Abstract
Background
The FEZ (fasciculation and elongation protein zeta) family designation was purposed by Bloom and Horvitz by genetic analysis of C. elegans unc-76. Similar human sequences were identified in the expressed sequence tag database as FEZ1 and FEZ2. The unc-76 function is necessary for normal axon fasciculation and is required for axon-axon interactions. Indeed, the loss of UNC-76 function results in defects in axonal transport. The human FEZ1 protein has been shown to rescue defects caused by unc-76 mutations in nematodes, indicating that both UNC-76 and FEZ1 are evolutionarily conserved in their function. Until today, little is known about FEZ2 protein function.
Methodology/Principal Findings
Using the yeast two-hybrid system we demonstrate here conserved evolutionary features among orthologs and non-conserved features between paralogs of the FEZ family of proteins, by comparing the interactome profiles of the C-terminals of human FEZ1, FEZ2 and UNC-76 from C. elegans. Furthermore, we correlate our data with an analysis of the molecular evolution of the FEZ protein family in the animal kingdom.
Conclusions/Significance
We found that FEZ2 interacted with 59 proteins and that of these only 40 interacted with FEZ1. Of the 40 FEZ1 interacting proteins, 36 (90%), also interacted with UNC-76 and none of the 19 FEZ2 specific proteins interacted with FEZ1 or UNC-76. This together with the duplication of unc-76 gene in the ancestral line of chordates suggests that FEZ2 is in the process of acquiring new additional functions. The results provide also an explanation for the dramatic difference between C. elegans and D. melanogaster unc-76 mutants on one hand, which cause serious defects in the nervous system, and the mouse FEZ1 -/- knockout mice on the other, which show no morphological and no strong behavioural phenotype. Likely, the ubiquitously expressed FEZ2 can completely compensate the lack of neuronal FEZ1, since it can interact with all FEZ1 interacting proteins and additional 19 proteins.
Introduction
The FEZ (fasciculation and elongation protein zeta) family designation was first purposed by Bloom and colleagues in 1997 by genetic analysis of C. elegans unc-76. The 376 and 385 amino acid containing isoforms of the UNC-76 protein arise by alternative splicing of the same gene and showed no strong similarity to any previously characterized proteins, but similar human sequences were identified in the expressed sequence tag database, dbEST [1], [2] and Bloom & Horvitz named the two identified human genes FEZ1 and FEZ2.
The unc-76 function is necessary for normal fascicle structure and is required specifically for axon-axon interactions in C. elegans. The human FEZ1 gene was able to restore partial locomotion and axonal fasciculation in the C. elegans unc-76 mutants in germ-line transformation experiments, indicating that both the function and the structure of the FEZ proteins have been conserved in evolution [1]. Loss of Drosophila Unc-76 function results in locomotion and axonal transport defects reminiscent of phenotypes observed in kinesin mutants and thereby suggesting that UNC-76 is required for kinesin-dependent axonal transport [3]. The FEZ1-deficient mice however, did not exhibit any obvious abnormal brain architecture, although they manifest slight behavioral abnormalities, including a hyperlocomotion phenotype and enhanced responsiveness to psychostimulants [4].
The homologous proteins UNC-76, FEZ1 and FEZ2, share a conserved predicted coiled-coil region at their C-terminal regions [1], [5], [6]. Coiled-coils are autonomous folding units consisting of two to four α-helices that wrap around each other with a slight left-handed super-helical twist [7] and thereby mediate sub-unit oligomerization [8] as well as protein-protein interactions [9]. Up to now 4 interactions were reported for C. elegans UNC-76, 45 interactions for human FEZ1 and 17 interactions for human FEZ2 according to BIND, HPRD, BioGRID data and previous research published by our group [10]. All of these interactions occur at the C-terminus with the predicted coiled-coil region. Some of these interactions implicate FEZ1 protein in the context of schizophrenia (DISC1) [11], [12], aspects of viral infection such as post-entry block of retroviral infection [13] and inhibition of the release of progeny virions (agnoprotein of the human polyomavirus JC virus) [14], and naturally in neuritogenesis (NBR1, PKCζ, DISC1) [5], [11], [15]. Furthermore, the association between FEZ1 and E4B was enhanced by co-expression of a constitutively active form of protein kinase Cζ, and phosphorylation of FEZ1 by this kinase and its subsequent ubiquitylation by E4B resulted in neurite extension in PC12 cells [4], [16]. FEZ1 also contributes to the polarization of hippocampal neurons by controlling mitochondrial motility [4], [17].
Here, we report that nucleotide sequences belonging to the FEZ family can be found in many eukaryotic species, but that most of the putative homologs are not only functionally uncharacterized but even remain unrecognized as FEZ family members in the databases. Our phylogenetic analysis of the FEZ family of proteins indicates that the ancestral gene of the FEZ family is found only in the animal kingdom. The gene duplication of unc-76 to FEZ1 and FEZ2 is predicted to have occurred during the two rounds of whole-genome duplication in the chordate ancestral line, after the divergence of cephalochordates but before the splitting of the teleosts and tetrapods taxa.
The most conserved region in FEZ proteins is its C-terminus, and we and others showed previously that this region is involved in the association with other proteins [10], [18]. Here, yeast two-hybrid assays with C. elegans UNC-76 (248–372), human FEZ1 (221–392) and human FEZ2 (207–353) as baits and human prey clones from cDNA libraries demonstrated that the pattern of protein-protein interactions (PPIs) is highly conserved between UNC-76 and FEZ1. These orthologues did however not interact with 19 proteins that interacted specifically with FEZ2, although FEZ2 interacted with all 40 proteins that interact with FEZ1/UNC-76. In summary, we found largely overlapping PPI patterns for FEZ1 and UNC-76 and extended PPIs for FEZ2. Interestingly, our data provide an explanation for the ability of FEZ1 to rescue the defects caused by unc-76 mutations in nematodes and to the lack of strong defects in FEZ1 deficient mice, since the continuous presence of the ubiquitous expressed paralogue FEZ2 may be able to compensate lack of FEZ1 because its PPI pattern contains all the interactions found for FEZ1.
Methods
Protein sequence analysis and multiple sequence alignments
Protein Psi-Blast [19] searches with the full length C. elegans UNC-76 sequence were performed at the NCBI Web site http://www.ncbi.nlm.nih.gov/BLAST/ using the non-redundant protein sequence database available at August 16, 2009. After six rounds of iteration, all UNC-76, FEZ1 and FEZ2 orthologs with an E-value of 0.005 or below were selected and all redundant sequences were excluded (except to B. floridae). All sequences were collected in FASTA format for further analysis as shown in Table S1. The identification and posterior naming of the protein sequences as either UNC-76, FEZ1 or FEZ2 is based on the phylogenetical analyses shown in Figure 1 and Figure S1. The sequences were aligned using the ClustalW2 Web Site http://www.ebi.ac.uk/Tools/clustalw2/index.html at default settings. The alignments were then shaded using the multiple sequence alignment editor GENEDOC http://www.nrbsc.org/gfx/genedoc/index.html. The identity and similarity were calculated using default settings by NPS@: Network Protein Sequence Analysis trough the alignments of human proteins FEZ1 and FEZ2 and C. elegans UNC-76 protein (http://npsa-pbil.ibcp.fr/cgi-bin/align_clustalw.pl) [20].
