Abstract
Slitrks are a family of structurally-related transmembrane proteins belonging to the leucine-rich repeat (LRR) superfamily. Six family members exist (Slitrk1–Slitrk6), and all are highly expressed in the central nervous system (CNS). Slitrks have been implicated in mediating basic neuronal processes ranging from neurite outgrowth and dendritic elaboration to neuronal survival. Recent studies in humans and genetic mouse models have led to the identification of Slitrks as candidate genes that may be involved in the development of neuropsychiatric conditions such as obsessive compulsive spectrum disorders and schizophrenia. While these system level approaches have suggested that Slitrks play prominent roles in CNS development, key questions remain regarding the molecular mechanisms through which Slitrks mediate neuronal signaling and connectivity.
SLITRK FAMILY
The Slitrk gene family is composed of six members (Slitrk1 - Slitrk6), which are highly expressed in the central nervous system (CNS) [1,2]. This family was identified in a screen for genes that were differentially expressed in mice with neural tube defects [1]. Slitrk5 had been previously discovered as a gene expressed in early hematopoietic progenitors but not in mature hematopoietic cells [3]. All Slitrks form single pass (type I) transmembrane proteins with an intracellular domain that varies in length [1]. At the extracellular domain, Slitrks contain two leucine-rich repeat (LRR) domains, which are each composed of 13 to 17 LRRs, flanked by cysteine rich domains (Glossary) [1]. Sequence analysis has revealed that the extracellular LRR domains of Slitrks resembled Slit proteins, and a conserved region in their intracellular carboxyl terminus (C-terminus) has a high degree of consensus with the last 16 amino acids of the neurotrophin receptor, tropomyosin-related kinase (Trk) [1]. Based on their similarity with Slits and Trks, these proteins were named Slitrks [1].
Slitrks are classified as members of the LRR superfamily, due to the presence of the LRR domains. The LRR superfamily is a class of molecules containing structural motifs of 20–29 amino acids with a defining sequence LxxLxLxxN/GxL (x being any amino acid) [4,5]. LRR domains are composed of tandem repeats of this LRR motif, and are usually involved in protein-protein interactions [3–5]. LRR-containing proteins are emerging as key regulators of nervous system development and function, playing important roles in processes such as neurite outgrowth, neuronal survival, synapse formation and dendritic morphogenesis (see Table 1 and [5–7]). Genes encoding human Slitrks are present at three different loci on chromosomes 3 (Slitrk3), 13 (Slitrk1, 5 and 6), and X (Slitrk2 and 4) [2]. This chromosomal organization is conserved in the mouse where Slitrks are located on chromosomes 3, 14 and X. All Slitrk genes have a single protein coding exon [2]. As we will discuss in this Review, a recent series of human and mouse genetic studies have identified Slitrks as candidate genes involved in the development of neuropsychiatric conditions. Thus, understanding their various functional roles in the CNS will have important implications for understanding not only normal CNS development, but may also provide valuable insights into mechanisms that go awry during the development of neuropsychiatric disorders.
Table 1.
Summary of LRR-containing proteins and their functions
| Protein | Expression | Functions | Refs |
|---|---|---|---|
| AMIGO 1/2 | Enriched in CNS | - TM protein that promotes prominent neurite extension in cultured hippocampal neurons | [6,99] |
| Amigo 2 | - Exhibits homophilic and heterophilic binding activity | ||
| ubiquitous | - Modulates depolarization and NMDA-dependent survival of cerebellar granule neurons | ||
| FLRT3 | Ubiquitous | - TM protein identified in a screen for genes upregulated after nerve injury | [100] |
| - Promotes neurite outgrowth in cultured DRG neurons | |||
| Lingo-1 | CNS | - TM protein that acts as co-receptor for Nogo receptor | [101–104] |
| - Over-expression enhances responsiveness to myelin-associated inhibitors | |||
| - Dominant negative form protects midbrain dopaminergic neurons against degeneration | |||
| - Pathologically upregulated in SN of PD patients | |||
| - Also interacts with EGFR and decreases its protein levels | |||
| Linx | DRG neurons | - TM protein that interacts with Trk and Ret receptors and augments neurotrophin and GDNF/Ret signaling |
[31] |
| - Knocking out Linx partially photocopies axonal projection defects in mice lacking NGF, TrkA, or Ret |
|||
| Lrig1 | Ubiquitous; | - TM protein that interacts with ErbB receptor family members (EGFR, ErbB2-4) and recruits cytoplasmic E3 ubiquitin ligases |
[105–108] |
| Ubiquitous | - Attenuates GDNF/Ret signaling in neuronal cells through its physical interaction with Ret | ||
| Lrig3 | - TM protein that interacts with Xenopus FGFR through extracellular domain | [109] | |
| - Inhibits FGF-dependent Erk phosphorylation | |||
| - Negatively regulates FGF expression levels | |||
| LRRTM2 | CNS, post- synaptic |
- TM post-synaptic partner for α and β Neurexins | [29,32–34] |
| - Promotes excitatory synapse formation | |||
| - Interacts with PSD-95 and regulates surface expression of AMPARs | |||
| NGL-1 | CNS: ST, CT, TCA, dendritic |
- TM protein that binds netrin G-1 through LRRs | [110,111] |
| - Stimulates outgrowth of embryonic thalamic axons | |||
| NGL-2 | CNS, HC, CT, post-synaptic |
- TM protein that induces presynaptic differentiation and clustering of postsynaptic proteins |
[35,36,110] |
| - Increases the number of excitatory synapses | |||
| NGL-3 | Brain | - TM protein that binds LAR through LRRs | [35] |
| - Induces presynaptic differentiation | |||
| CNS | - Promotes excitatory synapse formation | ||
| Omgp | - GPI-anchored protein that is expressed by neurons and oligodendrocytes | [112] | |
| - Mediates growth cone collapse and inhibition of neurite outgrowth | |||
| SALM 1–3 | CNS | - TM proteins that form homo and heteromeric complexes | [113–116] |
| - Promotes neurite outgrowth in young neurons (DIV 4–6) but not in older neurons (DIV 14– 16) |
|||
| - SALM1-3 contain a PDZ-binding motif | |||
| - SALM1 induces the dendritic clustering of NMDA receptors | |||
| - SALM2 increases the number of excitatory synapses and dendritic spines | |||
| - SALM3 & 5 induces excitatory and inhibitory pre-synaptic differentiation | |||
| Trks | CNS | - TM receptors that bind the neurotrophin family of growth factors | [14,15] |
| - Influences the proliferation, differentiation, plasticity and survival of neurons | |||
Abbreviations: Amigo, amphoterin-induced protein; FLRT3, fibronectin leucine rich transmembrane protein 3; Lingo-1, leucine rich repeat and Ig domain containing 1; Linx, immunoglobulin superfamily containing leucine-rich repeat 2; Lrig, leucine-rich repeats and immunoglobulin-like domains; NGL, Netrin G ligand; Omgp, oligodendrocyte myelin glycoprotein; SALM, Synaptic adhesion like molecule; TCA, thalamocortical axons; ST, striatum; CT, Cortex; PD, Parkinson’s disease; DA, dopamine; TM, transmembrane; DRG, dorsal root ganglia; GDNF, glial cell line-derived neurotrophic factor; SN, substantia nigra; TM, transmembrane; HC, hippocampus, AMPAR, α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor; FGF, fibroblast growth factor; ErbB/EGFR, epidermal growth factor receptor; LAR, leukocyte antigen-related; GPI, glycosylphosphatidylinisotol; DIV, developmental day in vitro; PSD-95, synapse associated protein 95 kDa
SLIT AND TRK PROTEINS
Slits are one of the most well known LRR-containing protein families [8,9], functioning as soluble ligands for Robo receptors [9]. They are involved in axonal guidance and repulsion, tangential neuronal migration, cytoskeletal dynamics, and modifying cell adhesion properties [10,11]. Despite the high degree of sequence homology between Slit and Slitrk LRR domains, recent findings indicate that these protein families are likely to serve divergent functions in the CNS. Firstly, a similar degree of sequence and structural similarity exists in the LRR domains of Slitrk and many other LRR family members. Secondly, Slits do not have any particular homology outside the LRR domains with Slitrks. Thirdly, Slit is a secreted protein, whereas Slitrks are transmembrane proteins with intracellular domains, suggestive of a functional role in regulating intracellular signaling events [1,10]. Lastly, Slitrks do not interact with Robo, suggesting that they likely signal via different receptor-ligand pairings [1]. Although a recent study reported that Slitrk1 can undergo extra-cellular domain shedding (by α-secretase activity) in vivo, it is unknown if the secreted domain plays a functional role [12]. Further information regarding the exact site of α-secretase cleavage within Slitrk1 will elucidate if such a mechanism is conserved amongst other Slitrk family members.
Trk receptors are a family of receptor tyrosine kinases that selectively bind the neurotrophin family of growth factors (eg. brain derived neurotrophic factor, BDNF, and nerve growth factor, NGF, are two neurotrophins from this family) [13,14]. Trk signaling influences the proliferation, differentiation, plasticity and survival of neurons, and has been implicated in higher-order cognitive activities such as learning and memory [13–15]. Neurotrophins are secreted ligands that bind to Trks leading to the phosphorylation and activation of their catalytic tyrosine kinase domains, which rapidly initiate intracellular signaling cascades (Figure 1) [16]. All Trk receptors exhibit high conservation in their intracellular domains, including the catalytic tyrosine kinase domain, the juxtamembrane NPxY motif, as well as the C-terminus domain, in particular, tyrosine 791 (Y791 in human TrkA) [13–15]. Upon phosphorylation, the NPxY motif serves as a binding site for adaptor proteins such as Shc (that initiates Ras and phosphoinositide 3 kinase downstream signaling) [16–18]. Phosphorylation of the tyrosine residue within this particular motif can also serve as signal for receptor endocytosis [19].
