Abstract
Cell adhesion molecules in Saccharomyces cerevisiae and Candida albicans contain amyloid-forming sequences that are highly conserved. We have now used site-specific mutagenesis and specific peptide perturbants to explore amyloid-dependent activity in the Candida albicans adhesin Als5p. A V326N substitution in the amyloid-forming region conserved secondary structure and ligand binding, but abrogated formation of amyloid fibrils in soluble Als5p and reduced cell surface thioflavin T fluorescence. When displayed on the cell surface, Als5p with this substitution prevented formation of adhesion nanodomains and formation of large cellular aggregates and model biofilms. In addition, amyloid nanodomains were regulated by exogenous peptides. An amyloid-forming homologous peptide rescued aggregation and biofilm activity of Als5pV326N cells, and V326N substitution peptide inhibited aggregation and biofilm activity in Als5pWT cells. Therefore, specific site mutation, inhibition by anti-amyloid peturbants, and sequence-specificity of pro-amyloid and anti-amyloid peptides showed that amyloid formation is essential for nanodomain formation and activation.
Introduction
Candida albicans is a human commensal fungus that is pathogenic when its growth becomes uncontrolled, especially in imunocompromised individuals. Under such conditions these eukaryotes can form biofilms that are resistant to a variety of environmental assaults, including antimicrobials [1], [2], [3]. The formation of biofilms is a developmental process with multiple steps including cell adhesion, extracellular matrix production and the formation of hyphae [4], [5].
Cell wall proteins called adhesins are critical for biofilm formation, and mediate adhesion of C. albicans to various substrates and each other. Among many adhesins, members of the Als family of glycoproteins are particularly active in cell aggregation, adhesion to endothelia and epithelia, formation of biofilms and pathogenesis in mouse models. There are eight ALS gene loci, with high heterozygosity, so Als adhesin sequences and binding specificities are diverse. However, all Als proteins have similar modularity and domain structure (Figure 1A) [6], [7]. Three N-terminal Ig-like domains determine substrate specificity [7], [8], [9]. A 103-residue Thr-rich T domain is highly conserved among paralogs, and contains a 7-residue sequence that forms amyloids under native–like conditions [10], [11]. The central region of the protein contains a variable number of tandem repeats (TR domains) that are 36 amino acids in length, and these repeats bind to each other and to substrates through the hydrophobic effect [12], [13]. A highly glycosylated, C-terminal, serine- threonine-rich (Ser/Thr) stalk precedes a GPI anchor, which is processed to form a covalent linkage to cell wall polysaccharide [14]. The overlapping binding specificities and variable expression of Als proteins make them difficult to study in C. albicans [6], [7], [9], [15]. Nevertheless, expression in Saccharomyces cerevisiae has shown that Als1p, Als3p and Als5p bind similarly to a variety of substrates and are involved in endothelial cell adhesion, fungal aggregation, and tissue invasion [7], [16], [17], [18], [19].
Figure 1. Effect of V326N mutation on Als5p expression and aggregation in Saccharomyces cerevisiae.
(A) Maps of Als5pWT and Als5pV326N. The open reading frame is 1419 amino acid residues long, and the mutation is near the N-terminus of the 103-residue T domain. (B) Immunofluorescence analysis shows expression of V5-tagged Als5pWT and Als5pV326N in S. cerevisiae. (C) Adherence and aggregation of yeast cells (gray) to heat denatured BSA-coated beads (brown-gold) for S. cerevisiae without Als5p (EV) or expressing Als5pWT or Als5pV326N. Images shown were visualized using bright field microscopy. The diameter of the beads is 2.8 µm.
Als-mediated adhesion to substrate is followed 5–20 min later by aggregation of Als-expressing yeast cells [20]. This aggregation is accompanied by a cell surface conformational change in the Als proteins to mediate stronger adhesive interactions [13], [21]. This activation is independent of cellular metabolism, because it occurs in heat-killed cells [13].
The T domains of Als proteins contain amyloid-forming sequences that are highly conserved [10]. Amyloids are insoluble fibrillar protein aggregates whose cores consist of crystalline arrays of identical sequences in many molecules of the amyloid protein [22], [23]. In the presence of amyloids, Congo red absorbance is red-shifted and increased, and the fluorescence emission of thioflavin T increases several fold [24], [25], [26]. At high concentrations, these dyes can perturb amyloid structure [27], [28], [29]. Some amyloid-forming bacterial adhesins can elicit cell-cell and cell-substrate adhesion leading to the formation of biofilms [30], [31], [32]. Other known roles for amyloid include amyloid-like stacking of residues in β-helices in viral spike proteins [33], curli in gram negative bacteria [30], [32], [34], sequestration of regulatory proteins in yeast [35], packing of pro-hormones in secretory vesicles [36], and as template activity for melanin assembly [37].
Previously, we showed that the amyloid sequence in the T region of Als proteins mediates amyloid formation, and that amyloid binding dyes can inhibit aggregation in the S. cerevisiae surface display model [8], [10], [11]. However, there is no direct data for the roles of Als amyloid sequences per se in vivo. We have therefore created a version of Als5p with its amyloid sequence disrupted by a single site substitution, and assayed its effects in the S. cerevisiae display model. We report the effects of this mutation on aggregation and biofilm formation in a model system. In addition, we have tested amyloid-forming and amyloid-inhibiting peptides for their effect in vivo in C. albicans. Finally, we provide direct evidence for the formation of amyloid adhesion nanodomains in C. albicans, using single-molecule atomic force microscopy (AFM) [38].
Results
Cells expressing a mutation in the amyloid sequence of Als5p exhibit less efficient aggregation
S. cerevisiae cells expressing Als5p form large aggregates, similar to those seen in C. albicans [8], [39]. This cell aggregation can be inhibited by amyloid binding dyes [8], [11]. A V326N mutation in the Als5p sequence reduces TANGO β-aggregation potential of the amyloid region from 93% to 4% [10]. To test the hypothesis that the amyloid forming sequence is critical for cell aggregation, we incorporated the V326N mutation into full length wild type Als5p, and expressed it in S. cerevisiae (Figure 1A). Immunofluorescence confirmed that that both wild type Als5p (Als5pWT) and the substitution sequence (Als5pV326N) were expressed on the yeast cell surface, with non-amyloid Als5pV326N expression slightly greater than Als5pWT (Figure 1B). In agreement with previous results, cells expressing Als5pWT bound to heat denatured BSA-coated beads and formed large aggregates (Figure 1C) [8], [11], [39]. Cells expressing Als5pV326N also bound to most of the beads, but formed much smaller aggregates. S. cerevisiae with vector alone did not bind to beads or form aggregates. Therefore, the V326N substitution had a small effect on binding BSA as a ligand, but a compromised the ability of Als5p to mediate formation of yeast aggregates.
Increased thioflavin T fluorescence is a characteristic test for amyloid formation [40]. Aggregated C. albicans or Als5pWT S.cerevisae cells stained brightly with 100 nM thioflavin T (Figure 2O and 2P), a concentration that had no effect on cell aggregation (Figure 2A–2D vs. 2I-2L). In contrast the small aggregates of cells expressing Als5pV326N showed little fluorescence under these conditions (Figure 2N). Therefore, thioflavin T fluorescence was associated with robust aggregation of cells expressing amyloid-forming adhesins.