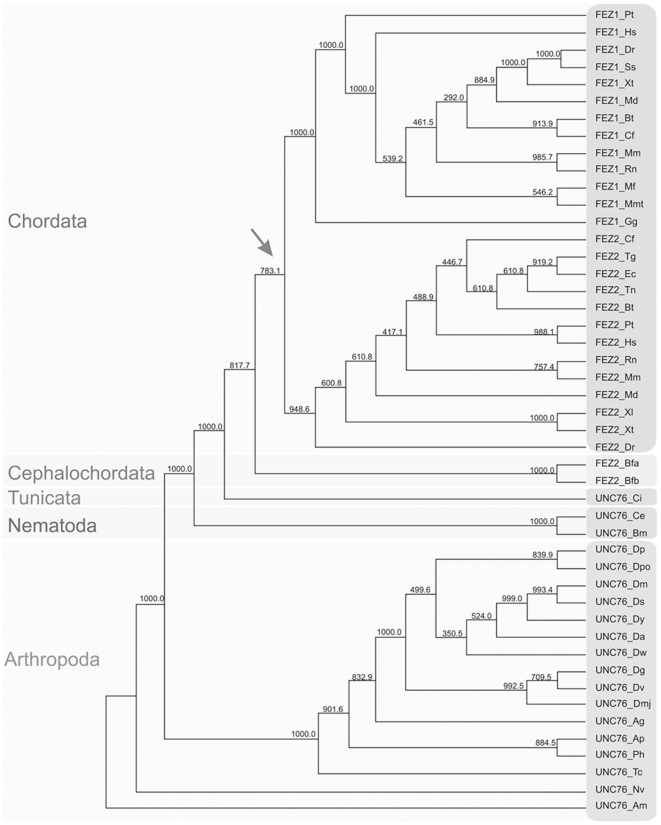
Figure 1. A phylogenetic tree of the FEZ protein family.
The tree was derived by the parsimony method (Phylip protpars). The bootstrap multidataset and resampling method was employed and the numbers on the branches indicate the number of times the partition of the species into the two sets which are separated by that branch occurred among the trees out of 999.96 trees. The arrow indicates the probable point of FEZ gene duplication. Ag – Anopheles gambiae, Am – Apis mellifera, Ap – Acyrthosiphon pisum, Bf – Branchiostoma floridae (Bfa and Bfb refer to two sequences [polymorphisms] of selected hypothetical genes in this species), Bm – Brugia malayi, Bt – Bos taurus, Ce – Caenorhabditis elegans, Cf – Canis familiaris, Ci – Ciona intestinalis, Da – Drosophila ananassae, Dg – Drosophila grimshawi, Dm – Drosophila melanogaster, Dmj – Drosophila mojavensis, Dp – Drosophila persimilis, Dpo – Drosophila pseudoobscura, Dr – Danio rerio, Ds – Drosophila sechellia, Dv – Drosophila virilis, Dw – Drosophila willistoni, Dy – Drosophila yakuba, Ec – Equus caballus, Gg –Gallus gallus, Hs – Homo sapiens, Md – Monodelphis domestica, Mf – Macaca fascicularis, Mm – Mus musculus, Mmt – Macaca mulatta, Nv – Nasonia vitripennis, Ph – Pediculus humanus, Pt – Pan troglodytes, Rn – Rattus norvegicus, Ss – Salmo salar, Tc – Tribolium castaneum, Tg –Taeniopygia guttata, Tn – Tetraodon nigroviridis, Xl – Xenopus laevis, Xt – Xenopus tropicalis.
Phylogenetical analyses
PHYLIP version 3.5c [21] was used for the phylogenetical analyses at the web site http://mobyle.pasteur.fr/cgi-bin/portal.py?jobs=http://mobyle.pasteur.fr/data/jobs/protpars. Parsimony analyses were performed using the protein alignment as input. Bootstrap values were obtained by using SEQBOOT and creating 1000 Bootstrap data sets. Analysis was then performed using PROTPARS with standard parameters. The “M” option was invoked for the analysis of the multiple data sets generated by SEQBOOT.
Plasmid constructions
The plasmid constructions of pBMT116-FEZ1 (221–392) (NM_005103.3) and pBTM-FEZ2 (207–353) (NM_005102.2) have been described previously [10]. The nucleotide sequence coding residues 248–372 of C. elegans UNC-76 (NM_074311.3) was PCR-amplified using a specific primer set (5′- GAATTCGATAATCTTCAAGAGCTCTCC-3′ , 5′- GTCGACCTAACACGATATATTTTTTGG-3′) as well as the template plasmid pSU001, that had been generously provided by Dr. Hengartner [4]. Subsequently the fragment was cloned in pGEM®-T vector (Promega), and subcloned via EcoRI and SalI restriction sites into the vector pBTM116. The orientation, frame, and correctness of sequence of each insert DNA was confirmed by restriction endonuclease analysis and automated DNA sequencing. These are no new cell lines but only cDNA clones obtained by in vitro experiments as described.
Yeast two-hybrid screen and DNA sequence analysis
Using FEZ1 as a bait in a screening, Assmann et al. (2006) [10] identified 16 proteins interacting with FEZ1 and all of them interacted with FEZ2 protein (Fig. 2A). In this work, yeast two-hybrid screens [22] of human fetal brain and bone marrow cDNA libraries (Clontech) were performed as described previously by using the yeast strain L40 (trp1-901, his3Δ200, leu2-3, ade2 LYS2::(lexAop)4-HIS3 URA3::(lexAop)8-lac GAL4) and human FEZ2 (207–353) as a bait in fusion with the LexA protein as encoded in the recombinant vector pBTM116 [23]. This fragment of FEZ2 does not auto-activate the yeast reporter genes (as FEZ proteins full length), neither did so the C-terminal constructs of FEZ1 and UNC-76 used in one-to-one confirmation. The autonomous activation test for HIS3 was performed in minimal medium plates without tryptophane and histidine but containing 10, 20, 30, or 50 mM of 3-amino-1,2,4-triazole (3-AT). Furthermore, the autonomous activation of LacZ was measured by the beta-galactosidase filter assay as described below. Yeast cells were transformed according to the protocols supplied by the cDNA library manufacturer (Clontech). The screening with FEZ2 (207–353) as bait was performed in minimal medium plates without tryptophan, leucine, and histidine and with addition of 10 mM of 3-AT to repress unspecific background growth.Recombinant pACT2 plasmids of positive clones were isolated and their insert DNAs were sequenced with a DNA sequencer model 377S (Applied Biosystems, Foster City, CA). The obtained DNA sequence data were compared with sequences in the NCBI data bank using the BLASTX 2.2.12 program [19]. The corresponding Accession numbers of the DNA sequences identified are given in the Table S1. As no new sequences have been obtained no new sequence data have been deposited in the GenBank. After identification, one representative clone of each prey protein was selected to yeast two-hybrid one-to-one confirmation.
Figure 2. Schematic representation summarizing the primary yeast two-hybrid screens with UNC-76, FEZ1 and FEZ2 as baits.