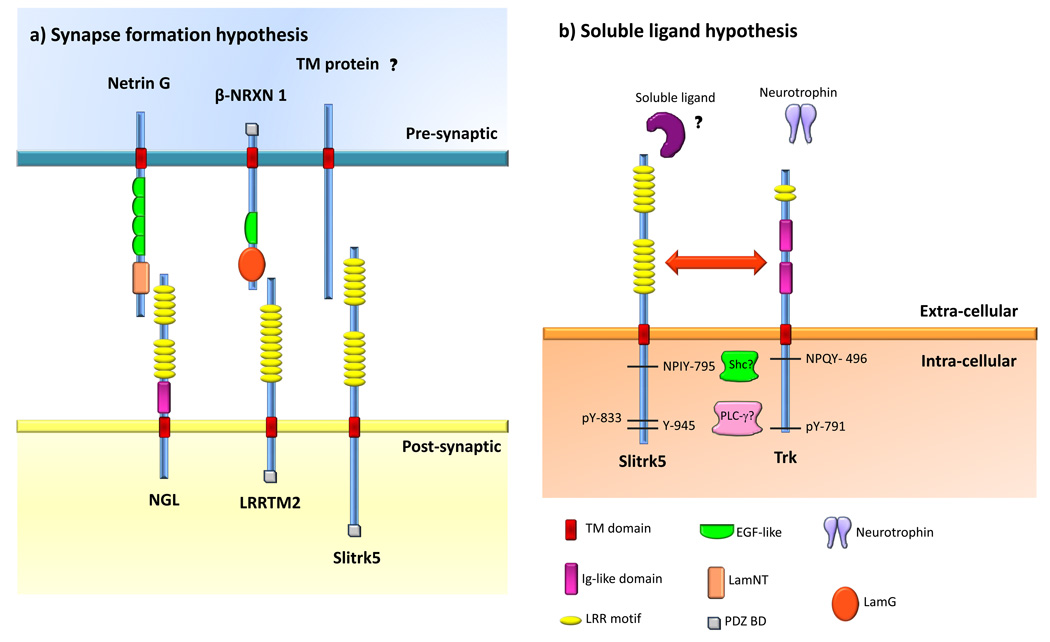
Figure 1. Possible models for the functions of Slitrks.
Slitrk5 was selected as the prototype of the Slitrk family and protein domains are shown according to the SMART database (http://smart.embl-heidelberg.de/). a) Synapse formation hypothesis for Slitrk function. According to this hypothesis, Slitrks could localize to the postsynaptic membrane and bind pre-synaptic membrane proteins. The identity of such binding partners remains unknown. This hypothesis is based on studies in culture whereby overexpression of Slitrk2 in fibroblasts induced excitatory synaptic differentiation in contacting neurons [29], as well as immunofluorescence studies demonstrating that Slitrk5 is localized at synaptic sites [28]. Other LRR-containing proteins that are known to play such a function in synapse formation are depicted. Netrin G-ligand (NGL) localizes to the post-synaptic cell, where it binds Netrin G through its LRR domains. LRRTM2 also localizes to the post-synaptic cell, where it binds both α and β Neurexins (NRXN) through its LRR domains. β-NRXN1 is shown as an example. b) Soluble ligand hypothesis. Slitrk1, 5 and 6 have been shown to play trophic roles in neurons [26–28]. Slitrk6 has been demonstrated to modulate neurotrophin signaling [26]. In PC12 cells, Slitrks modulate neurite outgrowth upon NGF treatment [1]. Thus, it is likely that Slitrks modulate RTK signaling. In this model, two possible scenarios are presented: one in which Slitrks act as receptors for soluble ligands that lead to signaling transduction events related to the neurotrophin system (ie. similar to how Trks and their neurotrophins interact). In a second scenario, Slitrks could physically interact with Trk receptors (and/or other RTKs) and directly modulate their function. The depicted Trk amino acid numbers correspond to human TrkA but similar residues and signaling components are found in TrkB and TrkC. Neurotrophin binding to Trks leads to receptor dimerization and subsequent activation and phosphorylation of their catalytic tyrosine kinase domains as well as phosphorylation of intracellular tyrosines such as the juxtamembrane NPXY domain and Y791 (in human TrkA). Y791 recruits PLC-γ. This enzyme subsequently hydrolyses phosphatidyl inositol to generate inositol tris-phosphate (IP3) and diacylglycerol (DAG), which ultimately leads to the release of Ca2+ from internal stores and protein kinase C (PKC) activation, respectively [13,16,20]. For simplicity, a single TrkA molecule is shown instead of the active dimmer. Y791 in human TrkA (and analogous tyrosines in TrkB and TrkC) show some degree of homology with the cytoplasmic tail of Slitrks [1], namely Y945 in human Slitrk5, raising the question if Slitrks are also implicated in PLC-γ signaling. Moreover, Slitrk2, 3 and 5 also contain an NPXY motif at the juxtamembrane region. Phosphorylated NPXY serves as a docking site for phosphotyrosine bindig proteins such as Shc that links the activated neurotrophin receptor to two separate intracellular signaling pathways: mitogen activated protein kinase (MAP kinase) and PI-3 kinase [17]. Slitrks might mediate similar signaling events through their conserved NPXY motif. Finally, it was found that Y833 could be phosphorylated in Slitrk5, however its function is still unknown [21]. Abbreviations: TM, transmembrane domain; Ig, immunoglobulin-like domain; LRR, leucine rich repeat; EGF, epidermal growth factor-like domain; LamNT, laminin N-terminal domain; LamG, laminin G domain.
Slitrk2, 3 and 5 share similarities with Trk receptors in that they contain an NPxY motif near their intracellular juxtamembrane region. The stretch of amino acids that spans the NPxY motif in human Slitrk5 shares 25% amino acid identity with the TrkA NPxY region, and higher than 60% with Slitrk2 and 3 suggesting they might also recruit Shc and other adaptor proteins that could initiate intracellular signaling cascades. Furthermore, Slitrk2–6 all contain a common C-terminal region and a conserved tyrosine with significant homology to Y791 in human TrkA [1]. With respect to the TrkA receptor, it is known that phosphorylation of Y791 leads to the recruitment and activation of the γ isoform of phospholipase C (PLC), resulting in the release of Ca2+ from internal stores and protein kinase C (PKC) activation [13,15,20]. It is currently unknown if this conserved tyrosine residue is phosphorylated in Slitrks, however, it is tempting to speculate that Slitrks may couple to PLC-γ signaling cascades, based on this domain similarity with the TrkA receptor. Slitrk5 has been shown to be phosphorylated at another intracellular tyrosine residue (Y833) in HeLa cells, but the function and conservation of this site in other Slitrks still needs to be clarified [21].
SLITRK EXPRESSION IN THE CNS
Slitrks are highly and broadly expressed in the mammalian CNS [1,2,22–24] and also in hematopoietic and leukemic cells [25]. Initially it was shown by Northern blot analysis that Slitrk1, 2 and 4 can be detected as early as embryonic day 11.5/12.5 (E11.5/12.5) [1]. Despite some overlapping expression, several key brain regions express different combinations of Slitrks suggesting that each member might play a particular role in the maturation of selective populations of neurons across different stages of development. Slitrk6 is the member that differs mostly from the rest of the family, with expression being restricted to a few areas of the nervous system (such as the thalamus, lateral geniculate nucleus and auditory and vestibular system of the ear) [1,22,26]. Slitrk6 is, however, also expressed in other organs, including the epidermis, limb bud, cochlea and tongue of mouse embryos [23].
Similarly as in the mouse, the expression of Slitrk mRNAs has been demonstrated to be enriched in human brain tissue [3]. In human fetal tissue, Slitrk1–5 were predominantly expressed in the brain, whereas Slitrk6 was also present in the lung and liver [2]. In the adult, Slitrk1 mRNA expression was highest in the frontal lobe and Slitrk5 was expressed in both occipital and frontal lobes [2]. Slitrk2 and 5 were the only members significantly expressed in the spinal cord and medulla [2]. Slitrk6 expression differs greatly from the other members, with no expression in the cortex but high levels of expression in the putamen [2].
A comparative study that analyzed Slitrk1 expression in mouse, rhesus monkey and humans revealed that regional and developmental expression is conserved amongst these species [24]. Slitrk1 was found in projecting neurons of the corticostriatal-thalamocortical circuits (CTC) compartmentalized in striosomes. Subcellularly, it is enriched in the somato-dendritic compartment and cytoplasmic vesicles of cortical pyramidal neurons [24]. In the adult striatum, the expression is confined to cholinergic interneurons [24].
Besides neurons, Slitrks can be found in brain tumors, embryonic stem cells, subsets of endothelial cells, hematopoietic stem cells, as well as leukemic and lymphoma cells [2,25]. Regarding brain tumors, Slitrk5 had the broadest expression amongst the various tumors analyzed, whereas gliomas mainly express Slitrk1–3 [2]. Thus, besides their roles in normal development, Slitrks might also be implicated in malignancy.
SLITRK FUNCTIONS IN THE CNS
Slitrks have been implicated in the modulation of neuronal process outgrowth [1,27,28] promoting neuronal survival [26,28], and in synapse formation [29]. Initial studies performed by over-expressing each of the Slitrk members in PC12 cells revealed that exposure to NGF resulted in a decreased number of neurites per cell in the case of all Slitrk members [1]. Furthermore, the neurite length was decreased in all cases, with the exception of Slitrk1 and 4 [1]. Subsequently, the neurite outgrowth function of Slitrk1 has been analyzed in mouse cortical neurons [12,27]. Cortical neurons cultured from mouse embryos previously electroporated with Slitrk1 cDNA showed increased dendritic length [27], suggesting that this Slitrk member plays an important role in promoting neurite outgrowth. More recently, it has been demonstrated that phosphorylation of Slitrk1 on Ser695 by casein kinase II is critical for the interaction of Slitrk1 with 14-3-3 proteins [12], which are ubiquitously expressed phosphorylation-binding proteins that regulate a number of important cellular processes including cell proliferation, neuronal migration and membrane excitability [12,30]. Mutation of this serine residue to an alanine residue abolished the interaction of Slitrk1 with 14-3-3 proteins as well as blocking the induction of neurite outgrowth in cultured mouse cortical neurons [12].
Recently, a Slitrk5 knockout mouse was generated that displayed reduced total brain volume, which was most prominent in the striatum [28]. In addition, there was a marked decrease in dendritic complexity of medium spiny neurons in the striatum of adult Slitrk5 null mice, as determined by Golgi tracing analyses [28].
