Abstract
Background
We are interested in identifying molecular markers that can aid in the diagnosis of adrenocortical carcinoma (ACC). The aim of this study was to identify microRNAs (miRNAs or miRs) that are differentially expressed in malignant adrenocortical tumors as compared to benign tumors and assess their potential as diagnostic predictors.
Methods
Differentially expressed miRNAs were identified using microarray profiling of adrenocortical tumors and validated by quantitative real-time RT-PCR.
Results
Microarray profiling in benign and primary malignant adrenocortical tumors revealed a number of significant differences between these histological groups. Using directed quantitative RT-PCR analysis on a subset of these differentially expressed miRNAs, we determined that miRs −100, −125b, and −195 were significantly downregulated and miR-483-5p was significantly upregulated in malignant as compared to benign tumors. Furthermore, our work shows that miR-483-5p expression can accurately categorize tumors as benign or malignant.
Conclusions
We identified four miRNAs that are dysregulated in adrenocortical carcinoma. The high expression of one of these, miR-483-5p, appears to be a defining characteristic of adrenocortical malignancies and can be used to accurately distinguish between benign and malignant adrenocortical tumors.
Keywords: adrenocortical carcinoma, adrenal, microRNA, diagnostic marker
Introduction
Adrenocortical carcinoma (ACC) is a rare but aggressive malignancy of the adrenal cortex. This cancer affects 1 to 2 people per million per year and accounts for 0.02–0.2% of all cancer deaths 1–3. Approximately half of all patients have metastatic disease at the time of diagnosis resulting in an average five-year survival of less than 10% 1, 3. Currently there is limited knowledge regarding the initiation and pathophysiology of ACC and a lack of effective therapies to treat this disease.
Benign adrenocortical tumors are a much more common occurrence with approximately 5% of people over 50 years old having at least one nodule 4. These tumors often share many imaging characteristics with their ACC counterparts and therefore determining if a tumor is benign or malignant is not always straightforward. Metastatic disease or local invasion is the only absolute indicator of malignancy. Masses without these features are assessed preoperatively based on size, and imaging characteristics, although the findings of these studies often are unable to definitively categorize the tumor as benign or malignant. After resection, tumor pathology is assessed based on several histologic criteria including cell morphology, cellular proliferation, and tumor invasiveness (Weiss criteria, 5). However for some tumors, with some suspicious features, a definitive diagnosis may not be possible. Accurate diagnosis is critical since the prognosis, follow up, and therapeutic strategy for ACC is much different than that for a benign tumor. Therefore there is a need for better diagnostic tools for assessing adrenocortical tumors, preoperatively and as an adjunct to routine histopathology.
MicroRNAs (miRNAs or miRs) are a class of short non-coding RNAs that post-transcriptionally regulate gene expression by directly targeting mRNAs and affecting their stability and/or translation (reviewed in 6). These regulators are important in a wide range of normal physiologic and pathologic processes. MiRNA expression profiling has been performed on many types of human cancers and indicates that malignancy involves miRNA dysregulation and that particular miRNA expression signatures could be useful for molecular diagnosis and/or prognosis. Additionally, functional studies suggest that aberrant miRNA expression contributes to cancer pathogenesis making these molecules potential targets for cancer therapy.
Given the potential of miRNAs as cancer markers and as targets for molecular therapy, we used genome-wide miRNA expression profiling of adrenocortical tumors (26 benign and 10 malignant) to identify a number of miRNAs that were dysregulated in ACC. Here we report a signature of low miR-100, −125b, and −195 and high miR-483-5p expression that is characteristic of ACC. Moreover, we found that the expression level of miR-483-5p alone can accurately diagnose a tumor as benign or malignant and validated this finding in an independent set of adrenocortical tumors (35 benign and 31 malignant).
Materials and Methods
Tissue samples
Adrenocortical tissue samples were procured at the time of surgery, snap frozen in liquid nitrogen, and stored at −80°C. Demographic, clinical, and pathologic information and tissue samples were collected under an Institutional Review Board (IRB) approved protocol. Patient and tumor characteristics used for miRNA microarray profiling are summarized in Table 1. Tumors were classified as adrenocortical carcinoma when gross local invasion and/or metastasis (lymph or distant) was present at the time of diagnosis or occurred during follow-up. Localized tumors with no evidence of recurrence at follow-up (mean 2.1 years) were classified as benign. An independent set of benign (n=35), locoregional ACC recurrences (n=2), and ACC metastases (n=29) tumor samples were used for validation.
Table 1.
Clinical Features of Microarray Samples
| Adrenocortical Carcinoma |
Benign Adrenocortical Tumor |
|
|---|---|---|
| Number of Patients | 10 | 26 |
| Age (Ave. ± St. Dev.) | 45.1 ± 21.3 | 50.1 ± 13.2 |
| Sex (Female/Male) | 7/3 | 19/7 |
| Syndrome: | ||
| Cushing’s | 7 | 8 |
| Subclinical Cushing’s | 0 | 2 |
| Conn’s | 0 | 4 |
| Nonfunctioning | 3 | 12 |
Normal adrenal glands were procured from healthy organ donors under an IRB approved protocol (n=21). Laser capture microdissection was used to isolate tissue from the cortex.
