin, N-terminally FLAG-tagged full-length Parkin cDNA was cloned in pcDNA3.1 (Invitrogen's modified vector lacking V5/His tags). For Mfn1 or Mfn2 knock-down, Hs_MFN1_5 or Hs_MFN2_5 validated siRNAs (both Qiagen) (final concentration 50 nM) were used and scramble siRNA (Silencer negative control 1 siRNA [Ambion]) (final concentration 50 nM) with no known mammalian homology served as negative control. All transfections of fibroblasts were performed using the Nucleofector Device (Lonza).
Analysis of the mitochondrial membrane potential
The mitochondrial membrane potential was analyzed using 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolylcarbocyanine iodide (JC-1; Invitrogen) according to the manufacturer's protocol [44].
Assessment of mitochondrial branching
The mitochondrial network in fibroblasts was stained with an anti-GRP75 antibody (Abcam, # ab53098) in combination with the zenon immunolabelling kit (Invitrogen) according to the manufacturer's protocol.
The morphology of the mitochondrial network was investigated using a fluorescence microscope equipped with an ApoTome and AxioVision software (all Zeiss). By means of ImageJ 1.42, raw images were binarized, mitochondrion area and outline were measured and the form factor was calculated [21]. Images of at least five randomly selected cells per individual were analyzed under basal conditions and after treatment with valinomycin.
Statistical analysis
For evaluation of the impact of stress on cells, a paired Student's t-test was used to determine differences before and after treatment. All p-values below 0.05 were considered indicative of a significant difference between measurements and are shown by an asterisk.
Supporting Information
Protein levels of Mfn1 after valinomycin treatment. (A) Fibroblasts from a healthy control, a homozygous PINK1 mutant and a homozygous Parkin mutant were cultured under basal conditions or treated with 1 µM valinomycin for 12 h. The protein levels of Mfn1 were investigated by means of Western blotting. Valinomycin exposure caused a drop in Mfn1 levels in controls, but not in PINK1- or Parkin-mutant cells. β-actin served as a loading control. (B) Mutant cells were transfected with scrambled siRNA, Mfn1 siRNA, Mfn2 siRNA or a combination of Mfn1 and Mfn2 siRNA for 40 h. Western blot analysis was performed with an antibody against Mfn1. The Mfn1 levels decreased only when Mfn1 siRNA was employed, confirming the specificity of the anti-Mfn1 antibody used in our study. Mfn1 – mitofusin 1; Mfn2 – mitofusin 2.
(TIFF)
Protein levels of Mfn2 after CCCP treatment. Fibroblasts from two healthy controls, two homozygous PINK1 mutants and one homozygous Parkin mutant were cultured under basal conditions or treated with 10 µM CCCP for 12 h. The protein levels of Mfn2 were investigated by means of Western blotting. CCCP exposure caused a decrease in Mfn2 levels in controls, but not in PINK1- or Parkin-mutant cells. β-actin served as a loading control. CCCP – cyanide m-chlorophenylhydrazone; Mfn2 – mitofusin 2.
(TIFF)
Immunoprecipitation with antibodies against Mfn2. Control, PINK1- and Parkin-mutant cells were treated with 1 µM valinomycin for 12 h. Cells were harvested and whole cell lysates were used for immunoprecipitation with antibodies against GRP75 or Mfn2. The resulting precipitates were analyzed by Western blotting using antibodies against Mfn2 and GRP75. An Mfn2 immunoreactive band of higher molecular weight was detected only in controls but not in either of the mutants. Immunoprecipitation with an antibody against the mitochondrial marker GRP75 served as a negative control. GRP75 – glucose-regulated protein 75; IgG – immunoglobulin G; Mfn2 – mitofusin 2; Ub-Mfn2 – ubiquitylated mitofusin 2.
(TIFF)
Mitochondrial localization of ubiquitylated Mfn1 and Mfn2 after valinomycin treatment. Control fibroblasts were cultured under basal conditions or treated with 1 µM valinomycin for 6 h. Cells were harvested, proteins of mitochondrial and cytosolic fractions were loaded on two SDS-PAGE gels and analyzed by Western blotting. The subcellular localizations of Mfn1 and Mfn2 were determined. Quality of cellular fractionation was confirmed using antibodies against VDAC1 and β-actin. The ubiquitylated forms of Mfn1 and Mfn2, which were observed only after valinomycin stress, are exclusively found in the mitochondrial fraction. Cyt – cytosolic fraction; Mit – mitochondrial fraction; Mfn1 – mitofusin 1; Mfn2 – mitofusin 2; VDAC1 – voltage-dependent anion channel 1.