Considering the more restricted distribution of Slitrk6 in the brain, with selectively high expression in the inner ear, a recent study performed a detailed analysis of this organ in a mouse model that ubiquitously lacks Slitrk6 expression [26]. Results from this study demonstrated that Slitrk6 promotes the survival and neurite outgrowth of sensory neurons of the inner ear [26]. Histological examinations revealed that vestibular innervation was markedly decreased and sometimes misguided in the Slitrk6 null mice. The mutant mice also showed significant cell death in the spiral and vestibular ganglia. Furthermore, cochlear sensory epithelia taken from these mice and co-cultured with wild-type spiral ganglion neurons were less effective in promoting neurite outgrowth of the spiral gangion neurons compared to co-cultures in which both types of cells were taken from wild-type mice [26]. Evidence for a trophic role of Slitrk6 was further strengthened by the observation that both BDNF and neurotrophin 3 (NT-3) mRNA levels, as well as their corresponding receptor protein levels, TrkB and TrkC respectively, were down-regulated in the inner ear of Slitrk6 null mice. Interestingly, this was observed at an early developmental stage of the cochlea, when cell death had not occurred [26]. This study suggests that Slitrk6 may exert trophic actions, in part, by modulation of the neurotrophin system. Several LRR containing proteins have been shown to interact and modulate receptor tyrosine kinase (RTK) signaling (summarized in Table 2), for example Linx, a leucine-rich repeat and immunoglobulin (LIG) family protein, physically interacts with both Trk and Ret receptors, resulting in increased neurotrophin and GDNF signaling, respectively [31]. Besides Slitrk6, it would be interesting to investigate if other Slitrks could also modulate Trk receptor signaling.
Table 2.
Summary of human genetic studies assessing associations between Slitrks and neuropsychiatric disorders OCD-spectrum disorders
| Slitrk gene | Disorder | Main conclusions of the studies | Refs | |
|---|---|---|---|---|
| Slitrk1 | GTS | • | Identified 3 patients (out of 174 unrelated individuals with GTS) carrying mutations in the Slitrk1 gene in 174; one patient had the varCDfs variant, while the other two expressed var321. These mutations were absent in more than 3600 control subjects. |
[27] |
| • | No mutations found in 6 patients belonging to a large Dutch family with GTS |
[62] | ||
| • | No association of varCDfs or var321 was reported in a cohort of 812 GTS patients from a Costa Rican and AJ population, however, over- representation of var321 in AJ population was found. |
[60] | ||
| • | Five subjects with GTS were identified in the AJ population who contained the var321, two of these did not transmit the mutation to affected child. One individual was found with var321 but did not have GTS. |
|||
| • | ||||
| • | var321 nor varCDfs were found in 82 Caucasian patients with GTS; a new mutation was found that neither changed amino acid nor altered mRNA splicing. |
[58] | ||
| • | ||||
| • | Did not find var321 nor varCDfs in 208 affected children, however, found significant association with 3 other novel SNPs in the Slitrk1 gene |
[64] | ||
| • | ||||
| • | No var321 or varCDfs found in 92 Austrian patients with GTS. A new 3’UTR variant was found in one patient and parents with GTS and was absent in 192 control individuals. |
[66] | ||
| GTS, CT | • | Large sequencing study did not find association between Slitrk1 and GTS or CT. Out of 1048 individuals with GTS or CT one individual carried var321; a second carrier of var321 had OCD. Neither transmitted the mutation to GTS offspring. Both of these carriers were of AJ descent. |
[61] | |
| GTS, CT, OCD | • | Provided additional information regarding the ethnicity of the two initially identified variants that are of European descent and found 4 additional affected individuals carrying var321 of Spanish, Eastern Europe and Caucasian. In addition, one AJ unaffected individual with var321 was identified. |
[63] | |
| OCD | • | Did not find var321 nor varCDfs in 322 subjects with OCD | [59] | |
| TTM | • | Two novel non-synonymous mutations were found in a cohort of 44 patients with TTM and which were absent in 3000 healthy controls. var321 and varCDfs were not found in this population. |
[53] | |
| Slitrk2 | SCZ | • | Systematic re-sequencing of X-chromosome genes found two novel missense variants in Slitrk2 gene in patients with SCZ and in their affected siblings |
[97] |
Abbreviations: CT, chronic tics; AJ, Ashkenazi Jew; SCZ, Schizophrenia;
Taken together, the results obtained with Slitrk1 overexpression in neurons [12,27] and the analyses of the Slitrk5 and 6 null mice [26,28], suggest that multiple Slitrk members play key roles in promoting neurite outgrowth. Even though when initially described, Slitrks were shown to play a negative role on neurite outgrowth when over-expressed in PC12 cells [1], it is likely that crucial binding partners involved in neurite outgrowth that are endogenously expressed in neurons are lacking in PC12 cells.
Slitrks have also been implicated in synapse formation. Slitrk5 has been demonstrated by immunocytochemical studies to localize to synaptic sites in cultured neurons [28]. A recent study that performed an expression screen for synaptogenic proteins revealed the abundance of LRR containing proteins capable of inducing synapse formation [29]. Slitrk2 was one of the LRR proteins identified in this screen, and it was shown to induce excitatory pre-synaptic neuronal differentiation in a cellular co-culture system, suggesting that it could be a postsynaptic protein [28]. Another LRR protein that was identified in the screen was LRRTM2 [29], a member of the brain enriched Leucine-rich repeat transmembrane neuronal protein (LRRTM) family [32]. LRRTM2 acts as a ligand for α and β Neurexin I and this binding is crucial for trans-synaptic cell adhesion, mediating excitatory synapse formation [33,34]. Besides LRRTM2, other LRR-containing proteins bind pre-synaptic membrane partners through their LRR domains [7], for example, Netrin G ligand 2 and 3 (NGL-2 and NGL-3) (Table 1) [35–37]. It is possible that Slitrks could interact with a pre-synaptic partner in a similar manner via their extracellular LRR domains and could recruit intracellular post-synaptic proteins (Figure 1). Identifying the intracellular binding partners for Slitrks would contribute significantly to understanding the molecular basis of Slitrk functions in the nervous system.
SLITRKS AND POTENTIAL LINKS TO NEUROPSYCHIATRIC DISORDERS
Considering the significant heritability of many psychiatric disorders, recent efforts have focused on finding susceptibility genes for these disorders [38–40]. Genome-wide association studies (GWAS) have allowed for the screening of known single nucleotide polymorphisms (SNPs) spanning the entire genome in large population samples [41,42]. However, most common variants found so far show low penetrance, thus do not contribute significantly towards disease risk [43]. In contrast to common polymorphisms, the effects of rare variants, whether SNP’s or copy number variants (CNV’s), on disease risk cannot be identified through GWAS approaches. The main reasons for this are twofold: firstly, such rare variants are not well tagged by the common SNP’s tested on whole-genome panels, and secondly, the statistical power of association analysis drops off sharply for rare variants [43]. Rather, the genetic effects of rare variants are generally identified through re-sequencing of candidate genes/loci in large subject and control samples and are often analyzed in aggregate to improve statistical power. In contrast to common polymorphisms, rare variants usually have high penetrance and therefore their detection greatly contributes to understanding the etiology of a disease. Furthermore, they are valuable as biomarkers for preventive strategies. The rare variant hypothesis has been strengthened by the recent findings that de novo mutations in synaptic genes such as Neuroligin 3 and 4 (NLGN3, NLGN4X) and SH3 and Multiple Ankyrin Repeat Domains 3 (SHANK3) are associated with autistic spectrum disorders in a small number of families [44–48]. Genetic studies have recently identified rare mutations in Slitrk genes as being associated with Obsessive-Compulsive Spectrum Disorders and other neuropsychiatric disorders, as discussed in more detail below.
Slitrk1 and OCD-spectrum disorders
Gilles de la Tourette's syndrome (GTS) is a common psychiatric disorder, present in ~1% of children between 5 and 16 years in USA and Europe [49,50]. It is characterized by persistent involuntary vocal and motor tics and it has comorbid occurrence with Obsessive-Compulsive disorder (OCD) and attention deficit hyperactivity disorder (ADHD) [49–53]. Family studies have suggested that genetic factors are involved in the manifestation of GTS, however finding susceptible genes for this disorder has been a daunting task [52,54]. The cortico-striatal thalamocortical circuit (CSTC) has been implicated in the pathogenesis of both GTS and OCD [55–57], however, the molecular and genetic mechanisms underlying these disorders are less clear. In a recent study, a de novo inversion of chromosome 13 was identified in one patient that suffered from GTS and ADHD but had no family history of any OCD-spectrum disorders [27] (Table 2). The 13q33.1 breakpoint mapped approximately 350kb from Slitrk1, and this gene was selected for re-sequencing in a cohort of 174 individuals with GTS. Two additional mutations were discovered in this screen: a single nucleotide deletion that led to a frame-shift and a prematurely truncated protein (varCDf) was found in one individual; and a missense mutation in the 3’ untranslated region (3’UTR) named variant 321 (var321) was detected in two unrelated individuals [27]. In two of these patients, the mothers were also carriers of the mutation and were affected. The truncated Slitrk1 human protein failed to promote dendritic growth when expressed in cultured mouse cortical neurons, in contrast to expression of wild type human Slitrk1 protein [27]. The var321 of Slitrk1 altered binding to the microRNA hsa-miR-189 and it negatively modulated Slitrk1 mRNA expression. These mutations were absent in over 3600 control samples, indicating that they were rare variants [27].
Subsequent studies were unable to find associations between these two reported variants in larger GTS populations [58–62], arguing against a segregation of Slitrk1 and GTS. Furthermore, it was reported that var321 was over-represented in the Ashkenazi Jew population, raising the possibility that population stratification might have led to a false positive result [60,61]. The largest screen for Slitrk1 and GTS performed so far, which involved sequencing 1048 samples from the Tourette Syndrome Association International Consortium for Genetics, did not find association between var321 and GTS [61]. Only two individuals were found who carried var321: one was diagnosed with GTS, OCD and Trichotillomania (TTM), and the other with OCD but not GTS. None transmitted the mutation to their affected offspring [61]. Additional studies provided more information regarding the ethnicity of the initially reported cases, and five additional subjects carrying var321 were reported [63]. Fine mapping of all seven carriers of the Slitrk1 region argued against population stratification confounding the original data [63].
