Abstract
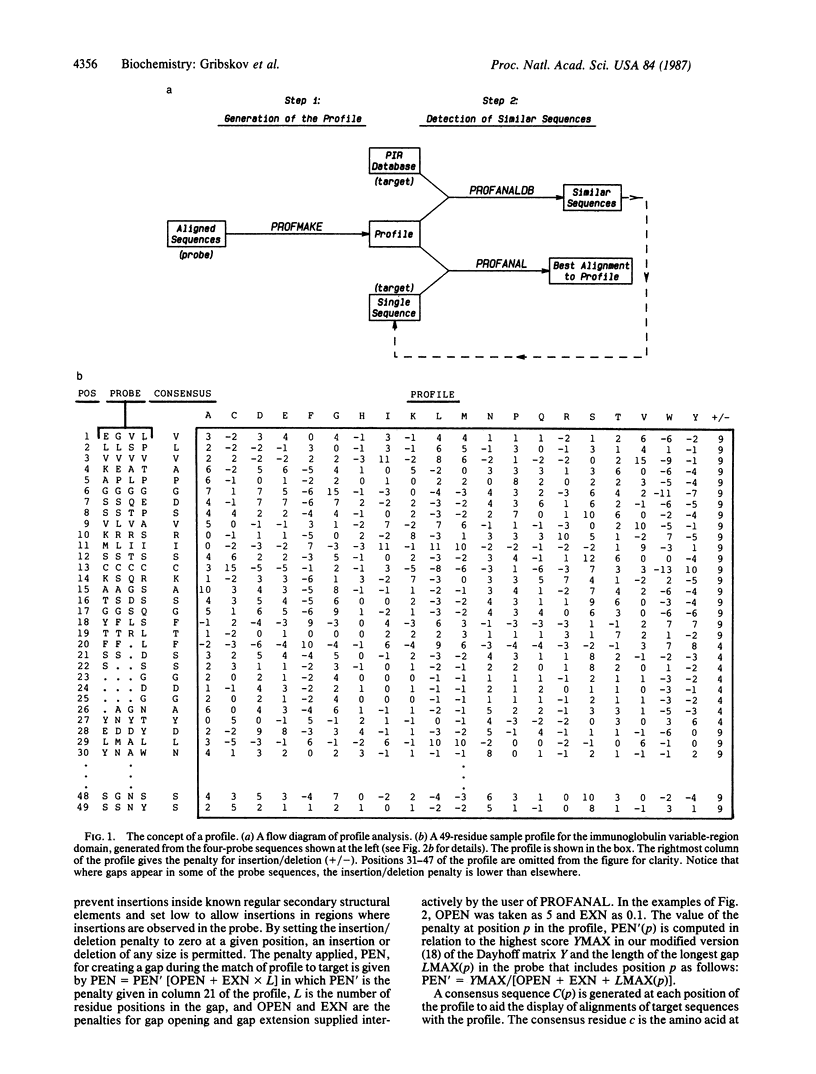
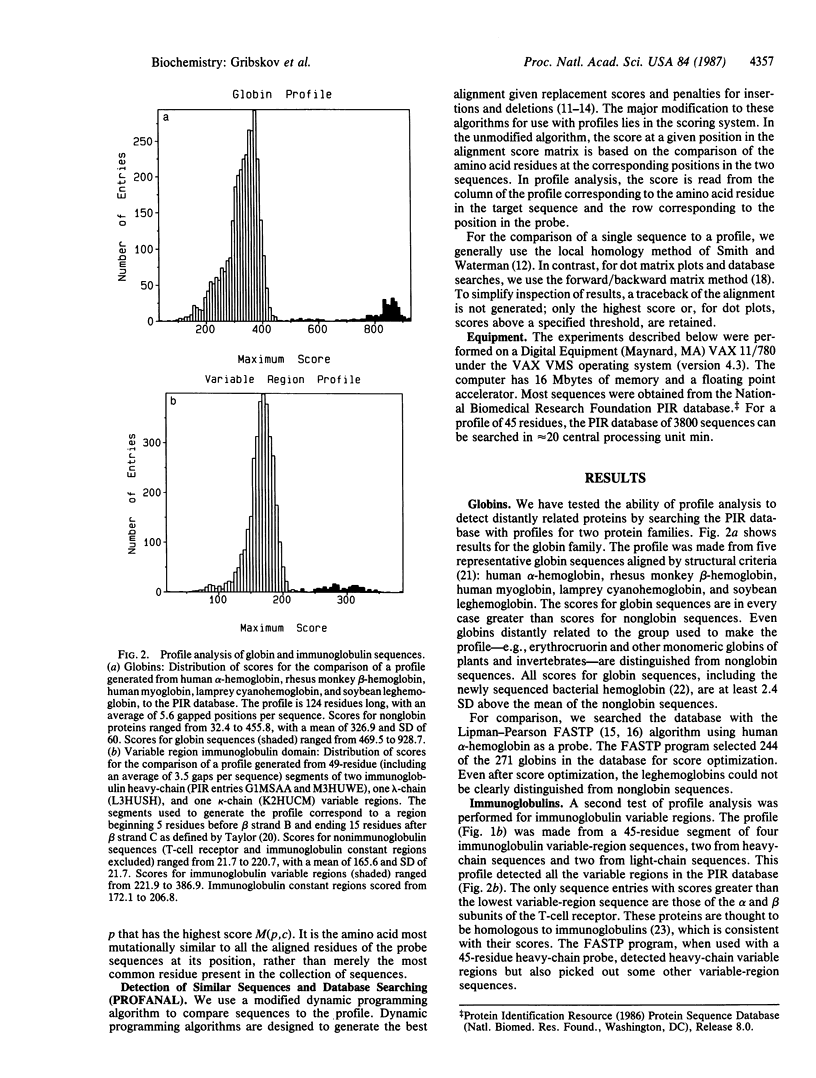
Profile analysis is a method for detecting distantly related proteins by sequence comparison. The basis for comparison is not only the customary Dayhoff mutational-distance matrix but also the results of structural studies and information implicit in the alignments of the sequences of families of similar proteins. This information is expressed in a position-specific scoring table (profile), which is created from a group of sequences previously aligned by structural or sequence similarity. The similarity of any other sequence (target) to the group of aligned sequences (probe) can be tested by comparing the target to the profile using dynamic programming algorithms. The profile method differs in two major respects from methods of sequence comparison in common use: (i) Any number of known sequences can be used to construct the profile, allowing more information to be used in the testing of the target than is possible with pairwise alignment methods. (ii) The profile includes the penalties for insertion or deletion at each position, which allow one to include the probe secondary structure in the testing scheme. Tests with globin and immunoglobulin sequences show that profile analysis can distinguish all members of these families from all other sequences in a database containing 3800 protein sequences.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blundell T., Sibanda B. L., Pearl L. Three-dimensional structure, specificity and catalytic mechanism of renin. Nature. 1983 Jul 21;304(5923):273–275. doi: 10.1038/304273a0. [DOI] [PubMed] [Google Scholar]
- Boswell D. R., McLachlan A. D. Sequence comparison by exponentially-damped alignment. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):457–464. doi: 10.1093/nar/12.1part2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Empirical predictions of protein conformation. Annu Rev Biochem. 1978;47:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle R. F. Similar amino acid sequences: chance or common ancestry? Science. 1981 Oct 9;214(4517):149–159. doi: 10.1126/science.7280687. [DOI] [PubMed] [Google Scholar]
- Fitch W. M. An improved method of testing for evolutionary homology. J Mol Biol. 1966 Mar;16(1):9–16. doi: 10.1016/s0022-2836(66)80258-9. [DOI] [PubMed] [Google Scholar]
- Gribskov M., Burgess R. R. Sigma factors from E. coli, B. subtilis, phage SP01, and phage T4 are homologous proteins. Nucleic Acids Res. 1986 Aug 26;14(16):6745–6763. doi: 10.1093/nar/14.16.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W., Sander C. On the use of sequence homologies to predict protein structure: identical pentapeptides can have completely different conformations. Proc Natl Acad Sci U S A. 1984 Feb;81(4):1075–1078. doi: 10.1073/pnas.81.4.