A) Yeast two-hybrid screen performed with FEZ1 (221–392) as a bait using a human fetal brain cDNA library retrieved 16 proteins that were confirmed to interact all with both FEZ1(221–392) and FEZ2 (207–353) in the one-to-one assay. B) Yeast two-hybrid screen performed with FEZ2 (207–353) as a bait using human fetal brain and bone marrow cDNA libraries retrieved 59 proteins as preys. In the one-to-one confirmation, 19 of these proteins interacted only with FEZ2 (207–353) protein, 40 with both FEZ2 (207–353) and FEZ1 (221–392) and 36 interacted with all FEZ2 (207–353), FEZ1 (221–392) and UNC-76 (242–378).
Yeast Two-hybrid assays one-to-one confirmation with the homologues FEZ1, FEZ2 and UNC-76
Yeast two-hybrid assays were performed with human FEZ1 (221–392) and C. elegans UNC-76 (248–372) as additional baits fused to LexA in vector pBTM116, as described above and previously [10]. The recombinant pACT2 plasmids of positives clones isolated from the library screens with bait FEZ2 (207–353) were used as preys and the interactions with FEZ1 and UNC-76 (and control FEZ2) were analyzed by growth in minimal medium plates and activation of the LacZ using the standard beta-galactosidase filter assay described in the following section (Fig. 2B).
Assay for beta-galactosidase activity in yeast cells
beta-Galactosidase activity in yeast cells was determined by the filter assay method. Yeast transformants (Leu+, Trp+, and His+) were transferred onto nylon membranes, permeabilized in liquid nitrogen, and placed on Whatman 3 MM paper previously soaked in Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM MgCl2, 50 mM 2-mercaptoethanol, pH 7.0) containing 1 mg/ml 5-bromo-4-chloro-3-indolyl-β-D-galactoside (X-gal). After incubation at 37°C for 1 h, the yeast cells forming dark blue colonies were taken from replica plates for further analysis (primary screen).
Results
Identification of members of the FEZ family and of conserved regions in their amino acid sequences
A database of sequences judged to be members of the FEZ protein family was compiled (Table S1, see Material and Methods section). Altogether 47 members for the FEZ family were identified by Psi-Blast searches with the C. elegans UNC-76 sequence in the non-redundant protein sequence database. FEZ1 family sequences were found in a variety of species ranging from Nematodes, via Arthropods to Mammals but not in Plants, Fungi or Protists. Multiple Clustal W alignments of sequences of the FEZ family identified a highly conserved region both at the N-terminus (ca. aa 160–180 in FEZ1) and at the C-terminus (Figure S1). The latter consists mainly in coiled-coil regions [10], [24], [25] located at the C-terminus which mediate the majority of the protein-protein interactions (ca. aa 230–307 in FEZ1) [10]. The alignment revealed, that the nuclear localization signal (NLS) (KKRRK, ca. aa 290–294 in FEZ1) [26], which consists of basic amino acids, is quite conserved in all FEZ1 genes that resulted from the putative unc-76 gene duplication (see details below), suggesting that new nuclear functions may have been acquired in FEZ1 afterwards. Although there is no striking NLS in FEZ2 sequences, they contain a conserved polybasic region (e.g. KKKKK) with conserved amino acid substitutions in the FEZ1 related NLS. The NLS is less well conserved in all UNC-76 sequences.
Phylogenetic analysis of the FEZ gene duplication
A phylogenetic tree (Fig. 1) was generated from the Clustal W alignment (Figure S1) of the all identified FEZ protein sequences (Table S1) using the parsimony method. Bootstrap options to resampling methods were defined to contain 1000 replications. The presence of FEZ1 and FEZ2 in all analyzed Actinopterygii (ray-finned fishes) genomes suggests that the FEZ1 and FEZ2 genes originated from the ancestral unc-76 by gene or whole genome duplications before the radiation of the Actinopterygii but after the divergence of the amphioxus branch and hence concomitant origin of the chordata. In fact, according to an amphioxus-human synteny analysis by Putnam [27] two rounds of whole-genome duplication seem to have occurred in the chordate stemline, after the divergence of the cephalochordates but before the split into teleosts and tetrapods (Fig. 1). Hence, the unc-76 gene seems to have duplicated in the chordate stemline into FEZ1 and FEZ2 genes.
Identification of proteins that interact with FEZ2
So far, only interactions for C-terminal of FEZ proteins are described, as by our screenings using C termini of FEZ proteins (due to auto-activation of full length and N-terminal constructs of the FEZ proteins as baits in these assays), as well as by other publications describing FEZ proteins recovered as prey in other screens (in this case, the interaction region was mapped to be the C-terminal region) [5], [14]. In a previous study, we had identified proteins that interact with human FEZ1 using its C-terminal region (221–392) as a bait in a yeast two-hybrid screen of a human fetal brain cDNA library [10]. We further found that all of the 16 interacting proteins identified were also able to interact with FEZ2 (207–353). The interacting proteins are functionally involved in transcriptional regulation (6 proteins), neuronal cell development (2 proteins), intracellular transport processes (3 proteins), apoptosis (2 proteins), or were of unknown function (3 proteins). Although these data suggested that FEZ1 and FEZ2 have largely overlapping protein interaction profiles we wanted to deepen our insights by performing another two-hybrid screen with the human paralogue FEZ2 as well as by introducing an evolutionary component by comparing the interaction profiles of both proteins with the orthologous C. elegans protein UNC-76, which may have preserved ancient features.
To this end we started by employing the yeast two-hybrid system [23] to screen humans fetal brain and bone marrow cDNA libraries. The truncated FEZ2 (207–353), which showed no auto-activation (see Materials and Methods), was used as a bait (Fig. 2). All grown colonies that showed a strong blue color in the subsequent β-galactosidase filter assay had their plasmid DNA extracted and sequenced. A total of 166 plasmid DNAs (69 from human bone marrow and 97 from human fetal brain cDNA libraries) from clones positive for both HIS3 and LacZ reporters were sequenced. 59 different proteins were identified using FEZ2 (207–353) as a bait (Table 1, Fig. 3). These can be organized into the following groups according the major described GO (Gene-Ontology) process attributed to them: transcription (10 proteins), translation (6 proteins), apoptosis (5 proteins), signal transduction (5 proteins), neuronal cell development (5 proteins), cytoskeleton/centrosome (3 proteins), unknown function (10 proteins) and other functions (15).
Table 1. Human FEZ2-interacting proteins identified by the yeast two-hybrid system screen.