(TIFF)
Mitochondrial localization of ubiquitylated Mfn2 after valinomycin treatment in SH-SY5Y cells. SH-SY5Y cells were cultured under basal conditions or treated with valinomycin for 6 h. Cells were harvested and mitochondrial and cytosolic fractions were analyzed by Western blotting. The subcellular localization of Mfn2 was determined. Quality of cellular fractionation was confirmed using antibodies against VDAC and β-actin. The ubiquitylated forms of Mfn2, which were observed only after valinomycin stress, are exclusively found in the mitochondrial fraction. Mfn2 – mitofusin 2; Ub-Mfn2 – ubiquitylated mitofusin 2; VDAC1 – voltage-dependent anion channel 1.
(TIFF)
Ubiquitylation of Mfn2 occurs within 1 h of valinomycin treatment. Fibroblasts from a healthy control were treated with 1 µM valinomycin alone (left panel) or with 1 µM valinomycin plus 10 µM MG132 (right panel). Proteins were extracted at different time points and analyzed by Western blotting. Valinomycin treatment initiated the ubiquitylation of Mfn2 after 1 h of incubation. This effect was prevented by simultaneous exposure to MG132. β-actin served as a loading control. An unspecific band is marked by an asterisk. Mfn2 – mitofusin 2; Ub-Mfn2 – ubiquitylated mitofusin 2.
(TIFF)
Treatment with valinomycin alone or in combination with epoxomicin causes a drop in mitochondrial membrane potential. Control fibroblasts were incubated with either 1 µM valinomycin alone or with 1 µM valinomycin plus 10 µM epoxomicin. The membrane potential was measured at different time points and corrected for protein concentration. Exposure to the proteasome inhibitor epoxomicin did not influence the membrane potential over time. In the graph mean values +/− standard deviation of three independent experiments are given. Δψ – mitochondrial membrane potential.
(TIFF)
No accumulation of Mfn2 after exposure to epoxomicin. Control fibroblasts were treated with 10 µM epoxomicin. Proteins were extracted at different time points and analyzed by Western blotting. Exposure to the proteasome inhibitor epoxomicin did not affect the expression of Mfn2 over time. The mitochondrial marker VDAC1 and the cytosolic marker β-actin served as loading controls. Mfn2 – mitofusin 2; VDAC1 – voltage-dependent anion channel 1.
(TIFF)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the Medical Faculty of the University of Lübeck [to A.G.], the Volkswagen Foundation [to C.K.], the Hermann and Lilly Schilling Foundation [to C.K.], the German Research Foundation (DFG) [GR 3731/1-1 to A.G.], the Hilde Ulrichs Foundation for Parkinson's Disease Research [to C.K.], the EU Grant GENEPARK [EU-LSHB-CT-2006-037544 to C.K.], and the German ‘Bundesministerium für Bildung und Forschung’ (NGFN plus) [PNP-01GS08135-3 to C.K.]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Klein C, Schlossmacher MG. Parkinson disease, 10 years after its genetic revolution: multiple clues to a complex disorder. Neurology. 2007;69:2093–2104. doi: 10.1212/01.wnl.0000271880.27321.a7. [DOI] [PubMed] [Google Scholar]
- 2.Palacino JJ, Sagi D, Goldberg MS, Krauss S, Motz C, et al. Mitochondrial dysfunction and oxidative damage in parkin-deficient mice. J Biol Chem. 2004;279:18614–18622. doi: 10.1074/jbc.M401135200. [DOI] [PubMed] [Google Scholar]
- 3.Flinn L, Mortiboys H, Volkmann K, Koster RW, Ingham PW, et al. Complex I deficiency and dopaminergic neuronal cell loss in parkin-deficient zebrafish (Danio rerio). Brain. 2009;132:1613–1623. doi: 10.1093/brain/awp108. [DOI] [PubMed] [Google Scholar]
- 4.Gegg ME, Cooper JM, Schapira AH, Taanman JW. Silencing of PINK1 expression affects mitochondrial DNA and oxidative phosphorylation in dopaminergic cells. PLoS One. 2009;4:e4756. doi: 10.1371/journal.pone.0004756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schapira AH, Cooper JM, Dexter D, Jenner P, Clark JB, et al. Mitochondrial complex I deficiency in Parkinson's disease. Lancet. 1989;1:1269. doi: 10.1016/s0140-6736(89)92366-0. [DOI] [PubMed] [Google Scholar]