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesk A. M., Chothia C. How different amino acid sequences determine similar protein structures: the structure and evolutionary dynamics of the globins. J Mol Biol. 1980 Jan 25;136(3):225–270. doi: 10.1016/0022-2836(80)90373-3. [DOI] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Maizel J. V., Jr, Lenk R. P. Enhanced graphic matrix analysis of nucleic acid and protein sequences. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7665–7669. doi: 10.1073/pnas.78.12.7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLachlan A. D. Analysis of gene duplication repeats in the myosin rod. J Mol Biol. 1983 Sep 5;169(1):15–30. doi: 10.1016/s0022-2836(83)80173-9. [DOI] [PubMed] [Google Scholar]
- Murata M., Richardson J. S., Sussman J. L. Simultaneous comparison of three protein sequences. Proc Natl Acad Sci U S A. 1985 May;82(10):3073–3077. doi: 10.1073/pnas.82.10.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman S. B., Wunsch C. D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970 Mar;48(3):443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Staden R. Computer methods to locate signals in nucleic acid sequences. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):505–519. doi: 10.1093/nar/12.1part2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet R. M., Eisenberg D. Correlation of sequence hydrophobicities measures similarity in three-dimensional protein structure. J Mol Biol. 1983 Dec 25;171(4):479–488. doi: 10.1016/0022-2836(83)90041-4. [DOI] [PubMed] [Google Scholar]
- Sweet R. M. Evolutionary similarity among peptide segments is a basis for prediction of protein folding. Biopolymers. 1986 Aug;25(8):1565–1577. doi: 10.1002/bip.360250813. [DOI] [PubMed] [Google Scholar]
- Taylor W. R. Identification of protein sequence homology by consensus template alignment. J Mol Biol. 1986 Mar 20;188(2):233–258. doi: 10.1016/0022-2836(86)90308-6. [DOI] [PubMed] [Google Scholar]
- Taylor W. R., Thornton J. M. Recognition of super-secondary structure in proteins. J Mol Biol. 1984 Mar 15;173(4):487–512. [PubMed] [Google Scholar]
- Wakabayashi S., Matsubara H., Webster D. A. Primary sequence of a dimeric bacterial haemoglobin from Vitreoscilla. 1986 Jul 31-Aug 6Nature. 322(6078):481–483. doi: 10.1038/322481a0. [DOI] [PubMed] [Google Scholar]
- Wierenga R. K., Terpstra P., Hol W. G. Prediction of the occurrence of the ADP-binding beta alpha beta-fold in proteins, using an amino acid sequence fingerprint. J Mol Biol. 1986 Jan 5;187(1):101–107. doi: 10.1016/0022-2836(86)90409-2. [DOI] [PubMed] [Google Scholar]
- Wilbur W. J., Lipman D. J. Rapid similarity searches of nucleic acid and protein data banks. Proc Natl Acad Sci U S A. 1983 Feb;80(3):726–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagi Y., Yoshikai Y., Leggett K., Clark S. P., Aleksander I., Mak T. W. A human T cell-specific cDNA clone encodes a protein having extensive homology to immunoglobulin chains. Nature. 1984 Mar 8;308(5955):145–149. doi: 10.1038/308145a0. [DOI] [PubMed] [Google Scholar]