| Gene | Acession no. | Protein Description1 | Coded protein residues (complete) | Coded protein residues (retrieved)2 | UNC763 | FEZ13 | FEZ23 | Biological Process (GO)4 |
| AATF | NP_036270 | apoptosis antagonizing transcription factor | 560 | 11–319 | + | + | + | apoptosis |
| ATP6V1H | NP_057078 | H(+)-transporting two-sector ATPase | 247 | 63–234 | + | + | + | ion transport |
| C1orf216 | NP_689587 | hypothetical protein LOC127703 | 229 | 20–229 | + | + | + | UNKNOWN |
| C10orf54 | Q9H7M9 | Platelet receptor Gi24 precursor | 311 | 215–311 | + | UNKNOWN | ||
| C10orf78 | NP_001002759 | hypothetical protein LOC119392 isoform a | 245 | 6–245 | + | + | + | UNKNOWN |
| C12orf51 | NP_001103132 | AF-1 specific protein phosphatase | 3996 | 2878–3161 | + | + | protein modification | |
| C14orf94 | CAD62584 | HAUS augmin-like complex, subunit 4 | 387 | 16–333 | + | + | + | centrosome organization |
| CAP1 | EAX07240 | CAP, adenylate cyclase-associated protein 1 (yeast) | 468 | 163–468 | + | + | + | cytoskeleton/cell polarity/signal transduction |
| CDKN1 | EAW96276 | cyclin-dependent kinase inhibitor 1B (p27, Kip1) | 53 | 1–53 | + | + | + | apoptosis/cell growth |
| CHERP | BAD92967 | calcium homeostasis endoplasmic reticulum protein | 399 | 301–399 | + | + | nervous development | |
| DEFA3 | AAA35753 | neutrophil peptide 3 precursor | 65 | 50–65 | + | + | + | xenobiotic metabolism |
| DNAJB11 | NP_057390 | DnaJ (Hsp40) homolog, subfamily B, member 11 precursor | 358 | 277–358 | + | protein folding | ||
| DNML1 | EAW88521 | dynamin 1-like, isoform CRA_c | 789 | 549–789 | + | + | + | endocytosis |
| DRAP1 | NP_006433 | DR1-associated protein 1 | 205 | 1–133 | + | + | + | transcription |
| DRG2 | BAD92577 | developmentally regulated GTP binding protein 2 variant | 259 | 51–259 | + | + | + | signal transduction |
| EEF1G | AAH21974 | H sapiens eukaryotic translation elongation factor 1 gamma | 355 | 19–346 | + | translation | ||
| EI2B5 | EAW78300 | eukaryotic translation initiation factor 2B, subunit 5 epsilon | 442 | 327–434 | + | + | + | translation/hormone mediated signaling |
| FEZ1 | EAW67636 | fasciculation and elongation protein zeta 1 (zygin I) | 392 | 131–371 | + | + | + | nervous development |
| FEZ1 | NP_072043 | zygin 1 isoform 2 | 104 | 1–89 | + | nervous development | ||
| GNAS | AAH89157 | GNAS complex locus | 380 | 293–380 | + | signal transduction | ||
| GOLPH3L | EAW53527 | golgi phosphoprotein 3-like | 299 | 159–299 | + | + | + | UNKNOWN |
| GPRASP2 | NP_612446 | G protein-coupled receptor associated sorting protein 2 | 838 | 496–812 | + | + | + | UNKNOWN |
| GTF2IRD2 | AAQ19673 | general transcription factor II i repeat domain 2 | 949 | 565–603 | + | transcription | ||
| GTPB6 | NP_036359 | pseudoautosomal GTP-binding protein-like protein | 403 | 98–347 | + | UNKNOWN | ||
| HSP90B1 | EAW97723 | heat shock protein 90 kDa beta (Grp94), member 1 | 367 | 1–287 | + | apoptosis/protein folding/muscular contraction | ||
| HSPA1A | BAD93055 | heat shock 70 kDa protein 1A variant | 709 | 75–163 | + | response to stress | ||
| IGL@ | AAH71804 | IGL@ protein | 236 | 1–236 | + | UNKNOWN | ||
| IK | EAW62028 | IK cytokine, down-regulator of HLA II | 557 | 119–333 | + | + | + | cell-cell signaling |
| INPP1 | NP_002185 | inositol polyphosphate-1-phosphatase | 399 | 355–399 | + | + | signal transduction | |
| INTS8 | NP_060334 | integrator complex subunit 8 | 995 | 508–816 | + | + | + | snRNA proccessing |
| MCM7 | NP_877577 | minichromosome maintenance complex component 7 | 543 | 410–543 | + | + | + | transcription/cell cycle |
| MED7 | NP_004261 | mediator complex subunit 7 | 233 | 116–233 | + | + | + | transcription |
| MLF1IP | NP_078905 | MLF1 interacting protein | 418 | 243–263 | + | + | + | transcription |
| NFS1 | BAD96959 | NFS1 nitrogen fixation 1 isoform a precursor variant | 457 | 417–457 | + | metabolic proccess | ||
| NQO2 | CAI23293 | NAD(P)H dehydrogenase, quinone 2 | 172 | 1–115 | + | oxidation reduction | ||
| OS9 | NP_006803 | amplified in osteosarcoma isoform 1 precursor | 667 | 389–635 | + | + | endoplasmatic reticum stress | |
| PDCD7 | AAI31705 | PDCD7 protein | 270 | 57–270 | + | + | + | apoptosis |
| PTN | EAW83871 | Pleiotrophin | 246 | 105–223 | + | + | + | nervous development |
| RARA | EAW60657 | retinoic acid receptor, alpha, isoform CRA_e | 520 | 111–327 | + | + | + | transcription |
| RPL27 | EAW60910 | ribosomal protein L27, isoform CRA_b | 80 | 53–80 | + | + | + | translation |
| RPL38 | NP_000990 | RPL38 ribosomal protein L38 | 70 | 1–70 | + | translation | ||
| RPS25 | NP_001019 | ribosomal protein S25 | 125 | 1–114 | + | translation | ||
| SAP30 | NP_003855 | Sin3A-associated protein, 30 kDa | 220 | 1–220 | + | + | + | transcription |
| SAP30L | NP_078908 | SAP30-like | 183 | 1–183 | + | + | + | transcription |
| SCOC | EAX05102 | short coiled-coil protein, isoform CRA_a | 122 | 42–122 | + | + | + | UNKNOWN |
| SERPINF1 | P36955 | Pigment epithelium-derived factor precursor (PEDF) | 418 | 16–267 | + | nervous development | ||
| SFRS8 | EAW98526 | splicing factor, arginine/serine-rich 8 | 951 | 263–511 | + | + | + | transcription |
| SHMT1 | BAD97272 | serine hydroxymethyltransferase 1 (soluble) isoform 1 variant | 483 | 364–483 | + | hormone mediated signaling | ||
| SLC25A37 | AAF71063 | PRO1584 | 81 | 1–58 | + | + | + | muscular constraction |
| SLTM | EAW77552 | SAFB-like, transcription modulator | 1168 | 389–723 | + | + | transcription/apoptosis | |
| SMC3 | BAF98736 | structural maintenance of chromosomes 3 | 1217 | 766–1059 | + | + | + | sister chromatides cohesion/signal transduction |
| STRA13 | NP_659435 | stimulated by retinoic acid 13 | 63 | 3–63 | + | + | + | UNKNOWN |
| TBC1D25 | NP_002527 | TBC1 domain family, member 25 | 688 | 498–677 | + | + | + | regulation Rab GTPase activity |
| TLOC1 | Q99442 | Translocation protein SEC62 | 399 | 2–95 | + | cotranslational protein targeting to membrane | ||
| TNNC2 | NP_003270 | fast skeletal muscle troponin C | 160 | 1–137 | + | muscular constraction | ||
| TSNAX | NP_005990 | translin-associated factor X | 290 | 1–276 | + | + | + | cell differentiation |
| TUBA1B | BAF82043 | tubulin, alpha 1b | 451 | 285–432 | + | cytoskeleton | ||
| WWC1 | EAW61508 | WW, C2 and coiled-coil domain containing 1 | 1018 | 775–1018 | + | + | + | UNKNOWN |
| “SIH” | A61065 | sucrase-isomaltase homolog – human | 48 | 1–44 | + | + | + | UNKNOWN |
Results obtained from BLASTX (GenBank);
It is depicted the mininum length of the retrieved sequences which could be visualized by forward DNA sequencing only,
Interaction confirmed (+) by yeast two hybrid system with UNC-76, FEZ1 or FEZ2 proteins;
Biological process based on the GO database (other functions may be known).