- 6.Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, et al. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- 7.Park J, Lee SB, Lee S, Kim Y, Song S, et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- 8.Yang Y, Gehrke S, Imai Y, Huang Z, Ouyang Y, et al. Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc Natl Acad Sci U S A. 2006;103:10793–10798. doi: 10.1073/pnas.0602493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greene JC, Whitworth AJ, Kuo I, Andrews LA, Feany MB, et al. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc Natl Acad Sci U S A. 2003;100:4078–4083. doi: 10.1073/pnas.0737556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pesah Y, Pham T, Burgess H, Middlebrooks B, Verstreken P, et al. Drosophila parkin mutants have decreased mass and cell size and increased sensitivity to oxygen radical stress. Development. 2004;131:2183–2194. doi: 10.1242/dev.01095. [DOI] [PubMed] [Google Scholar]
- 11.Poole AC, Thomas RE, Andrews LA, McBride HM, Whitworth AJ, et al. The PINK1/Parkin pathway regulates mitochondrial morphology. Proc Natl Acad Sci U S A. 2008;105:1638–1643. doi: 10.1073/pnas.0709336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng H, Dodson MW, Huang H, Guo M. The Parkinson's disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc Natl Acad Sci U S A. 2008;105:14503–14508. doi: 10.1073/pnas.0803998105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park J, Lee G, Chung J. The PINK1-Parkin pathway is involved in the regulation of mitochondrial remodeling process. Biochem Biophys Res Commun. 2009;378:518–523. doi: 10.1016/j.bbrc.2008.11.086. [DOI] [PubMed] [Google Scholar]
- 14.Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RL, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci U S A. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rakovic A, Grünewald A, Seibler P, Ramirez A, Kock N, et al. Effect of endogenous mutant and wild-type PINK1 on Parkin in fibroblasts from Parkinson disease patients. Hum Mol Genet. 2010;19:3124–3137. doi: 10.1093/hmg/ddq215. [DOI] [PubMed] [Google Scholar]
- 17.Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, et al. PINK1 Is Selectively Stabilized on Impaired Mitochondria to Activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poole AC, Thomas RE, Yu S, Vincow ES, Pallanck L. The mitochondrial fusion-promoting factor mitofusin is a substrate of the PINK1/parkin pathway. PLoS One. 2010;5:e10054. doi: 10.1371/journal.pone.0010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ziviani E, Tao RN, Whitworth AJ. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc Natl Acad Sci U S A. 2010;107:5018–5023. doi: 10.1073/pnas.0913485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gegg ME, Cooper JM, Chau KY, Rojo M, Schapira AH, et al. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum Mol Genet. 2010 doi: 10.1093/hmg/ddq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mortiboys H, Thomas KJ, Koopman WJ, Klaffke S, Abou-Sleiman P, et al. Mitochondrial function and morphology are impaired in parkin-mutant fibroblasts. Ann Neurol. 2008;64:555–565. doi: 10.1002/ana.21492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grünewald A, Breedveld GJ, Lohmann-Hedrich K, Rohe CF, Konig IR, et al. Biological effects of the PINK1 c.1366C>T mutation: implications in Parkinson disease pathogenesis. Neurogenetics. 2007;8:103–109. doi: 10.1007/s10048-006-0072-y. [DOI] [PubMed] [Google Scholar]
- 23.Hedrich K, Hagenah J, Djarmati A, Hiller A, Lohnau T, et al. Clinical spectrum of homozygous and heterozygous PINK1 mutations in a large German family with Parkinson disease: role of a single hit? Arch Neurol. 2006;63:833–838. doi: 10.1001/archneur.63.6.833. [DOI] [PubMed] [Google Scholar]
- 24.Moro E, Volkmann J, Konig IR, Winkler S, Hiller A, et al. Bilateral subthalamic stimulation in Parkin and PINK1 parkinsonism. Neurology. 2008;70:1186–1191. doi: 10.1212/01.wnl.0000307748.11216.03. [DOI] [PubMed] [Google Scholar]
- 25.Pramstaller PP, Schlossmacher MG, Jacques TS, Scaravilli F, Eskelson C, et al. Lewy body Parkinson's disease in a large pedigree with 77 Parkin mutation carriers. Ann Neurol. 2005;58:411–422. doi: 10.1002/ana.20587. [DOI] [PubMed] [Google Scholar]
- 26.Koshiba T, Detmer SA, Kaiser JT, Chen H, McCaffery JM, et al. Structural basis of mitochondrial tethering by mitofusin complexes. Science. 2004;305:858–862. doi: 10.1126/science.1099793. [DOI] [PubMed] [Google Scholar]
- 27.Langston JW, Ballard P, Tetrud JW, Irwin I. Chronic Parkinsonism in humans due to a product of meperidine-analog synthesis. Science. 1983;219:979–980. doi: 10.1126/science.6823561. [DOI] [PubMed] [Google Scholar]