Figure 2. Thioflavin T fluorescence of S. cerevisiae and C. albicans.
S. cerevisiae expressing no Als5p (EV), Als5pV326N, Als5pWT, or C. albicans were aggregated in the (A–H) absence or (I–P) presence of 100 nM thioflavin T. Aggregation state (upper panels) and thioflavin T fluorescence (lower panels) were monitored. The diameter of the beads is 2.8 µm, and all images are at the same magnification.
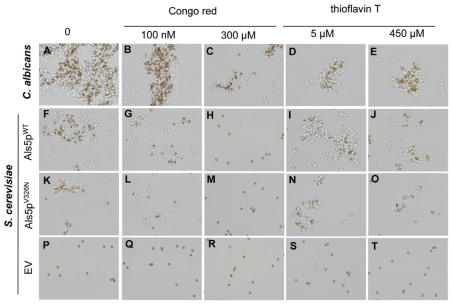
Greater concentrations of amyloid-binding dyes such as Congo red and thioflavin T inhibit aggregation of S. cerevisiae cells expressing Als proteins [11], [17]. If these dyes are acting through perturbation of the amyloid, they should not affect the adherence or residual aggregation of cells expressing Als5pV326N. We determined dye concentrations inhibiting cell aggregation and substrate binding for Congo red and thioflavin T (Figure 3). Both Als5pWT and C. albicans showed inhibition of aggregation in the presence of 100nM Congo red (Figures 3A vs. 3B and 3F vs. 3G). This concentration had no visual effect on the ability of the Als5pV326N to mediate formation of small aggregates (Figures 3K vs. 3L). Furthermore, this concentration is at least a thousand-fold lower than the concentrations that inhibit growth and fungal cell wall biogenesis [41]. A higher concentration of Congo red (300 µM) reduced the ability of both Als5pWT and Als5pV326N cells to bind to the beads and form aggregates (Figures 3H and 3M). Negative control cells did not aggregate with or without treatment (Figures 3P–3R).
Figure 3. Effect of Congo red and thioflavin T on aggregation.
(A–E) C. albicans, (F–J) Als5pWT in S. cerevisiae or (K–O) Als5pV326N in S. cerevisiae, and (P–T) empty vector (EV) cells were aggregated with beads coated with heat denatured BSA in the absence and in the presence of 100 nM or 300 µM Congo Red or 5 µM or 450 µM thioflavin T. Aggregates were observed by light microscopy. The diameter of the beads is 2.8 µm.
Thioflavin T (5 µM and 450 µM) also decreased cell-cell adhesion in C. albicans (Figure 3A vs. 3D and 3E) and S. cerevisiae expressing Als5pWT (Figure 3F vs. 3I and 3J). Thioflavin T had no effect on the smaller aggregates of S. cerevisiae Als5pV326N cells (Figure 3N and 3O) or on empty vector cells (Figure 3S and 3T). These data support the idea that most Als-mediated aggregation is dependent on formation of amyloids.
V326N soluble protein is deficient in amyloid formation and maintains native substrate binding activity
We expressed the V326N mutation in two soluble versions of the Als5p protein: Als5p1-431, including the Ig-like and Thr-rich amyloid region; and Als5p1-664, which also includes the tandem repeat region [8], [10], [12]. The proteins containing the V326N substitution were purified by published procedures and did not form amyloid fibers visible by electron microscopy (data not shown). When these soluble proteins were tested in modified ELISA assays for in vitro binding to fibronectin and polystyrene, the results were similar to those for the wild type Als5 protein within experimental error for both the 431-residue and 664-residue fragments. (Figure S1). Therefore, the in vitro binding properties of the proteins were not significantly altered by the substitution. Far UV Circular Dichroism spectra and secondary structure were also similar to those from the corresponding proteins with wild type sequences (Figure S2). These results demonstrated that the V326N substitution did not significantly affect ligand binding properties or secondary structure of Als5p.
An amyloid-forming peptide restores aggregation of non-amyloid-forming cells
A tridecapeptide consisting of the sequence of Als5pWT residues 322–334 (SNGIVIVATTRTV, amyloid sequence bolded) rapidly forms insoluble amyloids [10]. We reasoned that its strong amyloid-forming ability might increase amyloid formation in non-amyloid Als5pV326N by providing a stable amyloid template. S. cerevisiae expressing either form of Als5p and C. albicans were incubated without (Figure 4A–4H) or with this wild-type sequence tridecapeptide (2 µg/ml) (Figure 4I–4P) during the bead assay. This peptide greatly increased aggregate formation in S. cerevisiae expressing Als5pV326N (Figure 4B vs. 4J). The peptide had no detectable effect on the aggregation of empty vector, or Als5pWT-expressing S. cerevisiae or on C. albicans strains (Figure 4A vs. 4I, 4C vs. 4K and 4D vs. 4L). A scrambled-sequence peptide with the same amino acid composition as the Als5p amyloid-forming sequence (VITGVTNIRTSVA) did not induce aggregation, indicating that the observed effect was specific to the wild type sequence (Figure S3).
Figure 4. Effects of amyloid and non-amyloid peptides on cellular aggregation and thioflavin T fluorescence.
S. cerevisiae expressing no Als5p (EV), Als5pV326N, Als5pWT, or C. albicans were aggregated in the (A–H) absence, or (I–P) presence of amyloid forming or (Q–X) amyloid-disrupting peptides. The amyloid-forming peptide was SNGIVIVATTRTV, and the amyloid-disrupting peptide was SNGINIVATTRTV. Each vertical pair shows brightfield and thioflavin T fluorescence images of the same field. The diameter of the beads is 2.8 µm, and all images are at the same magnification.
To determine if the peptide-induced adhesion was accompanied by formation of amyloid-like interactions, cells were stained with thioflavin T (Figures 4E–4H vs. 4M–4P). The small Als5pV326N aggregates lacked intense thioflavin T fluorescence (Figure 4F). In contrast, aggregates formed from Als5pV326N-expressing cells in the presence of peptide exhibited more intense fluorescence (Figure 4N). In the presence of peptide, there was also increased fluorescence of the aggregates formed from C. albicans or Als5pWT-expressing S. cerevisiae (Figure 4G vs. 4O and 4H vs. 4P). Thus, exogenous amyloid-forming homologous sequence peptide induced increased aggregation in non-amyloid cells, and increased amyloid fluorescence in aggregating cells as well.
Non-amyloid V326N peptide blocks aggregation
Since the amyloid-forming peptide potentiated aggregation of the Als5pV326N-expressing strain, we hypothesized that the mutant peptide would block cell aggregation. All strains were incubated with the V326N peptide (SNGIN 326IVATTRTV; 200 µg/ml) during the bead assay (Figure 4). This peptide strongly inhibited aggregation, including Als5pWT-expressing S. cerevisiae and C. albicans (Figure 4C vs. 4S and 4D vs. 4T). The few remaining aggregates looked similar to those observed with Als5pV326N (Figure 4B). This peptide did not have an effect on either empty vector or Als5pV326N strains (Figures 4A vs. 4Q and 4B vs. 4R). A scrambled V326N peptide (VITGNTNIRTSVA) did not block aggregation indicating that the inhibitory effect is specific to the V326N sequence (Figure S3).