Figure 3. Protein-protein interaction pattern among homologs by beta-galactosidase assay.
Result of the yeast two-hybrid assay with the C-terminal of the three homologous proteins UNC-76 (242–378), FEZ1 (221–392) and FEZ2 (207–353) as baits. All 59 clones obtained here by our yeast two-hybrid library screens with the C-terminal of FEZ2 (207–353) were tested against all three bait constructs one-to-one. The pattern of interaction between the orthologous FEZ1 and UNC-76 was almost identical, indicating a conservation of the UNC-76 function in human FEZ1. The clones that showed specific interactions with FEZ2 only, are separated on the right side of the figure. The tests were performed in duplicate, but only one representative result is shown.
Most interestingly, of these 59 FEZ2 interactors only 40 interacted also with FEZ1 (Fig. 3, Table 1) and indeed 8 of the 40 were identical with proteins that had been identified in the screen with FEZ1 [10]. This result may suggest that FEZ2 gained in respect to FEZ1 additional 19 new interactors, within the set of preys analyzed, which are specific to FEZ2. Such an evolutionary gain of function is compatible with the histological expression patterns of both proteins. Whereas FEZ1 (as its ortholog UNC-76) is expressed only in the nervous system, FEZ2 shows a ubiquitous expression pattern. Thinking of the role of FEZ proteins as transport adaptor proteins, FEZ2 may have evolved to transport additional cargo proteins, which may be expressed in non-neuronal tissues.
UNC-76 and FEZ1 share a conserved protein-protein interaction pattern
Next we were interested to involve UNC-76 as a third component in our analysis, to try to understand how the protein-protein interaction pattern may have evolved during evolution of the FEZ family proteins. To this end we employed a C-terminal construct of UNC-76 (242–378) in a direct one-on-one analysis against all 59 FEZ2 interacting proteins (Fig. 3, Table 1). The cDNA for C. elegans UNC-76 was a kind gift from Dr. Hengartner.
To our great surprise, the pattern of interaction of UNC-76, in relation to the proteins identified in the screening assay of two-hybrid and FEZ2 as bait, was almost identical to that observed for FEZ1 (Table 1, Fig. 3). This suggests that the protein-protein interaction profile of the orthologs FEZ1 and UNC-76 is highly conserved. Only 4 of the 40 proteins that interacted with FEZ2 and FEZ1 did not interact with UNC-76. In conclusion the protein interaction pattern for both human FEZ1 and C. elegans UNC-76 seemed to be mostly conserved since the divergence of the deuterostomes (represented by the human FEZ1) from the “protostomes” (represented by C. elegans UNC-76), which occurred around 650 million years ago.
In this context it is worthwhile to differentiate between nuclear and cytoplasmic proteins that interact with the FEZ family proteins. A significant proportion of the interacting proteins are nuclear or predicted to be nuclear (18 of 59, 30.51%). Among these 18 nuclear proteins identified in the screen with FEZ2, a high proportion of 15 (ca. 83%) also interacted with FEZ1 (Fig. 4). This may be of potential relevance because we have shown in previous cell fractionation experiments that GFP-FEZ1 can be found in the nuclear fraction [26]. The exact role of FEZ1 in the nucleus remains however to be established. Interestingly, of the 41 cytoplasmic FEZ2 interacting proteins identified, a significant fraction of 16 (ca. 40%) interact exclusively with FEZ2. This again may suggest the acquisition of new interaction partners and thereby possibly the aggregation of new “cytoplasmic functions” for FEZ2 relative to FEZ1/UNC-76.
Figure 4. Schematic summary of the individual interactors of FEZ1, FEZ2 and UNC-76.
On top of the boxes, highlighted by larger print and in bold, are the bait proteins used in the yeast two-hybrid assays. Sets are represented by gray boxes. The darker the gray the larger the set of proteins. Nuclear proteins are indicated by an asterisk preceeding the official protein name.
In 2000, Walhout and collaborators [28] introduced the concept of an “interologue”, referring to a protein-protein interaction pattern that is conserved between pairs of orthologs. This notion is intuitive in molecular biology and has been documented by many interactions and may be conserved even in distantly related organisms, such as bacteria and animals. Our results show not only a high degree of preservation of the interactions between FEZ1, FEZ2 and UNC-76 but also the acquisition of new PPIs for FEZ2. Makino and coworkers [29] suggested that the rate of evolution of a protein that has more partners PPI is much smaller than the one that has few partners PPI. This is in good agreement with the high degree of sequence similarity among FEZ1/FEZ2/UNC-76, which is especially pronounced in the C-terminal coiled-coil region, which is essential for most protein-protein interactions (Figure S2). Only 12.8% of the amino acids in the coiled-coil regions of FEZ1 and FEZ2 are different.
Three different fates have been proposed after the duplication of a genes [29]. First, one of the copies can be silenced by the accumulation of deleterious mutations and eventually become indistinguishable from genomic non-coding neighboring regions, while the other copy retains the original function. Second, while one copy maintains the original function, the other acquires a new function possibly by aggregation of advantageous mutations. Third, both copies accumulate mutations that alter the original function, but the original function is retained and compensated cooperatively. Our results seem to strongly suggest that for the FEZ family the second pathway is in progression. All interactors identified for FEZ1 in our laboratory have been confirmed to interact with FEZ2, too [10], but only 60% of the interactions identified for FEZ2 were confirmed for FEZ1 and UNC-76. Thus, FEZ2 seems to have acquired additional new functions but still preserved the majority of interactions found for FEZ1/UNC-76.
In this context it is also interesting to analyze the origin of the proteins fished with the FEZ2 bait from either bone marrow or fetal brain cDNA libraries and to correlate this with the supposed expression patterns (FEZ1: brain, FEZ2: ubiquitous) and function (FEZ1: neuron specific, FEZ2: ubiquitous) (Table 1). Of the 17 proteins that were of origin from either bone marrow or brain (not from both, like 2 additonal proteins) and that were specific to interact with FEZ2, 13 were from bone marrow and only 4 from brain. Of the 52 proteins that were of origin from either bone marrow or brain only (7 additional proteins were fished from both libraries!) and that interacted with both FEZ1 and FEZ2, 32 were from the bone marrow library and 20 from the brain. Thus, FEZ1 interactions are relatively enriched in clones from the brain, while FEZ2 interactions are of more ubiquitous origin. In fact the ratio bone marrow proteins/brain protein for FEZ2 is almost three times higher (3.25; 13/4) than for FEZ1 (1.19; 19/16). This tendency may already indicate the acquisition of additional interacting partners and functions in the more ubiquitously expressed FEZ2 relative to the brain specific FEZ1 since the candidate cargo proteins for transport of FEZ1 can be mostly found in brain tissue, whereas those of FEZ2 preferentially in other tissues, including bone marrow.