- 28.Tanaka A, Cleland MM, Xu S, Narendra DP, Suen DF, et al. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J Cell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lutz AK, Exner N, Fett ME, Schlehe JS, Kloos K, et al. Loss of parkin or PINK1 function increases DRP1-dependent mitochondrial fragmentation. J Biol Chem. 2009 doi: 10.1074/jbc.M109.035774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeyaraju DV, Xu L, Letellier MC, Bandaru S, Zunino R, et al. Phosphorylation and cleavage of presenilin-associated rhomboid-like protein (PARL) promotes changes in mitochondrial morphology. Proc Natl Acad Sci U S A. 2006;103:18562–18567. doi: 10.1073/pnas.0604983103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaiser P, Huang L. Global approaches to understanding ubiquitination. Genome Biol. 2005;6:233. doi: 10.1186/gb-2005-6-10-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- 33.Verma R, Deshaies RJ. Assaying degradation and deubiquitination of a ubiquitinated substrate by purified 26S proteasomes. Methods Enzymol. 2005;398:391–399. doi: 10.1016/S0076-6879(05)98032-4. [DOI] [PubMed] [Google Scholar]
- 34.Cohen MM, Leboucher GP, Livnat-Levanon N, Glickman MH, Weissman AM. Ubiquitin-proteasome-dependent degradation of a mitofusin, a critical regulator of mitochondrial fusion. Mol Biol Cell. 2008;19:2457–2464. doi: 10.1091/mbc.E08-02-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Twig G, Hyde B, Shirihai OS. Mitochondrial fusion, fission and autophagy as a quality control axis: the bioenergetic view. Biochim Biophys Acta. 2008;1777:1092–1097. doi: 10.1016/j.bbabio.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawajiri S, Saiki S, Sato S, Sato F, Hatano T, et al. PINK1 is recruited to mitochondria with parkin and associates with LC3 in mitophagy. FEBS Lett. 2010;584:1073–1079. doi: 10.1016/j.febslet.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 37.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen H, Chan DC. Mitochondrial dynamics–fusion, fission, movement, and mitophagy–in neurodegenerative diseases. Hum Mol Genet. 2009;18:R169–176. doi: 10.1093/hmg/ddp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Piccoli C, Ripoli M, Quarato G, Scrima R, D'Aprile A, et al. Coexistence of mutations in PINK1 and mitochondrial DNA in early onset parkinsonism. J Med Genet. 2008;45:596–602. doi: 10.1136/jmg.2008.058628. [DOI] [PubMed] [Google Scholar]
- 40.Rothfuss O, Gasser T, Patenge N. Analysis of differential DNA damage in the mitochondrial genome employing a semi-long run real-time PCR approach. Nucleic Acids Res. 2010;38:e24. doi: 10.1093/nar/gkp1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suen DF, Narendra DP, Tanaka A, Manfredi G, Youle RJ. Parkin overexpression selects against a deleterious mtDNA mutation in heteroplasmic cybrid cells. Proc Natl Acad Sci U S A. 2010;107:11835–11840. doi: 10.1073/pnas.0914569107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanki T, Klionsky DJ. Mitophagy in yeast occurs through a selective mechanism. J Biol Chem. 2008;283:32386–32393. doi: 10.1074/jbc.M802403200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Almeida A, Medina JM. Isolation and characterization of tightly coupled mitochondria from neurons and astrocytes in primary culture. Brain Res. 1997;764:167–172. doi: 10.1016/s0006-8993(97)00453-8. [DOI] [PubMed] [Google Scholar]
- 44.Grünewald A, Gegg ME, Taanman JW, King RH, Kock N, et al. Differential effects of PINK1 nonsense and missense mutations on mitochondrial function and morphology. Exp Neurol. 2009;219:266–273. doi: 10.1016/j.expneurol.2009.05.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Protein levels of Mfn1 after valinomycin treatment. (A) Fibroblasts from a healthy control, a homozygous PINK1 mutant and a homozygous Parkin mutant were cultured under basal conditions or treated with 1 µM valinomycin for 12 h. The protein levels of Mfn1 were investigated by means of Western blotting. Valinomycin exposure caused a drop in Mfn1 levels in controls, but not in PINK1- or Parkin-mutant cells. β-actin served as a loading control. (B) Mutant cells were transfected with scrambled siRNA, Mfn1 siRNA, Mfn2 siRNA or a combination of Mfn1 and Mfn2 siRNA for 40 h. Western blot analysis was performed with an antibody against Mfn1. The Mfn1 levels decreased only when Mfn1 siRNA was employed, confirming the specificity of the anti-Mfn1 antibody used in our study. Mfn1 – mitofusin 1; Mfn2 – mitofusin 2.