Given that the V326N peptide blocked formation of aggregates in C. albicans and Als5pWT-expressing S. cerevisiae, we tested whether it prevented formation of amyloid-like interactions. In these strains, thioflavin T fluorescence was strongly reduced in the presence of V326N peptide (Figure 4G and 4H vs. 4W and 4X). The peptide had no effect on the fluorescence of the empty vector and Als5pV326N aggregates (Figure 4U and 4V). These results show that the V326N peptide blocked amyloid formation and aggregation mediated by Als5p in S. cerevisiae, as well as aggregation in C. albicans.
The amyloid sequence of Als5p is critical for cell-cell association and cell–substrate adhesion to polystyrene
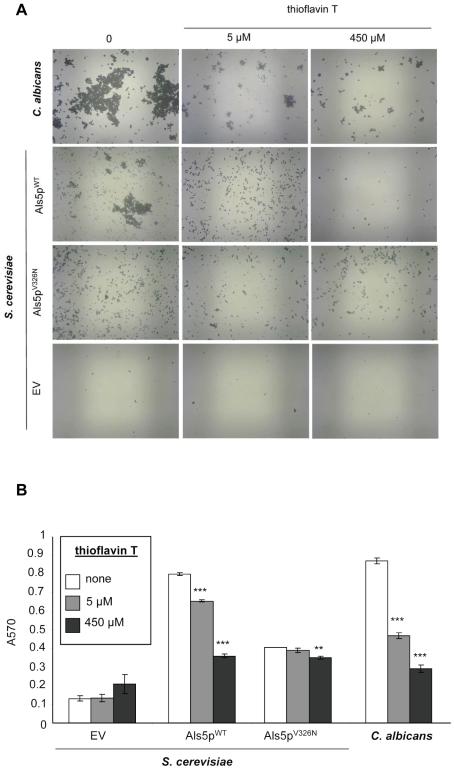
We also tested to determine whether amyloid formation was important for a model biofilm. S. cerevisiae expressing Als5pWT or C. albicans adhered to the polystyrene surface of a 96-well plate in Tris-EDTA buffer. The non-adherent cells and buffer were removed, and the adhering cells incubated in medium overnight [42]. Microscopy and quantitative crystal violet staining of the wells revealed that Als5pWT-expressing S. cerevisiae and C. albicans strains bound to the surface in aggregates (Figure 5A). Thioflavin T (5 µM) partially dispersed the C. albicans aggregates and fully dispersed the Als5pWT-mediated aggregates of S. cerevisiae. This thioflavin T treatment reduced adherence of Als5p-expressing S. cerevisiae by about 16%, and C. albicans adherence by 45% (Figure 5B). A 90-fold higher concentration of dye reduced adherence to the plastic by 79% and 90% respectively. Congo red had similar effects: 100 nM disrupted aggregates and partially inhibited adherence; 300 µM blocked almost all adhesion (Figure S4).
Figure 5. Effects of thioflavin T on binding and aggregation on a polystyrene surface.
C. albicans or S. cerevisiae expressing Als5pWT, Als5pV326N, or empty vector (EV) strains were adhered to a polystyrene surface in the absence and presence of 5 µM or 450 µM thioflavin T for 1.5 h. Adherent cells were grown overnight, washed, stained with 1% crystal violet and imaged. (A) The bottoms of wells were imaged. (B) Triplicate biofilms were quantified with crystal violet. Mean and standard deviation are shown. Significance was determined by Student's T-test with p values relative to untreated strains (p≤0.05(*), p≤0.01(**), and p≤0.001(***)).
In contrast to cells expressing Als5pWT, Als5pV326N cells bound to the surface as dispersed cells, rather than as aggregates (Figure 5A). The lower concentrations of the dyes did not affect the residual adherence to the plastic surface, and the higher concentrations partially blocked adherence. Taken together, these results show that adherence to polystyrene is followed by aggregation to form a biofilm, and is mediated by the amyloid-forming region of Als5p in the S. cerevisiae model. The similar behavior in C. albicans further supports the hypothesis that amyloid sequences in Als or other adhesins are important in biofilm formation.
Effects of an amyloid-forming peptide and an amyloid inhibitory peptide on model biofilms
Since a mutation in the Als5p amyloid-forming sequence disrupted adherence and aggregation on plastic, we hypothesized that the V326N and amyloid-forming Als5p peptides would also affect adhesion on polystyrene. The cells were incubated without or with amyloid forming peptide SNGIV326IVATTRTV or the V326N non-amyloid peptide. Microscopy and quantification revealed that the amyloid forming peptide rescued S. cerevisiae cells expressing Als5pV326N. In the presence of the amyloid-inducing peptide, these cells formed large aggregates like C. albicans and S. cerevisiae cells expressing wild type protein (Figure 6A). Conversely, the V326N peptide effectively blocked adherence and aggregation on plastic for each type of cell.
Figure 6. Effects of wild type and V326N peptides on polystyrene biofilms.
(A) C. albicans or S. cerevisiae expressing Als5pWT, Als5pV326N, or no Als5p (EV) strains were adhered to a polystyrene surface in the absence and presence of the wild-type peptide (2 ug/ml) or V326N peptide (200 ug/ml) for 1.5 h. Adherent cells were grown overnight, stained with 1% crystal violet and imaged. (B) Crystal violet quantification of binding. Peptide-treated strains with p values relative to untreated strains were determined as significant by Student's T-test (p≤0.05(*), p≤0.01(**), and p≤0.001(***)).
Quantification of adhesion to the polystyrene confirmed that Als5pWT-S. cerevisiae and C. albicans cells that were treated with amyloid-forming peptide bound better than untreated cells (Figure 6B). Additionally, Als5pV326N cells that were treated with the wild type peptide exhibited a nearly two-fold increase in adherence relative to cells not incubated with peptide. These results show that a peptide inhibitor of amyloid formation can abrogate adherence to plastic and biofilm-like aggregation, and conversely that an activator of amyloid formation facilitates biofilm formation.
AFM resolves Als adhesion nanodomains in S. cerevisae and C. albicans cells
Lastly, we used single-molecule AFM to probe the distribution of Als proteins in living C. albicans cells, with the aim to determine whether they are initially evenly distributed and clustered following application of force, as are Als proteins on S. cerevisiae [43](Figure 7). Yeast cells were trapped into porous polymer membranes, and analyzed using topographic imaging and spatially-resolved force spectroscopy. We first analyzed the S. cerevisiae surface display model expressing V5-tagged Als5p proteins using AFM tips bearing anti-V5 antibodies (Figure 7A–7L). Consistent with earlier work in S. cerevisiae [43] the initial distribution of Als5pWT was random (Figure 7B). Pulling on single adhesins with the AFM tip induced the formation of adhesion domains of 100-500 nm size (Figure 7C). In addition, the force-induced nanodomains propagated over the entire cell surface, since remote areas showed similar nanodomains (Figure 7D). Als5pWT remodelling was independent of cellular metabolic activity since heat-killed cells show the same behavior as live cells (Figure 7E–7H). Remarkably, Als5p clustering properties were almost completely abolished in the V326N mutant (Figure 7I–7L), indicating that amyloid interactions play a key role in clustering.