In addition to these two variants, a recent study testing for association of common tag SNP’s spanning SIitrk1 and GTS identified an association to one specific tag SNP (rs959383) as well as two three-marker haplotypes, suggesting there may be a common GTS risk factor of low penetrance in linkage disequilibrium (LD) with the associated marker/haplotypes [64]. Furthermore, novel mutations in the Slitrk1 gene have been found that co-segregate with OCD-spectrum disorders [64–66]. Two non-synonymous mutations in the Slitrk1 extracellular region were discovered in two independent individuals of European descent, in a set of 44 families with TTM, and were absent in a group of almost 3000 healthy controls [65]. TTM is an OCD-spectrum disorder thought to be genetically associated with GTS [67,68]. Moreover, a mutation in the 3’UTR of Slitrk1 gene was found in a group of 92 Austrian patients, that segregated in two additional family members with tic disorders, and which was absent in 192 control subjects [66] (Table 2).
Slitrk1 null mice have been generated and studied with the aim of better clarifying the potential role that Slitrk1 may play in neuropsychiatric disorders [69]. It has been demonstrated that these mutant mice display increased anxiety-like behavior (as measured by the elevated plus maze test; see Box 1) as well as depressive-like behaviors (as assessed in a forced swim task). Neurochemical analysis revealed that Slitrk1 null mice had increased levels of norepinephrine in the prefrontal cortex, striatum, and nucleus accumbens [69] which is consistent with the pathophysiology of GTS since patients with this disorder have been reported to have high concentrations of norepinephrine in their cerebrospinal fluid [70]. Although these mice did not recapitulate the hallmark motor characteristics of human GTS, anxiety and depressive disorders are highly comorbid with GTS [51,53], and administration of clonidine (an α2-adrenergic agonist commonly used to treat GTS patients [71–73]) was able to rescue the anxiety-like behavior of Slitrk1 null mice [69]. These findings add support to a likely role of Slitrk1 in neuropsychiatric disorders. The generation of future mouse models for Slitrk1, especially those which conditionally knock-down the gene in a spatially and/or temporally-specific manner, will help to further delineate the exact roles that this protein plays, and will help to avoid potential developmental compensation from other Slitrk family members that may occur with traditional knockout mouse models. Furthermore, it would be interesting to directly assess the consequences of the introduction of human Slitrk1 variants (eg. var321 or varCDfs) in future knock-in mouse models, and to assess these mice for their subsequent neuroanatomical and behavioral phenotypes. Such studies will help to directly assess the in vivo consequences of specific mutants in the Slitrk1 gene.
Box1. Behavioral tasks commonly used to study endophenotypes of psychiatric disorders in mice
Elevated plus maze
A behavioral test to assess anxiety-like behaviors in rodents. The experimental apparatus consists of a plus-shaped platform with two open and two closed arms that is elevated above the floor. This test is based on the aversion of open space in rodents. Anxiety–like behavior is indicated by a decrease in the proportion of time spent and entries into the open arms.
Open field test
Assesses locomotor activity and anxiety-like behavior in rodents. Subjects are allowed to explore a novel and bright arena; distance travelled as well as percent time spent in the center of the arena is recorded. Generally rodents avoid bright and aversive areas such as the center of the arena; anxious rodents have this behavior accentuated.
Marble burying test
A task that measures repetitive, stereotypical and anxiety-related behaviors in rodents. The experimental setting consists of a novel cage with multiple marbles spaced out regularly on the bedding. The number of marbles buried is measured. Anxious rodents tend to bury more marbles compared to control animals.
Fear conditioning test
Measures associative learning in rodents and it can have two components: contextual and cued-fear learning. A rodent is placed in a novel environment, where an unconditioned stimulus, usually a foot shock, is paired with a conditioned stimulus, usually a tone. Rodents can learn to associate the context/tone with the foot shock, and exhibit freezing behavior when exposed to the same environment (contextual fear) or the tone (cued fear). Contextual fear is hippocampal-dependent, whereas cued-fear is amygdala-dependent.
Forced swim test
A behavioral despair paradigm that reveals depressive-like behavior in rodents. Animals are subjected to trials in which they are forced to swim in a glass cylinder filled with water. The time that the animal spends immobile in the water is recorded. Mice with depressive-like behaviors spend more time immobile.
Tail suspension test
Similar to the forced swimming test, the tail suspension test measures depressive-like behavior in rodents. In this test, a mouse is suspended by its tail, and movements of the mouse are recorded. Mice with depressive-like behaviors spend more time immobile.
Slitrk5 and OCD-spectrum disorders
A recent study of a mouse model in which the coding region of the Slitrk5 gene was replaced by the β-galactosidase gene (lacZ) reporter gene has revealed further evidence in support of an association between Slitrk family members and the development of neuropsychiatric disorders [28]. Slitrk5 null mice initially developed increased anxiety-like behaviors (as assessed by the open field test), and subsequently displayed repetitive and excessive self-grooming behaviors, which led to the formation of severe facial skin lesions and hair loss [28]. Treatment with chronic fluoxetine, a selective serotonin reuptake inhibitor (SSRI) that is one of the major pharmacological agents used to treat depression as well as OCD [74,75], alleviated excessive grooming behavior in the mutant mice. These mice also showed selective over-activation of the orbitofrontal cortex [26]. Activation levels in this brain region relative to other areas of the brain was determined by measuring the protein levels of FosB, a transcription factor that is routinely used to assess neuronal activity in a variety of paradigms [28,76]. Overactivation of the orbitofrontal-subcortical circuits has been observed in functional imaging studies of human subjects with OCD, and is posited to be due to an imbalance in the basal ganglia pathways [77–79]. Thus, the Slitrk5 null mouse recapitulates an important aspect of the human disease.
Slitrk5 null mice also exhibit anatomical deficits in the striatum, such as reduced striatal volume and decreased dendritic complexity, as well as neurophysiological alterations in cortico-striatal transmission, which may have resulted from alterations in glutamate receptor expression levels in the striatum [28]. In this context, it has recently been postulated that striatal dysfunction, in the presence of orbitofrontal cortex overactivation, could lead to deficits in thalamic filtering, or an imbalance in the direct and indirect pathways of the basal ganglia [80,81]. Taken together, the Slitkr5 mouse model recapitulates several key aspects of OCD: (i) increased anxiety-like behavior that is followed by (ii) repetitive behaviors that are ameliorated by administration of a SSRI, as well as (iii) orbitofrontal cortical overactivation, and 4) alterations in basal ganglia circuitry and function.
A question that arises from this study is, given the wide distribution of Slitrk5 throughout the brain [1,22,28], why do these mutant mice reflect a predominantly basal ganglia-dependent phenotype? On the one hand, it is currently unknown if these mice also exhibit higher order cognitive defects, such as impaired learning and memory, since the results of such experiments have not yet been reported. On the other hand, it may be the case that Slitrk5 is the most highly expressed Slitrk family member in the corticostriatal circuitry. Studies comparing the protein expression levels between all members of the Slitrk family have not yet been performed, however, a recent report that compared levels of Slitrk1 protein in different murine brain regions showed very little expression of Slitrk1 in the striatum, as compared with the amygdala, frontal cortex or hippocampus [69]. Therefore, lack of developmental compensation by another Slitrk protein in the striatum of Slitrk5 null mice might be one possible explanation for the observed phenotype involving the basal ganglia circuitry. Considering the particular expression pattern of Slitrk5 within the striatum, together with the behavioral phenotype of these mice, it would be of interest to sequence the Slitrk5 gene within human subjects, to assess whether rare mutations or variants within this gene are associated with OCD and/or OCD-spectrum disorders.
It is striking that many of the behavioral and circuit phenotypes observed in the Slitrk5 null mice have been reported in a previously described genetic mouse model, which lacks the synapse-associated protein 90/ postsynaptic density-95-associated protein 3 (SAPAP3) [82]. SAPAP3 is an intracellular scaffolding molecule that localizes to the post-synaptic density (PSD) of excitatory synapses, and is crucial for maintenance of synaptic structure [83,84]. Similar to Slitrk5 null mice, SAPAP3 null mice displayed excessive grooming that led to the formation of facial lesions, increased anxiety-like behaviors, defects in cortico-striatal transmission and altered expression of glutamate receptors in the striatum [82]. One of the main differences observed between these two models is that the behavioral defects are also observed in the haploinsufficient mouse in the case for Slitrk5, but not in the case of SAPAP3 [26, 73]. It is interesting to speculate whether Slitrk5 and SAPAP3 form part of a common signaling complex that is involved in synaptic maintenance/function in the cortico-striatal circuit. The discovery of Slitrk5 binding partners would help to elucidate these mechanisms. With respect to SAPAP3, recent genetic analyses have led to the identification of a number of rare non-synonymous mutations that have been associated with OCD, GTS and TTM, however larger population analysis will be necessary to understand the precise involvement of SAPAP3 mutations in these disorders [85–87].
In addition to the SAPAP3 null mouse, there are a number of other genetic mouse models which display repetitive behaviors similar to the Slitrk5 null mouse (for recent reviews, see [88,89]). One such model which is of direct relevance to the Slitrk5 null mouse is the Hoxb8 null mouse [90–93] (Table 3). The Hoxb8 gene is a member of the mammalian Homeo-box containing complex (Hox), an important family of transcription factors. Hoxb8 null mice were initially described to develop large skin lesions on their bodies [93]; excessive grooming was later found as the cause of the lesions [92]. This phenotype had 100% penetrance in the offspring of Hoxb8 null mice and it was present in two different genetic backgrounds [92].
Table 3.
Summary of the Slitrk5, SAPAP3 and Hoxb8 genetic mouse models for OCD
| Genetic Model |
Behavioral Phenotype | Circuit Phenotype | Rescue (of overgrooming behavior) |
Refs |
|---|---|---|---|---|
| Slitrk5−/− |
 ObF cortical activity ObF cortical activity |
[28] | ||
 Repetitive grooming (head) Repetitive grooming (head) |
||||
 Striatal volume Striatal volume |
Fluoxetine (21 days) | |||
 Anxiety-like behaviors Anxiety-like behaviors |
||||
 Corticostriatal transmission Corticostriatal transmission |
||||
| Sapap3−/− | Fluoxetine (6 days) | [82] | ||
 Repetitive grooming (head) Repetitive grooming (head) |
 Corticostriatal transmission Corticostriatal transmission |
|||
 Anxiety-like behaviors Anxiety-like behaviors |
Genetic (overexpression of Sapap3 in ST) |
|||
| Hoxb8−/− |
 Repetitive grooming (body) Repetitive grooming (body) |
Bone marrow transplant | [91] | |
 Repetitive grooming Repetitive grooming(cagemates) |
Subcutaneous lidocaine (acute) |
[90] | ||
Abbreviations: ObF, Orbitofrontal cortex; ST, striatum.