Discussion
The large number of proteins that were identified to interact FEZ1, led to its classification as a hub protein [10]. Most proteins initially characterized as hubs tend to maintain this status after additional studies [30]. Indeed, here we were able to confirm the hub status for human FEZ1 and even extend it to the entire family of FEZ proteins, since both human FEZ2 as well as C. elegans UNC-76 interacted also with a large number of proteins. Genome-wide studies have shown that deletions of hub protein encoding genes are three times more likely to be lethal than deletions of non-hubs, a phenomenon known as the centrality-lethality rule [31]. This is confirmed in C. elegans mutations in the unc-76 gene, which although not lethal, cause deleterious abnormalities in the elongation of neuronal axons along other axonal surfaces (but not over non-neuronal surfaces) during the animals development, which results in a “paralyzed” phenotype, where the worm has severly reduced body movements (therefore the designation unc for uncoordinated movements). These abnormalities can be complemented by the expression of human FEZ1 [1]. This result already shows the high functional conservation which we confirmed and detailed here: the protein interaction profiles of FEZ1 and UNC-76 are highly overlapping, since 36 of 40 FEZ1 interactors also interact with UNC-76 (90%).
In this context it is also interesting to note that although the knockout of UNC-76 had severe and deleterious effects in the worm a recently described knockout mice for FEZ1 showed no dramatic changes in the brains anatomy or, the morphology of neuronal cell bodies, dendrites or axons, nor of the general locomotion or behavior of the animals [32]. Only after new social interactions or under stress conditions a slight hyper locomotion behavior was observed as well as a greater response to certain psycho-stimulants [32]. Our comparative analysis of the FEZ1 and FEZ2 interactomes becomes interesting at this point, because in mammals, and chordates in general, we have two FEZ family gene members in contrast to the C. elegans genome, which contains only unc-76. The duplication of the unc-76 gene occurred most likely in the stemline of the chordates (Table S1, Fig. 1) [6]. Our current interpretation of the lack of any greater neurophysiological or behavioural abnormalities in the FEZ1-/- mice, may be due to a compensatory expression of the ubiquitously expressed FEZ2, which - as we demonstrated here - not only acquired additional new interactors in respect to FEZ1, but at the same time also maintained all of the interactors of FEZ1. From our point of view it is therefore not at all surprising that the FEZ1-/- mice had such a mild phenotype in comparison to the UNC-76 -/- worm [32]. It may even be that expression of FEZ2 in the FEZ1 k.o. mice, by hitherto unknown mechanisms, may be up-regulated in comparison to wild-type mice. It is interesting to note that in Figure S1A of the article of Sakae and co-workers, we can observe a new band at the height of the FEZ1 protein in extracts from the stomach of FEZ1-/- but not wild type mice (anti-FEZ1 Western blot). Since FEZ2 has almost the same MW as FEZ1, this band may represent up-regulated FEZ2 protein, in response to the lack of FEZ1. The observed relatively mild phenotype in the FEZ -/- mice may be as well attributed to additional proteins interacting with FEZ2, which may interfere in the way of a dominant negative in FEZ1 specific functions, as to specific but not yet identified PPI for FEZ1.
Although FEZ2 has more interactors than FEZ1 there is not a 100% functional redundancy. This would explain why both genes are still maintained in the gene pool (from a population genetics point of view). The reasons why both are maintained may be manifold, including dosage effects or a promoter specific regulation that must be maintained in the neurons (FEZ1). Data from the literature suggest that FEZ1 expression is increased upon NGF treatment, a neuronal tissue specific factor [6], [33]. It may be even speculated that FEZ1 is somehow involved in neurogenesis per se. FEZ1 mRNA expression shows a peak that coincides with neurogenesis in rats, and decreases with the progress of adulthood. The mRNA expression FEZ2, although with higher levels in embryogenesis, is relatively constant, always less than that of FEZ1. Additional data on FEZ2 however are not available so far but one could speculate that its more ubiquitous expression depends on internal genetic programs rather than on external stimuli, like in the case of FEZ1.
Many of the proteins identified by our screen with FEZ2 interact with proteins that also interact with each other, increasing therefore the likelihood that these are true interactions of biological relevance. We like to highlight here the interactions of pleiotrophin (PTN), protein tyrosine phosphatase, receptor type S (PTPRS) ([18], and retinoic acid receptor alpha (RARA). In 2005, Stelzl et al. [18] performed a yeast two-hybrid screen on a large scale, reporting more than 3000 interactions between human proteins. The human protein FEZ1 interacted with 21 proteins. Of these 21 proteins, 4 interact with pleiotrophin (general transcription factor IIF, protocollagen hydroxylase, neurokinin beta, translocase of outer mitochondrial membrane 20 homolog). FEZ1 interacted further with a receptor protein tyrosine phosphatase (PTP) (sigma) of the same family of receptors of pleiotrophin (beta and zeta) [34]. Pleiotrophin is also known as neurite growth-promoting factor 1 (NEGF1), has mitotic activity and influences the growth of neuritis. It acts in the developing nervous system [35] and signals via the inactivation of a receptor-dependent ligand of RPTP beta/zeta (or PTPRS). It increases the level of tyrosine phosphorylated signaling proteins in the cell, promoting thereby downstream signaling events [34]. In the nervous system, PTN is expressed in both neurons and glia cells [36] as well as in neuronal stem cells [37]. SAGE analysis indicated a 10-fold elevation of PTN transcripts in response to long-term NGF treatment of PC12 cells. It is of interest, that in P19 neuroprogenitor cells, PTN was also induced by retinoic acid during neuronal differentiation [38], [39]. Retinoic acid is essential for the differentiation and morphogenesis of various tissues, including the nervous system [40]. Silencing of FEZ1 by RNA interference (RNAi) strongly inhibited the NGF-induced differentiation and efficiently reduced the anterograde transport of mitochondria in PC12 cells, suggesting that FEZ1 is essential for NGF-induced neuronal differentiation of PC12 cells [33], [41].
Retinoic acid receptors (RARs) are transcriptional activators which are activated by binding of their ligands and act in concert with a combination retinoid X type receptors (RXR alpha, beta and gamma). Even in the absence of ligand, RAR-RXR heterodimers bind to DNA sequences known as response elements for RA, but in this state they recruit co-repressors. These co-repressors mediate the negative modulation of transcription by recruitment of the histone deacetylase complex and the transfer of methyl radicals to the DNA bound histones, thereby stabilizing the nucleosome. The binding of the ligand retinoic acid causes however conformational changes in the binding domain of RAR, which in return promote the release of the co-repressors and the recruitment of co-activators. While some co-activators interact with factors of the basal transcription machinery, others induce chromatin remodeling and specific transcriptional activation. Data from our previous two-hybrid and pull-down assays confirmed the interaction of FEZ1 and FEZ2 proteins related to activation of transcription (BAF60a) and the repression of transcription (SAP30L) [10]. Furthermore, RXR/RAR interacts with N-Cor, thereby recruiting the complex of repression of transcription [42]. It is known that SAP30 also interacts with N-Cor [43] promoting the compaction of chromatin. These data gain potential relevance by the observation that both RAR and FEZ1/FEZ2 interact with protein complexes of chromatin remodeling, and with each other. Thus, FEZ1 may be also involved in the regulation of gene transcription mediated by transcription factors.