Figure 7. Force-induced adhesion nanodomains in single live cells.
(A) AFM topographic image (scale bar: 2 µm), in buffer, showing wild-type S. cerevisiae cells expressing V5-tagged Als5pWT proteins. (B) Adhesion force map (1 µm ×1 µm) recorded with an anti-V5 tip on a given target area of the native cell that was never subjected to force (maps #1, recorded on the square shown in (A). Blue and red pixels correspond to forces smaller and larger than 150 pN, respectively, thus to V5-tagged Als5pWT recognition and unfolding. (C) Second adhesion force map (1 µm × 1 µm) recorded on the same target area (map #1′). The heterogeneous distribution of coloured pixels, which represents the detection of single Als5pWT molecules documents the formation of nanoscale clusters (outlined in white). (D) Adhesion force map (1 µm × 1 µm) recorded on a remote area (map #2) localized several hundred nanometers away from the first map (see squares in A). (E–H) Same sequence of data as in A–D obtained on heat-killed S. cerevisiae cells expressing Als5pWT. (I–L) Same sequence of data as in A–D obtained on S. cerevisiae expressing ALS5pV326N. (M) AFM topographic image of a C. albicans cell (scale bar: 2 µm). (N) Adhesion force map (1 µm × 1 µm) recorded with an Als5p1-431-derivatized tip [44] on a given target area of the native cell (map #1, recorded on the square in M; red pixels correspond to unfolding forces that are in the 0-300 pN range). (O) Second adhesion force map (1 µm × 1 µm) recorded on the same target area (maps #1′). (P) Adhesion force map (1 µm × 1 µm) recorded on a remote area (map #2) localized several hundred nanometers away from the first map (see squares in M). (Q) Confocal imaging of punctate fluorescent nanodomains (white arrows) on aggregated C. albicans treated with Als5p or Als5pV326N peptide and stained with 100 nM thioflavin-T. (R) Cartoon model of force-induced amyloid-dependent clustering of Als5p. Cell walls are shown as heavy greent lines. Als5p adhesion molecules have green Ig-like binding domains, a red amyloid sequence, and a black line representing the TR and stalk domains. Application of pulling force in the AFM or mixing in the aggregation assays causes formation of amyloid-like arrays on the cell surface. The formation of these arrays is blocked in the presence of Congo red, high concentrations of thioflavin T, or by V326N peptide or mutation.
Because amyloid regions also appear in C. albicans (Figures 2– 6) and are accompanied by development of strong adhesion and surface birefringence [8], [11], [20], we tested to see if there was similar force-induced formation of adhesin nanodomains in C. albicans (Figure 7M–7P). In this case, living C. albicans yeast cells were probed with AFM tips bearing Als5p1-433 fragments (Supplemental Figure S5A) since these bind to all Als adhesins [13], [16], [43], [44]. As expected, the initial force mapping showed a random pattern of Als proteins (colored pixels), with a surface density being greater than in the S. cerevisiae surface display model (Figure 7N vs. 7B). The adhesive forces were typical for Als-Als interactions (Figure S5B; [44]). Force-extension curves showed patterns similar to those reported earlier for Als5p-Als5p interactions [44], except that the number of unfolding Tandem Repeat domains varied from 3–33, the range known for Als alleles (Supplemental Figure S5C; [6]). Upon remapping of the same region (Figure 7, map1') or a remote region on the same cell (Figure 7, map 2), the adhesin molecules were clustered, as in the S. cerevisiae model. These observations lead us to conclude that nanodomains form and propagate in response to force in C. albicans, just as they do in Als5pWT-expressing S. cerevisiae.
Adhesion nanodomains are also visualized by confocal microscopy of thioflavin T-stained cells. Therefore we tested the effects of the peptides on nanodomain formation in C. albicans. As in the Als5p S. cerevisiae surface display cells, the native sequence peptide potentiated nanodomain formation and the V326N substitution peptide inhibited nanodomains. The sequence specificity of nanodomain potentiation and inhibition implies that Als proteins are major components of surface amyloids in C. albicans.
Discussion
Amyloid-forming sequences are widespread in fungal adhesion proteins that form cellular aggregates [11]. That being said, there remains the question of whether amyloid formation per se is the function of these sequences in situ on the cell surface. We and others previously reported that anti-amyloid treatments disrupt Als protein-mediated aggregation [8], [10], [11], [17]. We have also demonstrated that a V326N mutation in Als5p prevents formation of surface nanodomains [43], but have not previously shown the functional consequences of this mutation. Therefore we set out to test directly the hypothesis that cell surface assembly of amyloids causes nanodomain formation and activation of adhesion. These new direct tests showed: first, that the V326N substitution in the amyloid core region of the intact cell surface-localized protein abrogated activation of adhesion; second, that the substitution did not affect secondary structure of the protein or its affinity for ligands; third, in agreement with the previous point, initial binding of cells to substrate was not affected by the substitution (Figures. 1, S1 and S2). We also took an independent and novel approach of monitoring effects of sequence-specific pro- and anti-amyloid peptides on nanodomain formation and adhesion activity. The effects were supportive of the hypothesis: the anti-amyloid peptide inhibited both nanodomain formation and adhesion activity, and the pro-amyloid peptide activated, even in cells expressing the V326N substituted protein. We also demonstrated for the first time that there are force-induced nanodomains on the surface of C. albicans and that the peptides perturb them. Finally, we showed that the peptides act similarly in a model biofilm, as do anti-amyloid dyes, a result also recently reported for Als3p by Nobbs et al. [17]. Together, these approaches confirm the hypothesis, showing that adhesion activity is increased by amyloid assembly and nanodomain formation. The widespread occurrence and the conservation of amyloid sequences in fungal adhesins are consistent with amyloid-mediated nanodomain formation being a general mechanism for activation of robust cell-cell aggregation.
We have tested this hypothesis by a detailed determination of phenotype for single site substitution V326N in the 1419 amino-acid Als5p sequence. This mutation is in the seven residue amyloid-forming sequence I325VIVATT331 in the highly conserved T domain of Als5p. The mutation reduces the TANGO β-aggregation potential, which is related to amyloid-forming ability, from >95% to 4% [45]. A peptide with this sequence change did not form amyloids in vitro [10]. Although 431-, and 664-residue fragments of Als5pWT form amyloid fibers [10], [11], the corresponding soluble versions of Als5pV326N did not (data not shown).
Therefore, the V326N mutation abrogated amyloid formation in vitro and also altered the activity of cell-bound Als5p. The mutation did not significantly affect cell wall localization of the protein (Figure 1), secondary structure (Figure S2), or binding of soluble Als5p to polystyrene or fibronectin (Figure S1). These results imply that the mutated protein folded to its native structure and retained its binding activity. Furthermore Als5pV326N cell surface expression was consistent with proper folding and processing. Nevertheless, cells expressing Als5pV326N bound to polystyrene or to heat denatured BSA-coated beads somewhat less efficiently than cells expressing the wild type protein (Figures 1, 5, and 6). However, there was a severe inhibition of cellular aggregation ability for cells expressing Als5pV326N. Thus the cellular consequences were great, despite the observation that the mutation did not affect in vitro activities of the soluble proteins. These consequences can be most easily explained as a failure of the mutated protein to form functional amyloid regions required for cell surface activation through adhesion nanodomain formation.