In a subsequent study, a Hoxb8 cell lineage mouse model was developed where expression of Hoxb8 could be analyzed by expression of yellow fluorescent protein (YFP) [91]. Hoxb8 mRNA was determined to be broadly expressed in the brain, including in the corticostriatal circuit [92], however, this mouse model unexpectedly revealed that Hoxb8 was not expressed in neurons, but rather in a subpopulation of microglia cells [91]. Consistent with the hypothesis that microglia cells derive from the bone marrow where Hoxb8 is strongly expressed [94,95], a set of bone marrow transplants were performed to evaluate if the over-grooming phenotype could be rescued. Interestingly, transplanting wild type mouse bone marrow into lethally irradiated Hoxb8 mutant mice partially rescued the excessive grooming behavior in a subset of mice (~60%), whereas transplantation of bone marrow from mutant Hoxb8 mice into lethally irradiated wild-type mice led to the development of overgrooming in ~20% of the mice [91]. The excessive grooming phenotype of Hoxb8 mutant mice was reported in a parallel study to be rescued by subcutaneous application of the local anesthetic, lidocaine [90], however, these experiments were not replicated in the subsequent report [91]. It was recently reported that microglia cells derive from primitive (embryonic) macrophages and that in adulthood these cells are maintained independently of definitive haematopoiesis [96]. In light of these conflicting studies, the association of Hoxb8 with OCD-spectrum disorders remains questionable and future studies are necessary to address key questions remaining. For example, it is currently unknown if brain regions that are affected in OCD are anatomically and functionally normal in the Hoxb8 null mice. It will also be of importance to determine if pharmacological treatments (such as SSRIs) can rescue the overgrooming phenotype of the Hoxb8 mutant mouse, as was the case with the behavioral phenotypes exhibited by the Slitrk1, Slitrk5 and SAPAP3 knockout mice [28,69,82]. Furthermore, it would be interesting to investigate whether there is any functional overlap between Hoxb8 and Slitrks. Slitrk5 is highly expressed in immature hematopoietic cells, and it is downregulated upon differentiation [25], as is the case for Hoxb8. Finally, determining if patients with OCD-spectrum disorders carry mutations in Hoxb8 gene will be an important line of investigation in linking this gene to disease processes.
Slitrk2 and psychiatric disorders
Slitrk2 is another member of the Slitrk family that has recently been associated with a neuropsychiatic disorder, namely schizophrenia [97]. A list of synaptic genes located on the X-chromosome was used as candidates for a genetic screen of subjects with schizophrenia [97]. Several new synonymous and non-synonymous mutations were found in genes that have not been previously associated with this disorder, including an amino acid change (V89M) in the second LRR domain of Slitrk2 [97]. Once the function of Slitrk2 is elucidated, it would be interesting to analyze if this point mutation disrupts normal Slitrk2 function.
While preliminary, this study further supports the notion that Slitrks play crucial roles in the CNS and that mutations in these genes can contribute to enhanced susceptibility to neuropsychiatric disorders. As shown by the examples of the Slitrk1, 5 and 6 knockout mice studies [26,28,69], generation of mutant mouse models for Slitrk2 will likely be highly informative for the human genetic studies, in terms of validating the specific biological impact of Slitrk2 on CNS function and in relation to neuropsychiatric disorders.
CONCLUSIONS
Slitrks represent a relatively newly discovered family of structurally-related transmembrane proteins that have a predominant expression in the CNS. Studies so far have shown that Slitrks play trophic roles in the CNS promoting neurite outgrowth and neuronal survival [12,26–28]. Human genetic variants and mouse genetic knockout models have suggested an association of these genes with neuropsychiatric disorders, in particular, OCD-spectrum disorders [27,28,64,65,98]. However, additional large population studies are needed to clarify these findings. Furthermore, the detailed cellular and molecular mechanisms by which Slitrks transmit intracellular signals are largely unknown. An important question to be addressed in future studies will be to define the main functions of the Slitrk family (Box 2). Furthering our understanding of the role of Slitrks in terms of intracellular signaling as well as neuronal connectivity will not only be informative for understanding their roles in normal CNS development but also will provide the foundation for subsequent vertically integrated studies to link Slitrks to the development of neuropsychiatric disorders.
Box2. Outstanding Questions
-
What are the main intra- and extra- cellular binding partners for Slitrks?
Do all Slitrk members function in a similar way or are several scenarios and unique binding partners possible based on divergent intra- and/or extra- cellular amino acid sequences?
Do Slitrks play structural roles at the synapse by binding other pre-synaptic transmembrane proteins, similar to neuroligins and neurexins?
Do Slitrks act as a receptor for an unknown secreted ligand and trigger signaling cascades with their intracellular domains?
Do Slitrks modulate RTK signaling directly by interacting with these membrane receptors or is the modulation indirect?
Are there specific mutations in the Slitrk5 gene that contribute to OCD-spectrum disorders in humans?
Is Slitrk1 broadly associated with OCD-spectrum disorders and not specifically with GTS as initially described?
Are other Slitrk members associated with neuropsychiatric conditions?
Besides the brain, are Slitrks playing key roles in the hematopoietic system or cancers such as leukemia?
Acknowledgments
We thank Harel Weinstein, Jufang Shan, and Spencer S. Ericksen for helpful discussions and Charles E Glatt, Kevin Bath, Pedro Ramos and Matias Okawa for helpful comments on drafts of the manuscript.
Glossary
- Direct and indirect pathways
two opposing circuit loops in the basal ganglia. The direct pathway originates in the frontal cortex and projects to the striatum. Medium spiny neurons in the striatum, expressing D1 dopamine receptors, project to the globus pallidus interna/substantia nigra pars reticulata (GPi/SNr) complex, which projects to the thalamus, that in turn has reciprocal, excitatory projections to and from the cortical site of origin. The indirect pathway also originates in the frontal cortex and projects to medium spiny neurons expressing D2 dopamine receptors in the striatum, then projects to the globus pallidus externa (GPe), then to the subthalamic nucleus, then back to GPi/SNr, before returning to the thalamus and finally back to frontal cortex. The direct pathway contains two excitatory and two inhibitory connections, resulting in a net positive circuit back to the cortex. The indirect pathway has three inhibitory connections resulting in a net negative feedback loop to the cortex
- Genome-wide association study (GWAS)
involves sequencing and comparing the genomes of individuals to search for variations and mutations that associate with a disease of interest
- Linkage disequilibrium
in a population, co-occurrence of a specific DNA marker and a disease at a higher frequency than would be predicted by random chance
- LRR
Leucine-rich repeat is a protein structural motif that is 20–29 amino acids in length and contains a signature sequence, LxxLxLxxN/GxL, with x being any amino acid. An individual LRR motif has a β strand-turn-α helix structure. Many LRR motifs often assemble into a functional domain called a LRR domain which typically has a horseshoe shape and is amenable for protein-protein interactions
- Obsessive-compulsive disorder (OCD)
is a psychiatric disorder characterized by intrusive, persistent thoughts (obsessions) and/or repetitive, intentional behaviors (compulsions) that significantly affect normal life. OCD is usually a chronic anxiety disorder and affects ~2–3% of the population.
- Penetrance
the probability that a given genotype manifests itself in a given phenotype. Conditions with high penetrance imply that most individuals carrying the genetic variant in question reflect the associated phenotype. These conditions are rare. Low penetrance conditions are more common and imply that the proportion of individuals with a specific genotype that manifest in an associated phenotype is low
- Gilles de la Tourette’s syndrome (GTS)
is a familial OCD-spectrum disorder with childhood-onset characterized by involuntary motor and vocal tics. It affects ~1% of the children between 5 and 16 years and its symptoms usually improve by adulthood. Approximately 75% of individuals with GTS have comorbid psychiatric conditions such as OCD ADHD and TTM
- Trichotillomania (TTM)
is an OCD-spectrum chronic disorder characterized by a compulsive urge to pull out one’s hair leading to noticeable hair loss. It can be triggered by stress and has high comorbidity with anxiety disorders as well as GTS
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors declare no competing financial interests.