(TIFF)
Protein levels of Mfn2 after CCCP treatment. Fibroblasts from two healthy controls, two homozygous PINK1 mutants and one homozygous Parkin mutant were cultured under basal conditions or treated with 10 µM CCCP for 12 h. The protein levels of Mfn2 were investigated by means of Western blotting. CCCP exposure caused a decrease in Mfn2 levels in controls, but not in PINK1- or Parkin-mutant cells. β-actin served as a loading control. CCCP – cyanide m-chlorophenylhydrazone; Mfn2 – mitofusin 2.
(TIFF)
Immunoprecipitation with antibodies against Mfn2. Control, PINK1- and Parkin-mutant cells were treated with 1 µM valinomycin for 12 h. Cells were harvested and whole cell lysates were used for immunoprecipitation with antibodies against GRP75 or Mfn2. The resulting precipitates were analyzed by Western blotting using antibodies against Mfn2 and GRP75. An Mfn2 immunoreactive band of higher molecular weight was detected only in controls but not in either of the mutants. Immunoprecipitation with an antibody against the mitochondrial marker GRP75 served as a negative control. GRP75 – glucose-regulated protein 75; IgG – immunoglobulin G; Mfn2 – mitofusin 2; Ub-Mfn2 – ubiquitylated mitofusin 2.
(TIFF)
Mitochondrial localization of ubiquitylated Mfn1 and Mfn2 after valinomycin treatment. Control fibroblasts were cultured under basal conditions or treated with 1 µM valinomycin for 6 h. Cells were harvested, proteins of mitochondrial and cytosolic fractions were loaded on two SDS-PAGE gels and analyzed by Western blotting. The subcellular localizations of Mfn1 and Mfn2 were determined. Quality of cellular fractionation was confirmed using antibodies against VDAC1 and β-actin. The ubiquitylated forms of Mfn1 and Mfn2, which were observed only after valinomycin stress, are exclusively found in the mitochondrial fraction. Cyt – cytosolic fraction; Mit – mitochondrial fraction; Mfn1 – mitofusin 1; Mfn2 – mitofusin 2; VDAC1 – voltage-dependent anion channel 1.
(TIFF)
Mitochondrial localization of ubiquitylated Mfn2 after valinomycin treatment in SH-SY5Y cells. SH-SY5Y cells were cultured under basal conditions or treated with valinomycin for 6 h. Cells were harvested and mitochondrial and cytosolic fractions were analyzed by Western blotting. The subcellular localization of Mfn2 was determined. Quality of cellular fractionation was confirmed using antibodies against VDAC and β-actin. The ubiquitylated forms of Mfn2, which were observed only after valinomycin stress, are exclusively found in the mitochondrial fraction. Mfn2 – mitofusin 2; Ub-Mfn2 – ubiquitylated mitofusin 2; VDAC1 – voltage-dependent anion channel 1.
(TIFF)
Ubiquitylation of Mfn2 occurs within 1 h of valinomycin treatment. Fibroblasts from a healthy control were treated with 1 µM valinomycin alone (left panel) or with 1 µM valinomycin plus 10 µM MG132 (right panel). Proteins were extracted at different time points and analyzed by Western blotting. Valinomycin treatment initiated the ubiquitylation of Mfn2 after 1 h of incubation. This effect was prevented by simultaneous exposure to MG132. β-actin served as a loading control. An unspecific band is marked by an asterisk. Mfn2 – mitofusin 2; Ub-Mfn2 – ubiquitylated mitofusin 2.
(TIFF)
Treatment with valinomycin alone or in combination with epoxomicin causes a drop in mitochondrial membrane potential. Control fibroblasts were incubated with either 1 µM valinomycin alone or with 1 µM valinomycin plus 10 µM epoxomicin. The membrane potential was measured at different time points and corrected for protein concentration. Exposure to the proteasome inhibitor epoxomicin did not influence the membrane potential over time. In the graph mean values +/− standard deviation of three independent experiments are given. Δψ – mitochondrial membrane potential.
(TIFF)
No accumulation of Mfn2 after exposure to epoxomicin. Control fibroblasts were treated with 10 µM epoxomicin. Proteins were extracted at different time points and analyzed by Western blotting. Exposure to the proteasome inhibitor epoxomicin did not affect the expression of Mfn2 over time. The mitochondrial marker VDAC1 and the cytosolic marker β-actin served as loading controls. Mfn2 – mitofusin 2; VDAC1 – voltage-dependent anion channel 1.
(TIFF)