If the loss of amyloid formation is the key defect in Als5pV326N, then chemical inhibition of amyloid formation should show similar effects, and the residual activity of the Als5pV326N protein should be resistant to the inhibitors. In fact these results were observed in cellular assays of binding to bead-bound ligand (Figure 3), and in the model biofilm study (Figures 5 and 6), the V326N mutant phenotype was mimicked by low concentrations of amyloid-perturbing dyes Congo red and thioflavin T. In keeping with this interpretation, the residual activity of Als5pV326N was not inhibited by these dye concentrations, a result consistent with the remaining activity being amyloid independent.
Thioflavin T at sub-inhibitory concentrations is standardly used to monitor formation of amyloids in vitro [46], [47], and thioflavin T was highly useful to monitor the cell surface amyloid levels in intact cells (Figures 3, 4, and 7). Unlike wild type cells, Als5pV326N cells showed minimal thioflavin T fluorescence, consistent with the idea that the mutation compromised amyloid formation. For cells expressing wild type Als5p, the fluorescence increased on aggregation, implying that cell-cell contact resulted in increased amyloid formation at the cell surface, both in the S. cerevisiae display model and in C. albicans itself, results consistent with an increase in cell surface birefringence [8], [11]. We could also use the technique to monitor peptide-induced modulation of cell surface amyloid levels (Figures 3, 4, and 7).
The finding that peptides modulated both aggregation of cells and cell surface amyloid levels in parallel was also consistent with amyloid dependence of aggregation. Amyloids are highly sequence specific, and form stacked β-sheets of identical sequences in many molecules of the same protein [22], [23]. Therefore, amyloid formation is highly sensitive to addition of amyloid-forming or amyloid-interfering peptides with sequences identical to or slightly changed from the amyloid-forming region of the protein [48], [49]. An amyloid-forming peptide rescued the S. cerevisiae cells expressing Als5pV326N, as well as increasing surface amyloid levels in cells expressing Als5pWT and in C. albicans (Figure 4). We propose that the wild type sequence peptide reinforces amyloids with homologous sequence, and is able to form a “seed” that forces amyloid–like interactions in Als5pV326N. The converse experiment was also informative: excess V326N peptide inhibited amyloid formation and aggregation in cells expressing wild type adhesins. These effects were sequence-specific, since sequence-scrambled peptides of the same composition had no effects on aggregation or biofilm formation (Figure S3). Therefore, the peptide studies confirmed a specific role for amyloid formation in cellular aggregation.
That the V326N peptide inhibited aggregation and nanodomain formation in C. albicans also implies that Als1p or Als5p is the major adhesin being assayed. Als1p, Als3p, and Als5p have identical amyloid sequences, and Als1p is the major adhesin expressed on C. albicans in the yeast form [6], [50]. Therefore, the peptides would likely affect aggregation caused by Als1p as well as that caused by Als5p or Als3p.
Our results are consistent with a role for amyloid formation in causing robust aggregation of cells expressing amyloid-forming adhesins. Recent AFM studies show that the amyloid-forming sequence in Als5p mediates force-induced clustering of the adhesins on the cell surface [43]. Such clustering increases the strength of adherence because it increases the chance that ligands remain bound for long times: random dissociation of a ligand is followed by immediate rebinding to a nearby adhesin. The clustering lowers the macroscopic dissociation constant KD [51]. This effect is known from immunology as antibody avidity, the increase in binding constant of intact antibody with multiple binding sites relative to the dissociation constant of a single monomeric FAb'. The clustering is absent in cells expressing Als5pV326N, a result consistent with amyloid formation being important for adhesin clustering (Figure 7). Like increased aggregation and amyloid formation, clustering is independent of cellular metabolism and protein synthesis [8], [39], [43], [52]. The Als adhesins cluster even though they are anchored to the cell wall polysaccharide [14], [53]. This clustering is facilitated by the length of the extended molecules, the longest of which can extend to almost 500 nm, giving a radius of gyration of almost 1 µm across the cell surface (Figure 7; Figure S5; [43]).
Thus, we find that amyloid-dependent clustering of Als5p to form adhesion nanodomains would account for increased avidity and robust aggregation (Figure 7Q). Amyloid formation at the cell surface is accompanied by conformational shifts in pre-existing cell surface proteins, and is co-temporal with development of robust aggregation. Three different modulators of amyloid forming ability prevented formation of thioflavin fluorescent amyloid nanodomains and abrogated strong aggregation: namely mutation, chemical perturbants, and an amyloid-disrupting peptide. Furthermore, adhesion was enhanced after exogenous application of an amyloid-forming Als5p homologous sequence peptide. The enhancement was accompanied by increased amyloid fluorescence, and peptides with scrambled sequences had no effect. Thus all of our results are consistent with amyloid formation itself being an essential part of cellular aggregation. We cannot think of other mechanisms consistent with the data.
Formation of amyloid-dependent adhesion nanodomains has broad implications as a mechanism for yeast cell-cell adhesion in general, as well as in mats and biofilms [15], [42]. Amyloid sequences are present in most yeast adhesins [11]. Furthermore, for C. albicans Als proteins, and S. cerevisiae Flo1p and Muc1p/Flo11p, adhesins, activity is inhibited by amyloid-perturbing dyes. These results, together with our discovery of the effects of specific sequence peptides promise new approaches to understanding and manipulation of cell-cell interactions.
Materials and Methods
Strains and media
C. albicans strain Day 286 was a gift from J. Rauceo (John Jay College, CUNY) and was grown in YPED with 80 mg/L uridine at 30°C. S. cerevisiae strain W303-1B (Rodney Rothstein, Columbia U.) was grown in CSM with galactose at 24°C. Cells harboring empty vector (pJL1-EV), or expressing Als5pWT or Als5pV326N were grown in CSM with galactose lacking Trp.
Generation of V5-tagged Als5pWT and Als5pV326N
V5-tagged Als5pWT was generated by restriction digestion of pGK114 with BamHI and XhoI which released the Als5p sequence from the vector backbone [39]. This backbone was then ligated to: an oligonucleotide, which contains sequences for the invertase secretion signal, V5 epitope tag, flanked by a 5′-BamHI and 3′-NotI restriction sites [54]. The coding region of Als5p between a 5′ NotI site and a 3′XhoI site was generated by PCR and was ligated to the modified vector to make pJL1. The resulting construct was verified by sequencing (GeneWiz, South Plainfield, NJ).