In summary, we have described here the molecular expansion of the FEZ protein family in the chordate stemline, most likely after two rounds of whole-genome duplication. We have identified many new members of the FEZ family from different species and a highly conserved C-terminal domain, which is responsible to mediate the majority of the protein-protein interactions. Our analyses indicate that the ancestral UNC-76 protein function and protein interaction profile is surprisingly conserved in animals, evolutionary distinct as vertebrates and nematodes. The analysis of the phylogeny and the detailed protein interaction profiles uncovered a likely functional divergence between FEZ1 and FEZ2, since FEZ2 acquired in respect to FEZ1/UNC-76 additional new interactors and is ubiquitously expressed and not restricted to neuronal cells, as is FEZ1/UNC-76. Our data provide an explanation for the ability of human FEZ1 to rescue the defects caused by unc-76 mutations in nematodes, since the PPI pattern of FEZ1 and UNC-76 are highly similar. Furthermore, no strong defects are caused in mice lacking the FEZ1, probably due to the compensatory presence of its paralogue FEZ2, which interacted with all FEZ1 interacting proteins, and additional new interactors that are FEZ2 specific. Our new data will hopefully stimulate and facilitate further studies on the functional role of proteins of the FEZ family in the development and function of both neuronal and non-neuronal cells. Furthermore, studies such as ours may contribute to our ever growing understanding of how protein networks function and specially how they came about and were modified and adapted during evolution. In the future we may devise new and more complete ways to visualize and comprehend how these networks were shaped throughout time.
Supporting Information
Amino acid sequence alignment of the members of the FEZ family. Names of the sequences are given in the Table S1. The residues in the alignment are shaded light grey, grey, or black to indicate shared identity in 40%, 70% or 100% of the analyzed sequences, respectively. The bars indicate regions predicted to form coiled-coil.
(PDF)
Identities and similarities between human proteins FEZ1 and FEZ2 and C. elegans UNC-76. A general scheme of the FEZ family proteins is shown at the top. The identity and similarity comparisons were made of two-by-two proteins both for the complete protein alignment as well as for local alignment of FEZ fragments by NPS@ (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_clustalw.html). I = identity, S = strongly similar, W = weakly similar, D = different.
(TIF)
FEZ family proteins used for amino acid sequence alignment.
(DOC)
Acknowledgments
We thank Maria Eugenia R. Camargo for technical assistance and Prof. Dr. Michael O. Hengartner for the kind gift of the pSU001 plasmid (containing unc-76).
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was financially supported by Fundação de Amparo à Pesquisa do Estado São Paulo, the Conselho Nacional de Pesquisa e Desenvolvimento and the LNBio-CNPEM. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bloom L, Horvitz HR. The Caenorhabditis elegans gene unc-76 and its human homologs define a new gene family involved in axonal outgrowth and fasciculation. Proc Natl Acad Sci U S A. 1997;94:3414–3419. doi: 10.1073/pnas.94.7.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boguski MS, Lowe TM, Tolstoshev CM. dbEST - database for “expressed sequence tags”. Nat Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- 3.Gindhart JG, Chen J, Faulkner M, Gandhi R, Doerner K, et al. The kinesin-associated protein UNC-76 is required for axonal transport in the Drosophila nervous system. Mol Biol Cell. 2003;14:3356–3365. doi: 10.1091/mbc.E02-12-0800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Su CW, Tharin S, Jin Y, Wightman B, Spector M, et al. The short coiled-coil domain-containing protein UNC-69 cooperates with UNC-76 to regulate axonal outgrowth and normal presynaptic organization in Caenorhabditis elegans. J Biol. 2006;5:9. doi: 10.1186/jbiol39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuroda S, Nakagawa N, Tokunaga C, Tatematsu K, Tanizawa K. Mammalian homologue of the Caenorhabditis elegans UNC-76 protein involved in axonal outgrowth is a protein kinase C zeta-interacting protein. J Cell Biol. 1999;144:403–411. doi: 10.1083/jcb.144.3.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujita T, Ikuta J, Hamada J, Okajima T, Tatematsu K, et al. Identification of a tissue-non-specific homologue of axonal fasciculation and elongation protein zeta-1. Biochem Biophys Res Commun. 2004;313:738–744. doi: 10.1016/j.bbrc.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 7.Cohen C, Parry DA. Alpha-helical coiled coils and bundles: how to design an alpha-helical protein. Proteins. 1990;7:1–15. doi: 10.1002/prot.340070102. [DOI] [PubMed] [Google Scholar]
- 8.Burkhard P, Stetefeld J, Strelkov SV. Coiled coils: a highly versatile protein folding motif. Trends Cell Biol. 2001;11:82–88. doi: 10.1016/s0962-8924(00)01898-5. [DOI] [PubMed] [Google Scholar]
- 9.Strauss HM, Keller S. Pharmacological interference with protein-protein interactions mediated by coiled-coil motifs. Handb Exp Pharmacol. 2008:461–482. doi: 10.1007/978-3-540-72843-6_19. [DOI] [PubMed] [Google Scholar]
- 10.Assmann EM, Alborghetti MR, Camargo ME, Kobarg J. FEZ1 dimerization and interaction with transcription regulatory proteins involves its coiled-coil region. J Biol Chem. 2006;281:9869–9881. doi: 10.1074/jbc.M513280200. [DOI] [PubMed] [Google Scholar]
- 11.Miyoshi K, Honda A, Baba K, Taniguchi M, Oono K, et al. Disrupted-In-Schizophrenia 1, a candidate gene for schizophrenia, participates in neurite outgrowth. Mol Psychiatry. 2003;8:685–694. doi: 10.1038/sj.mp.4001352. [DOI] [PubMed] [Google Scholar]
- 12.Hennah W, Porteous D. The DISC1 pathway modulates expression of neurodevelopmental, synaptogenic and sensory perception genes. PLoS One. 2009;4:e4906. doi: 10.1371/journal.pone.0004906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naghavi MH, Hatziioannou T, Gao G, Goff SP. Overexpression of fasciculation and elongation protein zeta-1 (FEZ1) induces a post-entry block to retroviruses in cultured cells. Genes Dev. 2005;19:1105–1115. doi: 10.1101/gad.1290005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suzuki T, Okada Y, Semba S, Orba Y, Yamanouchi S, et al. Identification of FEZ1 as a protein that interacts with JC virus agnoprotein and microtubules: role of agnoprotein-induced dissociation of FEZ1 from microtubules in viral propagation. J Biol Chem. 2005;280:24948–24956. doi: 10.1074/jbc.M411499200. [DOI] [PubMed] [Google Scholar]
- 15.Whitehouse C, Chambers J, Howe K, Cobourne M, Sharpe P, et al. NBR1 interacts with fasciculation and elongation protein zeta-1 (FEZ1) and calcium and integrin binding protein (CIB) and shows developmentally restricted expression in the neural tube. Eur J Biochem. 2002;269:538–545. doi: 10.1046/j.0014-2956.2001.02681.x. [DOI] [PubMed] [Google Scholar]
- 16.Okumura F, Hatakeyama S, Matsumoto M, Kamura T, Nakayama KI. Functional regulation of FEZ1 by the U-box-type ubiquitin ligase E4B contributes to neuritogenesis. J Biol Chem. 2004;279:53533–53543. doi: 10.1074/jbc.M402916200. [DOI] [PubMed] [Google Scholar]