References
- 1.Aruga J, Mikoshiba K. Identification and characterization of Slitrk, a novel neuronal transmembrane protein family controlling neurite outgrowth. Mol Cell Neurosci. 2003;24:117–129. doi: 10.1016/s1044-7431(03)00129-5. [DOI] [PubMed] [Google Scholar]
- 2.Aruga J, Yokota N, Mikoshiba K. Human SLITRK family genes: genomic organization and expression profiling in normal brain and brain tumor tissue. Gene. 2003;315:87–94. doi: 10.1016/s0378-1119(03)00715-7. [DOI] [PubMed] [Google Scholar]
- 3.Shmelkov SV, Visser JW, Belyavsky AV. Two-dimensional gene expression fingerprinting. Anal Biochem. 2001;290:26–35. doi: 10.1006/abio.2001.4969. [DOI] [PubMed] [Google Scholar]
- 4.Kobe B, Deisenhofer J. The leucine-rich repeat: a versatile binding motif. Trends Biochem Sci. 1994;19:415–421. doi: 10.1016/0968-0004(94)90090-6. [DOI] [PubMed] [Google Scholar]
- 5.Kobe B, Kajava AV. The leucine-rich repeat as a protein recognition motif. Curr Opin Struct Biol. 2001;11:725–732. doi: 10.1016/s0959-440x(01)00266-4. [DOI] [PubMed] [Google Scholar]
- 6.Chen Y, Aulia S, Li L, Tang BL. AMIGO and friends: an emerging family of brain-enriched, neuronal growth modulating, type I transmembrane proteins with leucine-rich repeats (LRR) and cell adhesion molecule motifs. Brain Res Rev. 2006;51:265–274. doi: 10.1016/j.brainresrev.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 7.Ko J, Kim E. Leucine-rich repeat proteins of synapses. J Neurosci Res. 2007;85:2824–2832. doi: 10.1002/jnr.21306. [DOI] [PubMed] [Google Scholar]
- 8.Kidd T, Bland KS, Goodman CS. Slit is the midline repellent for the robo receptor in Drosophila. Cell. 1999;96:785–794. doi: 10.1016/s0092-8674(00)80589-9. [DOI] [PubMed] [Google Scholar]
- 9.Brose K, Bland KS, Wang KH, Arnott D, Henzel W, Goodman CS, Tessier-Lavigne M, Kidd T. Slit proteins bind Robo receptors and have an evolutionarily conserved role in repulsive axon guidance. Cell. 1999;96:795–806. doi: 10.1016/s0092-8674(00)80590-5. [DOI] [PubMed] [Google Scholar]
- 10.Brose K, Tessier-Lavigne M. Slit proteins: key regulators of axon guidance, axonal branching, and cell migration. Curr Opin Neurobiol. 2000;10:95–102. doi: 10.1016/s0959-4388(99)00066-5. [DOI] [PubMed] [Google Scholar]
- 11.Ypsilanti AR, Zagar Y, Chedotal A. Moving away from the midline: new developments for Slit and Robo. Development. 2010;137:1939–1952. doi: 10.1242/dev.044511. [DOI] [PubMed] [Google Scholar]
- 12.Kajiwara Y, Buxbaum JD, Grice DE. SLITRK1 binds 14-3-3 and regulates neurite outgrowth in a phosphorylation-dependent manner. Biol Psychiatry. 2009;66:918–925. doi: 10.1016/j.biopsych.2009.05.033. [DOI] [PubMed] [Google Scholar]
- 13.Huang EJ, Reichardt LF. Trk receptors: roles in neuronal signal transduction. Annu Rev Biochem. 2003;72:609–642. doi: 10.1146/annurev.biochem.72.121801.161629. [DOI] [PubMed] [Google Scholar]
- 14.Lee FS, Kim AH, Khursigara G, Chao MV. The uniqueness of being a neurotrophin receptor. Curr Opin Neurobiol. 2001;11:281–286. doi: 10.1016/s0959-4388(00)00209-9. [DOI] [PubMed] [Google Scholar]
- 15.Reichardt LF. Neurotrophin-regulated signalling pathways. Philos Trans R Soc Lond B Biol Sci. 2006;361:1545–1564. doi: 10.1098/rstb.2006.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohmichi M, Decker SJ, Pang L, Saltiel AR. Nerve growth factor binds to the 140 kd trk proto-oncogene product and stimulates its association with the src homology domain of phospholipase C gamma 1. Biochem Biophys Res Commun. 1991;179:217–223. doi: 10.1016/0006-291x(91)91357-i. [DOI] [PubMed] [Google Scholar]
- 17.Songyang Z, Margolis B, Chaudhuri M, Shoelson SE, Cantley LC. The phosphotyrosine interaction domain of SHC recognizes tyrosine-phosphorylated NPXY motif. J Biol Chem. 1995;270:14863–14866. doi: 10.1074/jbc.270.25.14863. [DOI] [PubMed] [Google Scholar]
- 18.Obermeier A, Lammers R, Wiesmuller KH, Jung G, Schlessinger J, Ullrich A. Identification of Trk binding sites for SHC and phosphatidylinositol 3'-kinase and formation of a multimeric signaling complex. J Biol Chem. 1993;268:22963–22966. [PubMed] [Google Scholar]
- 19.Chen WJ, Goldstein JL, Brown MS. NPXY, a sequence often found in cytoplasmic tails, is required for coated pit-mediated internalization of the low density lipoprotein receptor. J Biol Chem. 1990;265:3116–3123. [PubMed] [Google Scholar]
- 20.Obermeier A, Halfter H, Wiesmuller KH, Jung G, Schlessinger J, Ullrich A. Tyrosine 785 is a major determinant of Trk--substrate interaction. EMBO J. 1993;12:933–941. doi: 10.1002/j.1460-2075.1993.tb05734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amanchy R, Kalume DE, Iwahori A, Zhong J, Pandey A. Phosphoproteome analysis of HeLa cells using stable isotope labeling with amino acids in cell culture (SILAC) J Proteome Res. 2005;4:1661–1671. doi: 10.1021/pr050134h. [DOI] [PubMed] [Google Scholar]
- 22.Beaubien F, Cloutier JF. Differential expression of Slitrk family members in the mouse nervous system. Dev Dyn. 2009;238:3285–3296. doi: 10.1002/dvdy.22160. [DOI] [PubMed] [Google Scholar]
- 23.Aruga J. Slitrk6 expression profile in the mouse embryo and its relationship to that of Nlrr3. Gene Expr Patterns. 2003;3:727–733. doi: 10.1016/s1567-133x(03)00141-8. [DOI] [PubMed] [Google Scholar]
- 24.Stillman AA, Krsnik Z, Sun J, Rasin MR, State MW, Sestan N, Louvi A. Developmentally regulated and evolutionarily conserved expression of SLITRK1 in brain circuits implicated in Tourette syndrome. J Comp Neurol. 2009;513:21–37. doi: 10.1002/cne.21919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Milde T, Shmelkov SV, Jensen KK, Zlotchenko G, Petit I, Rafii S. A novel family of slitrk genes is expressed on hematopoietic stem cells and leukemias. Leukemia. 2007;21:824–827. doi: 10.1038/sj.leu.2404525. [DOI] [PubMed] [Google Scholar]
- 26.Katayama K, Zine A, Ota M, Matsumoto Y, Inoue T, Fritzsch B, Aruga J. Disorganized innervation and neuronal loss in the inner ear of Slitrk6-deficient mice. PLoS One. 2009;4:e7786. doi: 10.1371/journal.pone.0007786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abelson JF, Kwan KY, O'Roak BJ, Baek DY, Stillman AA, Morgan TM, Mathews CA, Pauls DL, Rasin MR, Gunel M, et al. Sequence variants in SLITRK1 are associated with Tourette’s syndrome. Science. 2005;310:317–320. doi: 10.1126/science.1116502. [DOI] [PubMed] [Google Scholar]
- 28.Shmelkov SV, Hormigo A, Jing D, Proenca CC, Bath KG, Milde T, Shmelkov E, Kushner JS, Baljevic M, Dincheva I, et al. Slitrk5 deficiency impairs corticostriatal circuitry and leads to obsessive-compulsive-like behaviors in mice. Nat Med. 2010;16:598–602. doi: 10.1038/nm.2125. 591p following 602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linhoff MW, Lauren J, Cassidy RM, Dobie FA, Takahashi H, Nygaard HB, Airaksinen MS, Strittmatter SM, Craig AM. An unbiased expression screen for synaptogenic proteins identifies the LRRTM protein family as synaptic organizers. Neuron. 2009;61:734–749. doi: 10.1016/j.neuron.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bridges D, Moorhead GB. 14-3-3 proteins: a number of functions for a numbered protein. Sci STKE. 2005;2005 doi: 10.1126/stke.2962005re10. re10. [DOI] [PubMed] [Google Scholar]
- 31.Mandai K, Guo T, St Hillaire C, Meabon JS, Kanning KC, Bothwell M, Ginty DD. LIG family receptor tyrosine kinase-associated proteins modulate growth factor signals during neural development. Neuron. 2009;63:614–627. doi: 10.1016/j.neuron.2009.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lauren J, Airaksinen MS, Saarma M, Timmusk T. A novel gene family encoding leucine-rich repeat transmembrane proteins differentially expressed in the nervous system. Genomics. 2003;81:411–421. doi: 10.1016/s0888-7543(03)00030-2. [DOI] [PubMed] [Google Scholar]
- 33.de Wit J, Sylwestrak E, O'Sullivan ML, Otto S, Tiglio K, Savas JN, Yates JR, 3rd, Comoletti D, Taylor P, Ghosh A. LRRTM2 interacts with Neurexin1 and regulates excitatory synapse formation. Neuron. 2009;64:799–806. doi: 10.1016/j.neuron.2009.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ko J, Fuccillo MV, Malenka RC, Sudhof TC. LRRTM2 functions as a neurexin ligand in promoting excitatory synapse formation. Neuron. 2009;64:791–798. doi: 10.1016/j.neuron.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woo J, Kwon SK, Choi S, Kim S, Lee JR, Dunah AW, Sheng M, Kim E. Trans-synaptic adhesion between NGL-3 and LAR regulates the formation of excitatory synapses. Nat Neurosci. 2009;12:428–437. doi: 10.1038/nn.2279. [DOI] [PubMed] [Google Scholar]
- 36.Kim S, Burette A, Chung HS, Kwon SK, Woo J, Lee HW, Kim K, Kim H, Weinberg RJ, Kim E. NGL family PSD-95-interacting adhesion molecules regulate excitatory synapse formation. Nat Neurosci. 2006;9:1294–1301. doi: 10.1038/nn1763. [DOI] [PubMed] [Google Scholar]
- 37.Kwon SK, Woo J, Kim SY, Kim H, Kim E. Trans-synaptic adhesions between netrin-G ligand-3 (NGL-3) and receptor tyrosine phosphatases LAR, protein-tyrosine phosphatase delta (PTPdelta), and PTPsigma via specific domains regulate excitatory synapse formation. J Biol Chem. 2010;285:13966–13978. doi: 10.1074/jbc.M109.061127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gill M, Donohoe G, Corvin A. What have the genomics ever done for the psychoses? Psychol Med. 2010;40:529–540. doi: 10.1017/S0033291709991139. [DOI] [PubMed] [Google Scholar]
- 39.Kirov G, Gumus D, Chen W, Norton N, Georgieva L, Sari M, O'Donovan MC, Erdogan F, Owen MJ, Ropers HH, et al. Comparative genome hybridization suggests a role for NRXN1 and APBA2 in schizophrenia. Hum Mol Genet. 2008;17:458–465. doi: 10.1093/hmg/ddm323. [DOI] [PubMed] [Google Scholar]