Als5pV326N was generated by digestion of pGK114 with SphI and AleI to generate a 363bp fragment at nucleotide position 1242 to 1605, containing the target sequence to be mutated. This fragment was subcloned into pGEM-T vector and mutagenized using Quickchange (Agilent Technologies, Santa Clara, CA) with mutagenic primer 5′-GAA TAG TGA TGC CGG ATC TAA CGG TAT TAA CAT TGT TGC TAC AAC TAG AAC AGT TAC AGA CAG-3′. The correct mutation was verified by sequencing. The mutated fragment was released from the vector with the same enzymes used in its generation, and placed into the corresponding position of pJL1. The resulting product, pJL1V326N was verified by sequencing to determine the presence of the full-length Als5pV326N.
pJL-EV was produced by restriction digestion of pJL1 with BamHI and XhoI and ligating in the multiple cloning site from p414 (ATCC, Manassas, VA). The pJL plasmids were transformed into S. cerevisiae strain W303-1B.
Cell-bead assay
Cell aggregation was observed using a previously described method with minor modifications [8]. Briefly, M-280 tosyl-activated magnetic Dynabeads (Invitrogen, Carlsbad, CA) were coated overnight at 37°C with 1 mg/ml heat-denatured BSA according to the manufacturer's protocol. Cells were washed and resuspended in 10 mM Tris-HCl, 1 mM EDTA buffer, pH 7.0 (TE). Beads, 1×106, were added to 1×108 cells in 13×100 mm glass test tubes and the suspension was shaken at 200 rpm at 24°C 30-45 minutes. Adherent cells were separated and washed over a magnet, resuspended in TE buffer and observed by microscopy. Thioflavin T fluorescence was excited at 425 nm and monitored at 510 nm.
Polystyrene cell adhesion assay
Adhesion to polystyrene was assayed using a protocol previously described [42] with the following modifications. Cells were resuspended in TE alone or with dyes at 1×107 cells/ml. Cell suspension, 100 µl/well, was added to a polystyrene non-tissue culture plate with 96 wells. Each sample was done in triplicate. The cells adhered to the surface for 1.5 hours at room temperature. The polystyrene surface was washed with TE, sterile liquid media was added and the adherent cells were incubated overnight at 30°C. The next day the non-adherent cells were washed away with TE. Adherent cells were stained with crystal violet and observed by microscopy and imaged. Adhesion was quantified by solubilization of the crystal violet in 10% SDS for 1.5 hours and measuring the absorbance at 570 nm.
Atomic force microscopy
AFM measurements were performed at room temperature (20°C) in buffered solutions (sodium acetate; pH 4.75), using a Nanoscope IV Multimode AFM (Veeco Metrology Group, Santa Barbara, CA) and oxide sharpened microfabricated Si3N4 cantilevers (Microlevers, Veeco Metrology Group). Cells were immobilized by mechanical trapping into porous polycarbonate membranes (Millipore), with a pore size similar to the cell size. After filtering a concentrated cell suspension, the filter was gently rinsed with buffer, carefully cut (1 cm ×1 cm), attached to a steel sample puck (Veeco Metrology Group) and the mounted sample was transferred into the AFM liquid cell while avoiding dewetting. The spring constants of the cantilevers were measured using the thermal noise method (Picoforce, Veeco Metrology Group), yielding values ranging from 0.008 to 0.021 N/m. All force measurements were recorded with a loading rate of 10,000 pN/s. AFM tips were functionalized with anti-V5 antibodies or Als5p1-431 protein as described earlier [43], [44].
Supporting Information
ELISA assays of binding of Als5pV326N substitution proteins. The upper graph shows binding of increasing concentrations of proteins to different concentrations of fibronectin. The lower graph denotes binding of the proteins to polystyrene. The constructs shown are Ig-T-TR, wildtype (•) and V326N (▾), and Ig-T wildtype (○) and V326N (Δ). The protein concentration is 2.3±0.7μM for the fibronectin binding. The assays were carried out as previously described [11].
(TIF)
Far UV Circular Dichroism spectra of Als5p 1-664 protein. Wildtype protein is represented by the solid line and the V326N substation by the dashed line at 20°C.
(TIF)
Effects of scrambled V326N (VITGVTNIRTSVA) and wild type peptide (VITGNTNIRTSVA) on cellular aggregation. S. cerevisiae expressing no Als5p (EV), Als5pV326N, Als5pWT, or C. albicans were aggregated in the absence and presence of 2µg/ml scrambled wild type (S-wild type) or 200 µg/ml scrambled peptide (S-V326N). The diameter of the beads is 2.8 µm, and all images are at the same magnification.
(TIF)
Amyloid binding dye Congo red reduces binding and aggregation on polystyrene biofilms. C. albicans or S. cerevisiae expressing Als5pWT, Als5pV326N, or no Als5p (EV) adhered to a polystyrene surface in the absence and presence of 100 nM or 300 µM Congo red for 1.5 h. Adherent cells were grown overnight, stained with 1% crystal violet and imaged.
(TIF)
Detection and unfolding of single Als proteins in C. albicans. (A) Principle of the single-molecule detection experiment. C. albicans cells are probed, in buffer, using AFM tips derivatized with Als5p1-431. (B) Force extension curves obtained by stretching ALS proteins showed periodic features reflecting the sequential unfolding of the TR domains. (C) Plot of the rupture distances as a function of the number of unfolded TR.