- 17.Ikuta J, Maturana A, Fujita T, Okajima T, Tatematsu K, et al. Fasciculation and elongation protein zeta-1 (FEZ1) participates in the polarization of hippocampal neuron by controlling the mitochondrial motility. Biochem Biophys Res Commun. 2007;353:127–132. doi: 10.1016/j.bbrc.2006.11.142. [DOI] [PubMed] [Google Scholar]
- 18.Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 19.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Combet C, Blanchet C, Geourjon C, Deleage G. NPS@: network protein sequence analysis. Trends Biochem Sci. 2000;25:147–150. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- 21.Felsenstein J. PHYLIP – Phylogeny Inference Package (Version 3.2). Cladistics. 1989;5:164–166. [Google Scholar]
- 22.Chien CT, Bartel PL, Sternglanz R, Fields S. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci U S A. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bartel PL, Fields S. Analyzing protein-protein interactions using two-hybrid system. Methods Enzymol. 1995;254:241–263. doi: 10.1016/0076-6879(95)54018-0. [DOI] [PubMed] [Google Scholar]
- 24.Lanza DC, Meirelles GV, Alborghetti MR, Abrile CH, Lenz G, et al. FEZ1 interacts with CLASP2 and NEK1 through coiled-coil regions and their cellular colocalization suggests centrosomal functions and regulation by PKC. Mol Cell Biochem, 2010;338(1–2):35–45. doi: 10.1007/s11010-009-0317-9. [DOI] [PubMed] [Google Scholar]
- 25.Lanza DC, Silva JC, Assmann EM, Quaresma AJ, Bressan GC, et al. Human FEZ1 has characteristics of a natively unfolded protein and dimerizes in solution. Proteins. 2009;74(1):104–21. doi: 10.1002/prot.22135. [DOI] [PubMed] [Google Scholar]
- 26.Lanza DC, Trindade DM, Assmann EM, Kobarg J. Over-expression of GFP-FEZ1 causes generation of multi-lobulated nuclei mediated by microtubules in HEK293 cells. Exp Cell Res. 2008;314(10):2028–2039. doi: 10.1016/j.yexcr.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 27.Putnam NH, Butts T, Ferrier DE, Furlong RF, Hellsten U, et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- 28.Walhout AJ, Sordella R, Lu X, Hartley JL, Temple GF, et al. Protein interaction mapping in C. elegans using proteins involved in vulval development. Science. 2000;287:116–122. doi: 10.1126/science.287.5450.116. [DOI] [PubMed] [Google Scholar]
- 29.Makino T, Gojobori T. Evolution of protein-protein interaction network. Genome Dyn. 2007;3:13–29. doi: 10.1159/000107601. [DOI] [PubMed] [Google Scholar]
- 30.Fox A, Taylor D, Slonim DK. High throughput interaction data reveals degree conservation of hub proteins. Pac Symp Biocomput. 2009:391–402. doi: 10.1142/9789812836939_0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He X, Zhang J. Why do hubs tend to be essential in protein networks? PLoS Genet. 2006;2:e88. doi: 10.1371/journal.pgen.0020088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakae N, Yamasaki N, Kitaichi K, Fukuda T, Yamada M, et al. Mice lacking the schizophrenia-associated protein FEZ1 manifest hyperactivity and enhanced responsiveness to psychostimulants. Hum Mol Genet. 2008;17:3191–3203. doi: 10.1093/hmg/ddn215. [DOI] [PubMed] [Google Scholar]
- 33.Fujita T, Maturana AD, Ikuta J, Hamada J, Walchli S, et al. Axonal guidance protein FEZ1 associates with tubulin and kinesin motor protein to transport mitochondria in neurites of NGF-stimulated PC12 cells. Biochem Biophys Res Commun. 2007;361:605–610. doi: 10.1016/j.bbrc.2007.07.050. [DOI] [PubMed] [Google Scholar]
- 34.Meng K, Rodriguez-Pena A, Dimitrov T, Chen W, Yamin M, et al. Pleiotrophin signals increased tyrosine phosphorylation of beta beta-catenin through inactivation of the intrinsic catalytic activity of the receptor-type protein tyrosine phosphatase beta/zeta. Proc Natl Acad Sci U S A. 2000;97:2603–2608. doi: 10.1073/pnas.020487997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li YS, Milner PG, Chauhan AK, Watson MA, Hoffman RM, et al. Cloning and expression of a developmentally regulated protein that induces mitogenic and neurite outgrowth activity. Science. 1990;250:1690–1694. doi: 10.1126/science.2270483. [DOI] [PubMed] [Google Scholar]
- 36.Silos-Santiago I, Yeh HJ, Gurrieri MA, Guillerman RP, Li YS, et al. Localization of pleiotrophin and its mRNA in subpopulations of neurons and their corresponding axonal tracts suggests important roles in neural-glial interactions during development and in maturity. J Neurobiol. 1996;31:283–296. doi: 10.1002/(SICI)1097-4695(199611)31:3<283::AID-NEU2>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 37.Furuta M, Shiraishi T, Okamoto H, Mineta T, Tabuchi K, et al. Identification of pleiotrophin in conditioned medium secreted from neural stem cells by SELDI-TOF and SELDI-tandem mass spectrometry. Brain Res Dev Brain Res. 2004;152:189–197. doi: 10.1016/j.devbrainres.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 38.Brunet-de Carvalho N, Raulais D, Rauvala H, Souttou B, Vigny M. HB-GAM/Pleiotrophin and Midkine are differently expressed and distributed during retinoic acid-induced neural differentiation of P19 cells. Growth Factors. 2003;21:139–149. doi: 10.1080/08977190310001621014. [DOI] [PubMed] [Google Scholar]
- 39.Greene LA, Angelastro JM. You can't go home again: transcriptionally driven alteration of cell signaling by NGF. Neurochem Res. 2005;30:1347–1352. doi: 10.1007/s11064-005-8807-y. [DOI] [PubMed] [Google Scholar]
- 40.Zechel C. Requirement of retinoic acid receptor isotypes alpha, beta, and gamma during the initial steps of neural differentiation of PCC7 cells. Mol Endocrinol. 2005;19:1629–1645. doi: 10.1210/me.2004-0540. [DOI] [PubMed] [Google Scholar]
- 41.Maturana AD, Fujita T, Kuroda S. Functions of fasciculation and elongation protein zeta-1 (FEZ1) in the brain. ScientificWorldJournal. 10:1646–1654. doi: 10.1100/tsw.2010.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Privalsky ML. The role of corepressors in transcriptional regulation by nuclear hormone receptors. Annu Rev Physiol. 2004;66:315–360. doi: 10.1146/annurev.physiol.66.032802.155556. [DOI] [PubMed] [Google Scholar]
- 43.Laherty CD, Billin AN, Lavinsky RM, Yochum GS, Bush AC, et al. SAP30, a component of the mSin3 corepressor complex involved in N-CoR-mediated repression by specific transcription factors. Mol Cell. 1998;2:33–42. doi: 10.1016/s1097-2765(00)80111-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Amino acid sequence alignment of the members of the FEZ family. Names of the sequences are given in the Table S1. The residues in the alignment are shaded light grey, grey, or black to indicate shared identity in 40%, 70% or 100% of the analyzed sequences, respectively. The bars indicate regions predicted to form coiled-coil.
(PDF)
Identities and similarities between human proteins FEZ1 and FEZ2 and C. elegans UNC-76. A general scheme of the FEZ family proteins is shown at the top. The identity and similarity comparisons were made of two-by-two proteins both for the complete protein alignment as well as for local alignment of FEZ fragments by NPS@ (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_clustalw.html). I = identity, S = strongly similar, W = weakly similar, D = different.
(TIF)
FEZ family proteins used for amino acid sequence alignment.
(DOC)