- 40.Pauls DL. The genetics of obsessive compulsive disorder and Gilles de la Tourette's syndrome. Psychiatr Clin North Am. 1992;15:759–766. [PubMed] [Google Scholar]
- 41.Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 42.Roberts R, Wells GA, Stewart AF, Dandona S, Chen L. The genome-wide association study--a new era for common polygenic disorders. J Cardiovasc Transl Res. 2010;3:173–182. doi: 10.1007/s12265-010-9178-6. [DOI] [PubMed] [Google Scholar]
- 43.Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet. 2008;40:695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gauthier J, Spiegelman D, Piton A, Lafreniere RG, Laurent S, St-Onge J, Lapointe L, Hamdan FF, Cossette P, Mottron L, et al. Novel de novo SHANK3 mutation in autistic patients. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:421–424. doi: 10.1002/ajmg.b.30822. [DOI] [PubMed] [Google Scholar]
- 45.Jamain S, Quach H, Betancur C, Rastam M, Colineaux C, Gillberg IC, Soderstrom H, Giros B, Leboyer M, Gillberg C, et al. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, Nygren G, Rastam M, Gillberg IC, Anckarsater H, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moessner R, Marshall CR, Sutcliffe JS, Skaug J, Pinto D, Vincent J, Zwaigenbaum L, Fernandez B, Roberts W, Szatmari P, et al. Contribution of SHANK3 mutations to autism spectrum disorder. Am J Hum Genet. 2007;81:1289–1297. doi: 10.1086/522590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laumonnier F, Bonnet-Brilhault F, Gomot M, Blanc R, David A, Moizard MP, Raynaud M, Ronce N, Lemonnier E, Calvas P, et al. X-linked mental retardation and autism are associated with a mutation in the NLGN4 gene, a member of the neuroligin family. Am J Hum Genet. 2004;74:552–557. doi: 10.1086/382137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hornsey H, Banerjee S, Zeitlin H, Robertson M. The prevalence of Tourette syndrome in 13-14-year-olds in mainstream schools. J Child Psychol Psychiatry. 2001;42:1035–1039. doi: 10.1111/1469-7610.00802. [DOI] [PubMed] [Google Scholar]
- 50.Robertson MM. Diagnosing Tourette syndrome: is it a common disorder? J Psychosom Res. 2003;55:3–6. doi: 10.1016/s0022-3999(02)00580-9. [DOI] [PubMed] [Google Scholar]
- 51.Lombroso PJ, Scahill L. Tourette syndrome and obsessive-compulsive disorder. Brain Dev. 2008;30:231–237. doi: 10.1016/j.braindev.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O'Rourke JA, Scharf JM, Yu D, Pauls DL. The genetics of Tourette syndrome: a review. J Psychosom Res. 2009;67:533–545. doi: 10.1016/j.jpsychores.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carter AS, O'Donnell DA, Schultz RT, Scahill L, Leckman JF, Pauls DL. Social and emotional adjustment in children affected with Gilles de la Tourette's syndrome: associations with ADHD and family functioning. Attention Deficit Hyperactivity Disorder. J Child Psychol Psychiatry. 2000;41:215–223. [PubMed] [Google Scholar]
- 54.State MW. The genetics of child psychiatric disorders: focus on autism and Tourette syndrome. Neuron. 2010;68:254–269. doi: 10.1016/j.neuron.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Albin RL, Mink JW. Recent advances in Tourette syndrome research. Trends Neurosci. 2006;29:175–182. doi: 10.1016/j.tins.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 56.Modell JG, Mountz JM, Curtis GC, Greden JF. Neurophysiologic dysfunction in basal ganglia/limbic striatal and thalamocortical circuits as a pathogenetic mechanism of obsessive-compulsive disorder. J Neuropsychiatry Clin Neurosci. 1989;1:27–36. doi: 10.1176/jnp.1.1.27. [DOI] [PubMed] [Google Scholar]
- 57.Cummings JL. Frontal-subcortical circuits and human behavior. Arch Neurol. 1993;50:873–880. doi: 10.1001/archneur.1993.00540080076020. [DOI] [PubMed] [Google Scholar]
- 58.Deng H, Le WD, Xie WJ, Jankovic J. Examination of the SLITRK1 gene in Caucasian patients with Tourette syndrome. Acta Neurol Scand. 2006;114:400–402. doi: 10.1111/j.1600-0404.2006.00706.x. [DOI] [PubMed] [Google Scholar]
- 59.Wendland JR, Kruse MR, Murphy DL. Functional SLITRK1 var321, varCDfs and SLC6A4 G56A variants and susceptibility to obsessive-compulsive disorder. Mol Psychiatry. 2006;11:802–804. doi: 10.1038/sj.mp.4001848. [DOI] [PubMed] [Google Scholar]
- 60.Keen-Kim D, Mathews CA, Reus VI, Lowe TL, Herrera LD, Budman CL, Gross-Tsur V, Pulver AE, Bruun RD, Erenberg G, et al. Overrepresentation of rare variants in a specific ethnic group may confuse interpretation of association analyses. Hum Mol Genet. 2006;15:3324–3328. doi: 10.1093/hmg/ddl408. [DOI] [PubMed] [Google Scholar]
- 61.Scharf JM, Moorjani P, Fagerness J, Platko JV, Illmann C, Galloway B, Jenike E, Stewart SE, Pauls DL. Lack of association between SLITRK1var321 and Tourette syndrome in a large family-based sample. Neurology. 2008;70:1495–1496. doi: 10.1212/01.wnl.0000296833.25484.bb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Verkerk AJ, Cath DC, van der Linde HC, Both J, Heutink P, Breedveld G, Aulchenko YS, Oostra BA. Genetic and clinical analysis of a large Dutch Gilles de la Tourette family. Mol Psychiatry. 2006;11:954–964. doi: 10.1038/sj.mp.4001877. [DOI] [PubMed] [Google Scholar]
- 63.O'Roak BJ, Morgan TM, Fishman DO, Saus E, Alonso P, Gratacos M, Estivill X, Teltsh O, Kohn Y, Kidd KK, et al. Additional support for the association of SLITRK1 var321 and Tourette syndrome. Mol Psychiatry. 2010;15:447–450. doi: 10.1038/mp.2009.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miranda DM, Wigg K, Kabia EM, Feng Y, Sandor P, Barr CL. Association of SLITRK1 to Gilles de la Tourette Syndrome. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:483–486. doi: 10.1002/ajmg.b.30840. [DOI] [PubMed] [Google Scholar]
- 65.Zuchner S, Cuccaro ML, Tran-Viet KN, Cope H, Krishnan RR, Pericak-Vance MA, Wright HH, Ashley-Koch A. SLITRK1 mutations in trichotillomania. Mol Psychiatry. 2006;11:887–889. doi: 10.1038/sj.mp.4001898. [DOI] [PubMed] [Google Scholar]
- 66.Zimprich A, Hatala K, Riederer F, Stogmann E, Aschauer HN, Stamenkovic M. Sequence analysis of the complete SLITRK1 gene in Austrian patients with Tourette's disorder. Psychiatr Genet. 2008;18:308–309. doi: 10.1097/YPG.0b013e3283060f6f. [DOI] [PubMed] [Google Scholar]
- 67.Swedo SE, Leonard HL. Trichotillomania An obsessive compulsive spectrum disorder? Psychiatr Clin North Am. 1992;15:777–790. [PubMed] [Google Scholar]
- 68.Ferrao YA, Miguel E, Stein DJ. Tourette’s syndrome, trichotillomania, and obsessive-compulsive disorder: how closely are they related? Psychiatry Res. 2009;170:32–42. doi: 10.1016/j.psychres.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 69.Katayama K, Yamada K, Ornthanalai VG, Inoue T, Ota M, Murphy NP, Aruga J. Slitrk1-deficient mice display elevated anxiety-like behavior and noradrenergic abnormalities. Mol Psychiatry. 2010;15:177–184. doi: 10.1038/mp.2008.97. [DOI] [PubMed] [Google Scholar]
- 70.Leckman JF. Tourette's syndrome. Lancet. 2002;360:1577–1586. doi: 10.1016/S0140-6736(02)11526-1. [DOI] [PubMed] [Google Scholar]
- 71.Leckman JF, Hardin MT, Riddle MA, Stevenson J, Ort SI, Cohen DJ. Clonidine treatment of Gilles de la Tourette's syndrome. Arch Gen Psychiatry. 1991;48:324–328. doi: 10.1001/archpsyc.1991.01810280040006. [DOI] [PubMed] [Google Scholar]
- 72.Shapiro AK, Shapiro E, Eisenkraft GJ. Treatment of Gilles de la Tourette's syndrome with clonidine and neuroleptics. Arch Gen Psychiatry. 1983;40:1235–1240. doi: 10.1001/archpsyc.1983.01790100081011. [DOI] [PubMed] [Google Scholar]
- 73.Hedderick EF, Morris CM, Singer HS. Double-blind, crossover study of clonidine and levetiracetam in Tourette syndrome. Pediatr Neurol. 2009;40:420–425. doi: 10.1016/j.pediatrneurol.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 74.Rauch SL, Dougherty DD, Malone D, Rezai A, Friehs G, Fischman AJ, Alpert NM, Haber SN, Stypulkowski PH, Rise MT, et al. A functional neuroimaging investigation of deep brain stimulation in patients with obsessive-compulsive disorder. J Neurosurg. 2006;104:558–565. doi: 10.3171/jns.2006.104.4.558. [DOI] [PubMed] [Google Scholar]
- 75.Mataix-Cols D, Rauch SL, Manzo PA, Jenike MA, Baer L. Use of factor-analyzed symptom dimensions to predict outcome with serotonin reuptake inhibitors and placebo in the treatment of obsessive-compulsive disorder. Am J Psychiatry. 1999;156:1409–1416. doi: 10.1176/ajp.156.9.1409. [DOI] [PubMed] [Google Scholar]
- 76.McClung CA, Ulery PG, Perrotti LI, Zachariou V, Berton O, Nestler EJ. DeltaFosB: a molecular switch for long-term adaptation in the brain. Brain Res Mol Brain Res. 2004;132:146–154. doi: 10.1016/j.molbrainres.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 77.Menzies L, Chamberlain SR, Laird AR, Thelen SM, Sahakian BJ, Bullmore ET. Integrating evidence from neuroimaging and neuropsychological studies of obsessive-compulsive disorder: the orbitofronto-striatal model revisited. Neurosci Biobehav Rev. 2008;32:525–549. doi: 10.1016/j.neubiorev.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Whiteside SP, Port JD, Abramowitz JS. A meta-analysis of functional neuroimaging in obsessive-compulsive disorder. Psychiatry Res. 2004;132:69–79. doi: 10.1016/j.pscychresns.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 79.Saxena S, Rauch SL. Functional neuroimaging and the neuroanatomy of obsessive-compulsive disorder. Psychiatr Clin North Am. 2000;23:563–586. doi: 10.1016/s0193-953x(05)70181-7. [DOI] [PubMed] [Google Scholar]