(TIF)
Acknowledgments
We thank Roland Hosein for help with transmission electron microscopy, and Fred Naider and Steve Klotz for helpful discussions.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Work at Brooklyn College was supported by National Institutes of Health (NIH) grant SC1 GM 083756. Work at the Université catholique de Louvain was supported by the National Foundation for Scientific Research (FNRS), the Université catholique de Louvain (Fonds Spéciaux de Recherche), the Région Wallonne, the Federal Office for Scientific, Technical and Cultural Affairs (Interuniversity Poles of Attraction Programme), and the Research Department of the Communauté française de Belgique (Concerted Research Action). Y.F.D. and D.A. are Senior Research Associate and Research Fellow of the FRS-FNRS. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
References
- 1.Harriott MM, Noverr MC. Candida albicans and Staphylococcus aureus form polymicrobial biofilms: effects on antimicrobial resistance. Antimicrob Agents Chemother. 2009;53:3914–3922. doi: 10.1128/AAC.00657-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chandra J, Mukherjee PK, Leidich SD, Faddoul FF, Hoyer LL, et al. Antifungal resistance of candidal biofilms formed on denture acrylic in vitro. J Dent Res. 2001;80:903–908. doi: 10.1177/00220345010800031101. [DOI] [PubMed] [Google Scholar]
- 3.Hawser SP, Douglas LJ. Resistance of Candida albicans biofilms to antifungal agents in vitro. Antimicrob Agents Chemother. 1995;39:2128–2131. doi: 10.1128/aac.39.9.2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uppuluri P, Chaturvedi AK, Srinivasan A, Banerjee M, Ramasubramaniam AK, et al. Dispersion as an important step in the Candida albicans biofilm developmental cycle. PLoS Pathog. 2010;6:e1000828. doi: 10.1371/journal.ppat.1000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chandra J, Kuhn DM, Mukherjee PK, Hoyer LL, McCormick T, et al. Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J Bacteriol. 2001;183:5385–5394. doi: 10.1128/JB.183.18.5385-5394.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoyer LL, Green CB, Oh SH, Zhao X. Discovering the secrets of the Candida albicans agglutinin-like sequence (ALS) gene family—a sticky pursuit. Med Mycol. 2008;46:1–15. doi: 10.1080/13693780701435317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheppard DC, Yeaman MR, Welch WH, Phan QT, Fu Y, et al. Functional and structural diversity in the Als protein family of Candida albicans. J Biol Chem. 2004;279:30480–30489. doi: 10.1074/jbc.M401929200. [DOI] [PubMed] [Google Scholar]
- 8.Rauceo JM, Gaur NK, Lee KG, Edwards JE, Klotz SA, et al. Global cell surface conformational shift mediated by a Candida albicans adhesin. Infect Immun. 2004;72:4948–4955. doi: 10.1128/IAI.72.9.4948-4955.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoyer LL, Hecht JE. The ALS5 gene of Candida albicans and analysis of the Als5p N-terminal domain. Yeast. 2001;18:49–60. doi: 10.1002/1097-0061(200101)18:1<49::AID-YEA646>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 10.Otoo HN, Lee KG, Qiu W, Lipke PN. Candida albicans Als adhesins have conserved amyloid-forming sequences. Eukaryot Cell. 2008;7:776–782. doi: 10.1128/EC.00309-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramsook CB, Tan C, Garcia MC, Fung R, Soybelman G, et al. Yeast cell adhesion molecules have functional amyloid-forming sequences. Eukaryot Cell. 2010;9:393–404. doi: 10.1128/EC.00068-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frank AT, Ramsook CB, Otoo HN, Tan C, Soybelman G, et al. Structure and function of glycosylated tandem repeats from Candida albicans Als adhesins. Eukaryot Cell. 2010;9:405–414. doi: 10.1128/EC.00235-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rauceo JM, De Armond R, Otoo H, Kahn PC, Klotz SA, et al. Threonine-rich repeats increase fibronectin binding in the Candida albicans adhesin Als5p. Eukaryot Cell. 2006;5:1664–1673. doi: 10.1128/EC.00120-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dranginis AM, Rauceo JM, Coronado JE, Lipke PN. A biochemical guide to yeast adhesins: glycoproteins for social and antisocial occasions. Microbiol Mol Biol Rev. 2007;71:282–294. doi: 10.1128/MMBR.00037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nobile CJ, Schneider HA, Nett JE, Sheppard DC, Filler SG, et al. Complementary adhesin function in C. albicans biofilm formation. Curr Biol. 2008;18:1017–1024. doi: 10.1016/j.cub.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klotz SA, Gaur NK, De Armond R, Sheppard D, Khardori N, et al. Candida albicans Als proteins mediate aggregation with bacteria and yeasts. Med Mycol. 2007;45:363–370. doi: 10.1080/13693780701299333. [DOI] [PubMed] [Google Scholar]
- 17.Nobbs AH, Vickerman MM, Jenkinson HF. Heterologous Expression of Candida albicans Cell Wall-Associated Adhesins in Saccharomyces cerevisiae Reveals Differential Specificities in Adherence and Biofilm Formation and in Binding Oral Streptococcus gordonii. Eukaryot Cell. 2010;9:1622–1634. doi: 10.1128/EC.00103-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Silverman RJ, Nobbs AH, Vickerman MM, Barbour ME, Jenkinson HF. Infect Immun; 2010. Interaction of Candida albicans Cell-Wall Als3 Protein with Streptococcus gordonii SspB Adhesin Promotes Development of Mixed Species Communities. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.ten Cate JM, Klis FM, Pereira-Cenci T, Crielaard W, de Groot PW. Molecular and cellular mechanisms that lead to Candida biofilm formation. J Dent Res. 2009;88:105–115. doi: 10.1177/0022034508329273. [DOI] [PubMed] [Google Scholar]
- 20.Gaur NK, Klotz SA, Henderson RL. Overexpression of the Candida albicans ALA1 gene in Saccharomyces cerevisiae results in aggregation following attachment of yeast cells to extracellular matrix proteins, adherence properties similar to those of Candida albicans. Infect Immun. 1999;67:6040–6047. doi: 10.1128/iai.67.11.6040-6047.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaur NK, Klotz SA. Accessibility of the peptide backbone of protein ligands is a key specificity determinant in Candida albicans SRS adherence. Microbiology. 2004;150:277–284. doi: 10.1099/mic.0.26738-0. [DOI] [PubMed] [Google Scholar]
- 22.Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 23.Jahn TR, Makin OS, Morris KL, Marshall KE, Tian P, et al. The common architecture of cross-beta amyloid. J Mol Biol. 2010;395:717–727. doi: 10.1016/j.jmb.2009.09.039. [DOI] [PubMed] [Google Scholar]
- 24.Chiti F, Webster P, Taddei N, Clark A, Stefani M, et al. Designing conditions for in vitro formation of amyloid protofilaments and fibrils. Proc Natl Acad Sci U S A. 1999;96:3590–3594. doi: 10.1073/pnas.96.7.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng BY, Toyama BH, Wille H, Colby DW, Collins SR, et al. Small-molecule aggregates inhibit amyloid polymerization. Nat Chem Biol. 2008;4:197–199. doi: 10.1038/nchembio.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maezawa I, Hong HS, Liu R, Wu CY, Cheng RH, et al. Congo red and thioflavin-T analogs detect Abeta oligomers. J Neurochem. 2008;104:457–468. doi: 10.1111/j.1471-4159.2007.04972.x. [DOI] [PubMed] [Google Scholar]
- 27.Inbar P, Yang J. Inhibiting protein-amyloid interactions with small molecules: a surface chemistry approach. Bioorg Med Chem Lett. 2006;16:1076–1079. doi: 10.1016/j.bmcl.2005.10.067. [DOI] [PubMed] [Google Scholar]
- 28.Kim YS, Randolph TW, Manning MC, Stevens FJ, Carpenter JF. Congo red populates partially unfolded states of an amyloidogenic protein to enhance aggregation and amyloid fibril formation. J Biol Chem. 2003;278:10842–10850. doi: 10.1074/jbc.M212540200. [DOI] [PubMed] [Google Scholar]