- 80.Rauch SL, Wedig MM, Wright CI, Martis B, McMullin KG, Shin LM, Cannistraro PA, Wilhelm S. Functional magnetic resonance imaging study of regional brain activation during implicit sequence learning in obsessive-compulsive disorder. Biol Psychiatry. 2007;61:330–336. doi: 10.1016/j.biopsych.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 81.Rauch SL. Neuroimaging and neurocircuitry models pertaining to the neurosurgical treatment of psychiatric disorders. Neurosurg Clin N Am. 2003;14:213–223. doi: 10.1016/s1042-3680(02)00114-6. vii–viii. [DOI] [PubMed] [Google Scholar]
- 82.Welch JM, Lu J, Rodriguiz RM, Trotta NC, Peca J, Ding JD, Feliciano C, Chen M, Adams JP, Luo J, et al. Cortico-striatal synaptic defects and OCD-like behaviours in Sapap3-mutant mice. Nature. 2007;448:894–900. doi: 10.1038/nature06104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim E, Naisbitt S, Hsueh YP, Rao A, Rothschild A, Craig AM, Sheng M. GKAP, a novel synaptic protein that interacts with the guanylate kinase-like domain of the PSD-95/SAP90 family of channel clustering molecules. J Cell Biol. 1997;136:669–678. doi: 10.1083/jcb.136.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Takeuchi M, Hata Y, Hirao K, Toyoda A, Irie M, Takai Y. SAPAPs. A family of PSD-95/SAP90-associated proteins localized at postsynaptic density. J Biol Chem. 1997;272:11943–11951. doi: 10.1074/jbc.272.18.11943. [DOI] [PubMed] [Google Scholar]
- 85.Crane J, Fagerness J, Osiecki L, Gunnell B, Stewart SE, Pauls DL, Scharf JM. Family-based genetic association study of DLGAP3 in Tourette Syndrome. Am J Med Genet B Neuropsychiatr Genet. 2010 doi: 10.1002/ajmg.b.31134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zuchner S, Wendland JR, Ashley-Koch AE, Collins AL, Tran-Viet KN, Quinn K, Timpano KC, Cuccaro ML, Pericak-Vance MA, Steffens DC, et al. Multiple rare SAPAP3 missense variants in trichotillomania and OCD. Mol Psychiatry. 2009;14:6–9. doi: 10.1038/mp.2008.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bienvenu OJ, Wang Y, Shugart YY, Welch JM, Grados MA, Fyer AJ, Rauch SL, McCracken JT, Rasmussen SA, Murphy DL, et al. Sapap3 and pathological grooming in humans: Results from the OCD collaborative genetics study. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:710–720. doi: 10.1002/ajmg.b.30897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang L, Simpson HB, Dulawa SC. Assessing the validity of current mouse genetic models of obsessive-compulsive disorder. Behav Pharmacol. 2009;20:119–133. doi: 10.1097/FBP.0b013e32832a80ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ting JT, Feng G. Glutamatergic Synaptic Dysfunction and Obsessive-Compulsive Disorder. Curr Chem Genomics. 2008;2:62–75. doi: 10.2174/1875397300802010062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Holstege JC, de Graaff W, Hossaini M, Cano SC, Jaarsma D, van den Akker E, Deschamps J. Loss of Hoxb8 alters spinal dorsal laminae and sensory responses in mice. Proc Natl Acad Sci U S A. 2008;105:6338–6343. doi: 10.1073/pnas.0802176105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen SK, Tvrdik P, Peden E, Cho S, Wu S, Spangrude G, Capecchi MR. Hematopoietic origin of pathological grooming in Hoxb8 mutant mice. Cell. 2010;141:775–785. doi: 10.1016/j.cell.2010.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Greer JM, Capecchi MR. Hoxb8 is required for normal grooming behavior in mice. Neuron. 2002;33:23–34. doi: 10.1016/s0896-6273(01)00564-5. [DOI] [PubMed] [Google Scholar]
- 93.van den Akker E, Reijnen M, Korving J, Brouwer A, Meijlink F, Deschamps J. Targeted inactivation of Hoxb8 affects survival of a spinal ganglion and causes aberrant limb reflexes. Mech Dev. 1999;89:103–114. doi: 10.1016/s0925-4773(99)00212-9. [DOI] [PubMed] [Google Scholar]
- 94.Kaur C, Hao AJ, Wu CH, Ling EA. Origin of microglia. Microsc Res Tech. 2001;54:2–9. doi: 10.1002/jemt.1114. [DOI] [PubMed] [Google Scholar]
- 95.Ransohoff RM, Perry VH. Microglial physiology: unique stimuli, specialized responses. Annu Rev Immunol. 2009;27:119–145. doi: 10.1146/annurev.immunol.021908.132528. [DOI] [PubMed] [Google Scholar]
- 96.Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, Conway SJ, Ng LG, Stanley ER, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Piton A, Gauthier J, Hamdan FF, Lafreniere RG, Yang Y, Henrion E, Laurent S, Noreau A, Thibodeau P, Karemera L, et al. Systematic resequencing of X-chromosome synaptic genes in autism spectrum disorder and schizophrenia. Mol Psychiatry. 2010 doi: 10.1038/mp.2010.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Burton A. SLITRK1 trouble in Tourette's syndrome. Lancet Neurol. 2005;4:801. doi: 10.1016/s1474-4422(05)70242-8. [DOI] [PubMed] [Google Scholar]
- 99.Kuja-Panula J, Kiiltomaki M, Yamashiro T, Rouhiainen A, Rauvala H. AMIGO, a transmembrane protein implicated in axon tract development, defines a novel protein family with leucine-rich repeats. J Cell Biol. 2003;160:963–973. doi: 10.1083/jcb.200209074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Robinson M, Parsons Perez MC, Tebar L, Palmer J, Patel A, Marks D, Sheasby A, De Felipe C, Coffin R, Livesey FJ, et al. FLRT3 is expressed in sensory neurons after peripheral nerve injury and regulates neurite outgrowth. Mol Cell Neurosci. 2004;27:202–214. doi: 10.1016/j.mcn.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 101.Mi S, Lee X, Shao Z, Thill G, Ji B, Relton J, Levesque M, Allaire N, Perrin S, Sands B, et al. LINGO-1 is a component of the Nogo-66 receptor/p75 signaling complex. Nat Neurosci. 2004;7:221–228. doi: 10.1038/nn1188. [DOI] [PubMed] [Google Scholar]
- 102.Inoue H, Lin L, Lee X, Shao Z, Mendes S, Snodgrass-Belt P, Sweigard H, Engber T, Pepinsky B, Yang L, et al. Inhibition of the leucine-rich repeat protein LINGO-1 enhances survival, structure, and function of dopaminergic neurons in Parkinson's disease models. Proc Natl Acad Sci U S A. 2007;104:14430–14435. doi: 10.1073/pnas.0700901104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mi S, Sandrock A, Miller RH. LINGO-1 and its role in CNS repair. Int J Biochem Cell Biol. 2008;40:1971–1978. doi: 10.1016/j.biocel.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 104.Mosyak L, Wood A, Dwyer B, Buddha M, Johnson M, Aulabaugh A, Zhong X, Presman E, Benard S, Kelleher K, et al. The structure of the Lingo-1 ectodomain, a module implicated in central nervous system repair inhibition. J Biol Chem. 2006;281:36378–36390. doi: 10.1074/jbc.M607314200. [DOI] [PubMed] [Google Scholar]
- 105.Gur G, Rubin C, Katz M, Amit I, Citri A, Nilsson J, Amariglio N, Henriksson R, Rechavi G, Hedman H, et al. LRIG1 restricts growth factor signaling by enhancing receptor ubiquitylation and degradation. EMBO J. 2004;23:3270–3281. doi: 10.1038/sj.emboj.7600342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Laederich MB, Funes-Duran M, Yen L, Ingalla E, Wu X, Carraway KL, 3rd, Sweeney C. The leucine-rich repeat protein LRIG1 is a negative regulator of ErbB family receptor tyrosine kinases. J Biol Chem. 2004;279:47050–47056. doi: 10.1074/jbc.M409703200. [DOI] [PubMed] [Google Scholar]
- 107.Ledda F, Bieraugel O, Fard SS, Vilar M, Paratcha G. Lrig1 is an endogenous inhibitor of Ret receptor tyrosine kinase activation, downstream signaling, and biological responses to GDNF. J Neurosci. 2008;28:39–49. doi: 10.1523/JNEUROSCI.2196-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shattuck DL, Miller JK, Laederich M, Funes M, Petersen H, Carraway KL, 3rd, Sweeney C. LRIG1 is a novel negative regulator of the Met receptor and opposes Met and Her2 synergy. Mol Cell Biol. 2007;27:1934–1946. doi: 10.1128/MCB.00757-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhao H, Tanegashima K, Ro H, Dawid IB. Lrig3 regulates neural crest formation in Xenopus by modulating Fgf and Wnt signaling pathways. Development. 2008;135:1283–1293. doi: 10.1242/dev.015073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rajasekharan S, Kennedy TE. The netrin protein family. Genome Biol. 2009;10:239. doi: 10.1186/gb-2009-10-9-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lin JC, Ho WH, Gurney A, Rosenthal A. The netrin-G1 ligand NGL-1 promotes the outgrowth of thalamocortical axons. Nat Neurosci. 2003;6:1270–1276. doi: 10.1038/nn1148. [DOI] [PubMed] [Google Scholar]
- 112.Wang KC, Koprivica V, Kim JA, Sivasankaran R, Guo Y, Neve RL, He Z. Oligodendrocyte-myelin glycoprotein is a Nogo receptor ligand that inhibits neurite outgrowth. Nature. 2002;417:941–944. doi: 10.1038/nature00867. [DOI] [PubMed] [Google Scholar]
- 113.Wang PY, Seabold GK, Wenthold RJ. Synaptic adhesion-like molecules (SALMs) promote neurite outgrowth. Mol Cell Neurosci. 2008;39:83–94. doi: 10.1016/j.mcn.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang CY, Chang K, Petralia RS, Wang YX, Seabold GK, Wenthold RJ. A novel family of adhesion-like molecules that interacts with the NMDA receptor. J Neurosci. 2006;26:2174–2183. doi: 10.1523/JNEUROSCI.3799-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Seabold GK, Wang PY, Chang K, Wang CY, Wang YX, Petralia RS, Wenthold RJ. The SALM family of adhesion-like molecules forms heteromeric and homomeric complexes. J Biol Chem. 2008;283:8395–8405. doi: 10.1074/jbc.M709456200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mah W, Ko J, Nam J, Han K, Chung WS, Kim E. Selected SALM (synaptic adhesion-like molecule) family proteins regulate synapse formation. J Neurosci. 2010;30:5559–5568. doi: 10.1523/JNEUROSCI.4839-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]