- 29.Lorenzo A, Yankner BA. Amyloid fibril toxicity in Alzheimer's disease and diabetes. Ann N Y Acad Sci. 1996;777:89–95. doi: 10.1111/j.1749-6632.1996.tb34406.x. [DOI] [PubMed] [Google Scholar]
- 30.Cegelski L, Pinkner JS, Hammer ND, Cusumano CK, Hung CS, et al. Small-molecule inhibitors target Escherichia coli amyloid biogenesis and biofilm formation. Nat Chem Biol. 2009;5:913–919. doi: 10.1038/nchembio.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Romero D, Aguilar C, Losick R, Kolter R. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc Natl Acad Sci U S A. 2010;107:2230–2234. doi: 10.1073/pnas.0910560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X, Hammer ND, Chapman MR. The molecular basis of functional bacterial amyloid polymerization and nucleation. J Biol Chem. 2008;283:21530–21539. doi: 10.1074/jbc.M800466200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simkovsky R, King J. An elongated spine of buried core residues necessary for in vivo folding of the parallel beta-helix of P22 tailspike adhesin. Proc Natl Acad Sci U S A. 2006;103:3575–3580. doi: 10.1073/pnas.0509087103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Jong W, Wosten HA, Dijkhuizen L, Claessen D. Attachment of Streptomyces coelicolor is mediated by amyloidal fimbriae that are anchored to the cell surface via cellulose. Mol Microbiol. 2009;73:1128–1140. doi: 10.1111/j.1365-2958.2009.06838.x. [DOI] [PubMed] [Google Scholar]
- 35.Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maji SK, Perrin MH, Sawaya MR, Jessberger S, Vadodaria K, et al. Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science. 2009;325:328–332. doi: 10.1126/science.1173155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fowler DM, Koulov AV, Alory-Jost C, Marks MS, Balch WE, et al. Functional amyloid formation within mammalian tissue. PLoS Biol. 2006;4:e6. doi: 10.1371/journal.pbio.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muller DJ, Helenius J, Alsteens D, Dufrene YF. Force probing surfaces of living cells to molecular resolution. Nat Chem Biol. 2009;5:383–390. doi: 10.1038/nchembio.181. [DOI] [PubMed] [Google Scholar]
- 39.Gaur NK, Klotz SA. Expression, cloning, and characterization of a Candida albicans gene, ALA1, that confers adherence properties upon Saccharomyces cerevisiae for extracellular matrix proteins. Infect Immun. 1997;65:5289–5294. doi: 10.1128/iai.65.12.5289-5294.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsiao K, Chapman P, Nilsen S, Eckman C, Harigaya Y, et al. Correlative memory deficits, Abeta elevation, and amyloid plaques in transgenic mice. Science. 1996;274:99–102. doi: 10.1126/science.274.5284.99. [DOI] [PubMed] [Google Scholar]
- 41.Ram AF, Klis FM. Identification of fungal cell wall mutants using susceptibility assays based on Calcofluor white and Congo red. Nat Protocol. 2006;1:2253–6. doi: 10.1038/nprot.2006.397. [DOI] [PubMed] [Google Scholar]
- 42.Reynolds TB, Fink GR. Bakers' yeast, a model for fungal biofilm formation. Science. 2001;291:878–881. doi: 10.1126/science.291.5505.878. [DOI] [PubMed] [Google Scholar]
- 43.Alsteens D, Garcia MC, Lipke PN, Dufrene YF. Proc Natl Acad Sci U S A; 2010. Force-induced formation and propagation of adhesion nanodomains in living fungal cells. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alsteens D, Dupres V, Klotz SA, Gaur NK, Lipke PN, et al. Unfolding Individual Als5p Adhesion Proteins on Live Cells. ACS Nano. 2009;3:3063–3068. doi: 10.1021/nn900078p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fernandez-Escamilla A-M, Rousseau F, Schymkowitz J, Serrano L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat Biotech. 2004;22:1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- 46.Eisert R, Felau L, Brown LR. Methods for enhancing the accuracy and reproducibility of Congo red and thioflavin T assays. Anal Biochem. 2006;353:144–146. doi: 10.1016/j.ab.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 47.Lockhart A, Ye L, Judd DB, Merritt AT, Lowe PN, et al. Evidence for the presence of three distinct binding sites for the thioflavin T class of Alzheimer's disease PET imaging agents on beta-amyloid peptide fibrils. J Biol Chem. 2005;280:7677–7684. doi: 10.1074/jbc.M412056200. [DOI] [PubMed] [Google Scholar]
- 48.Granic I, Masman MF, Kees Mulder C, Nijholt IM, Naude PJ, et al. LPYFDa neutralizes amyloid-beta-induced memory impairment and toxicity. J Alzheimers Dis. 2010;19:991–1005. doi: 10.3233/JAD-2010-1297. [DOI] [PubMed] [Google Scholar]
- 49.Citron M. Alzheimer's disease: strategies for disease modification. Nat Rev Drug Discov. 2010;9:387–398. doi: 10.1038/nrd2896. [DOI] [PubMed] [Google Scholar]
- 50.Loza L, Fu Y, Ibrahim AS, Sheppard DC, Filler SG, et al. Functional analysis of the Candida albicans ALS1 gene product. Yeast. 2004;21:473–482. doi: 10.1002/yea.1111. [DOI] [PubMed] [Google Scholar]
- 51.Cantor CR, Schimmel PR. San Francisco: W. H. Freeman; 1980. The behavior of biological macromolecules. pp. 878–893. [Google Scholar]
- 52.Gaur NK, Smith RL, Klotz SA. Candida albicans and Saccharomyces cerevisiae expressing ALA1/ALS5 adhere to accessible threonine, serine, or alanine patches. Cell Commun Adhes. 2002;9:45–57. doi: 10.1080/15419060212187. [DOI] [PubMed] [Google Scholar]
- 53.Kapteyn JC, Hoyer LL, Hecht JE, Muller WH, Andel A, et al. The cell wall architecture of Candida albicans wild-type cells and cell wall-defective mutants. Mol Microbiol. 2000;35:601–611. doi: 10.1046/j.1365-2958.2000.01729.x. [DOI] [PubMed] [Google Scholar]
- 54.Gonzalez M, Goddard N, Hicks C, Ovalle R, Rauceo JM, et al. A screen for deficiencies in GPI-anchorage of wall glycoproteins in yeast. Yeast. 2010;27:583–596. doi: 10.1002/yea.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ELISA assays of binding of Als5pV326N substitution proteins. The upper graph shows binding of increasing concentrations of proteins to different concentrations of fibronectin. The lower graph denotes binding of the proteins to polystyrene. The constructs shown are Ig-T-TR, wildtype (•) and V326N (▾), and Ig-T wildtype (○) and V326N (Δ). The protein concentration is 2.3±0.7μM for the fibronectin binding. The assays were carried out as previously described [11].
(TIF)
Far UV Circular Dichroism spectra of Als5p 1-664 protein. Wildtype protein is represented by the solid line and the V326N substation by the dashed line at 20°C.
(TIF)
Effects of scrambled V326N (VITGVTNIRTSVA) and wild type peptide (VITGNTNIRTSVA) on cellular aggregation. S. cerevisiae expressing no Als5p (EV), Als5pV326N, Als5pWT, or C. albicans were aggregated in the absence and presence of 2µg/ml scrambled wild type (S-wild type) or 200 µg/ml scrambled peptide (S-V326N). The diameter of the beads is 2.8 µm, and all images are at the same magnification.
(TIF)
Amyloid binding dye Congo red reduces binding and aggregation on polystyrene biofilms. C. albicans or S. cerevisiae expressing Als5pWT, Als5pV326N, or no Als5p (EV) adhered to a polystyrene surface in the absence and presence of 100 nM or 300 µM Congo red for 1.5 h. Adherent cells were grown overnight, stained with 1% crystal violet and imaged.
(TIF)
Detection and unfolding of single Als proteins in C. albicans. (A) Principle of the single-molecule detection experiment. C. albicans cells are probed, in buffer, using AFM tips derivatized with Als5p1-431. (B) Force extension curves obtained by stretching ALS proteins showed periodic features reflecting the sequential unfolding of the TR domains. (C) Plot of the rupture distances as a function of the number of unfolded TR.
(TIF)