RNA extraction and quality control
Total RNA was extracted from frozen tissue as described previously 7. RNA integrity and quality was confirmed using an Agilent 2100 Bioanalyzer. All RNA samples used for miRNA profiling had a RNA integrity number > 5.
miRNA microarray expression profiling and data analysis
A total of 36 microarrays were run comparing either benign adrenocortical tumors (n=26) or ACCs (n=10) to a common reference pool of 21 normal adrenal cortices. For each array, 300 ng of total RNA (either tumor or reference) was labeled with Cy5 or Cy3 using the miRCURY LNA microRNA Array Labeling Kit (Exiqon, Denmark). Cy5 and Cy3 samples were combined such that all tumor samples were compared to normal pooled. In 12 samples, the reference normal pooled sample was labeled with Cy5 (dye swap). Fluorescently labeled RNA was hybridized to Exiqon miRCURY LNA miRNA arrays (v. 11.0) using SureHyb DNA microarray hybridization chambers and gasket slides (Agilent, Santa Clara, CA) for 18 hours at 56°C. Arrays were scanned on an Axon GenePix 4000B scanner (Molecular Devices, Sunnyvale, CA), and GenePix results files (GPR) containing fluorescence intensities were generated using GenePix Pro 6.0 software.
GPR files were loaded into R/Bioconductor using the marray package 8. Flagged spots were removed from subsequent analysis and the remaining probes were used for normalization and subsequent analyses. The log2 ratio of the intensity of Cy5 to Cy3 signals were calculated for each miRNA on every array (with no background subtraction) and normalized by print tip loess normalization 9, 10. Since individual miRNAs were represented by four probes on the array, the median of normalized log2 ratio of the replicate probes (for those with more than one unflagged probe) was used as the value for the miRNA. The summarized log2 ratios for each experiment were then used in moderated t-statistics and p-value calculation using the limma package in R/Bioconductor 11, 12 with adjustment for false discovery rate using the Benjamini-Hochberg method 13.
Real-Time quantitative RT-PCR analysis
MiRNAs that were found to be differentially expressed in the microarray experiments were validated using TaqMan quantitative real-time RT-PCR (Applied Biosystems, Foster City, CA). Single-stranded cDNA was synthesized from 5ng of total RNA using specific miRNA primers (TaqMan MicroRNA Assay, PN 4427975, Applied Biosystems) and the TaqMan MicroRNA Reverse Transcription Kit (PN 4366596, Applied Biosystems). 2ul of cDNA was used as a template in a 10ul PCR reaction. PCR products were amplified using specific primers (TaqMan MicroRNA Assay) and the TaqMan Universal PCR Master Mix (PN 4324018, Applied Biosystems) and detected using 7900HT Fast Real-Time PCR System (Applied Biosystems). PCR reactions for each sample were run in triplicate. Control reactions included cDNA synthesized without reverse transcriptase enzyme (RNA only) and no cDNA template. The following TaqMan MicroRNA Assays used in this study were obtained from Applied Biosystems: let-7g (002282), miR-26b (000407), miR-483-5p (002338), miR-214 (002306), miR-195 (000494), miR-193b (002367), miR-126 (002228), miR-125b (000449), miR-125a-5p (002198), miR-30b (000602), miR-34a (000426), and miR-100 (000437). RNU48 (001006), RNU6b (001093), U6 (001973), miR-34c (000428), miR-542-3p (001284), and miR-1285 (002822) were tested as possible endogenous controls for data normalization by measuring their expression in all of the samples. The prediction algorithm NormFinder was used to analyze expression stability of the possible controls 14.
For measuring IGF2 expression in patient samples, single-stranded cDNA was synthesized from 100ng of total RNA. TaqMan Real-time quantitative PCR was used to measure IGF2 mRNA expression level relative to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA expression. The TaqMan probes for the IGF2 (Hs01005970_m1) and GAPDH (Hs99999905_m1) were obtained from Applied Biosystems. All the PCR was performed in a final volume of 10 µL, with 2 µL of cDNA template, using TaqMan Universal Master Mix (PN 4440043, Applied Biosystems) on a 7900HT Fast Real-Time PCR System (Applied Biosystems).
MiRNA expression level was expressed as the difference between cycle threshold (Ct) for the miRNA of interest and that of RNU48 (ΔCt). IGF2 mRNA expression level was expressed as the difference between cycle threshold (Ct) for IGF2 and that of GAPDH (ΔCt). Since the sample sizes were small, the Mann-Whitney U test was used to assess statistical significance. A p-value of less than 0.05 was considered statistically significant.
Results
Identification of differentially expressed miRNAs
The global miRNA profiles were obtained for the samples listed in Table 1 which included 10 primary adrenocortical carcinomas (ACCs) and 26 benign adrenocortical tumors. A total of 36 microarrays were performed comparing the total RNA from each individual tumor sample to a common reference pool of RNA from 21 normal adrenal cortices. All 36 arrays were of adequate quality for analysis.
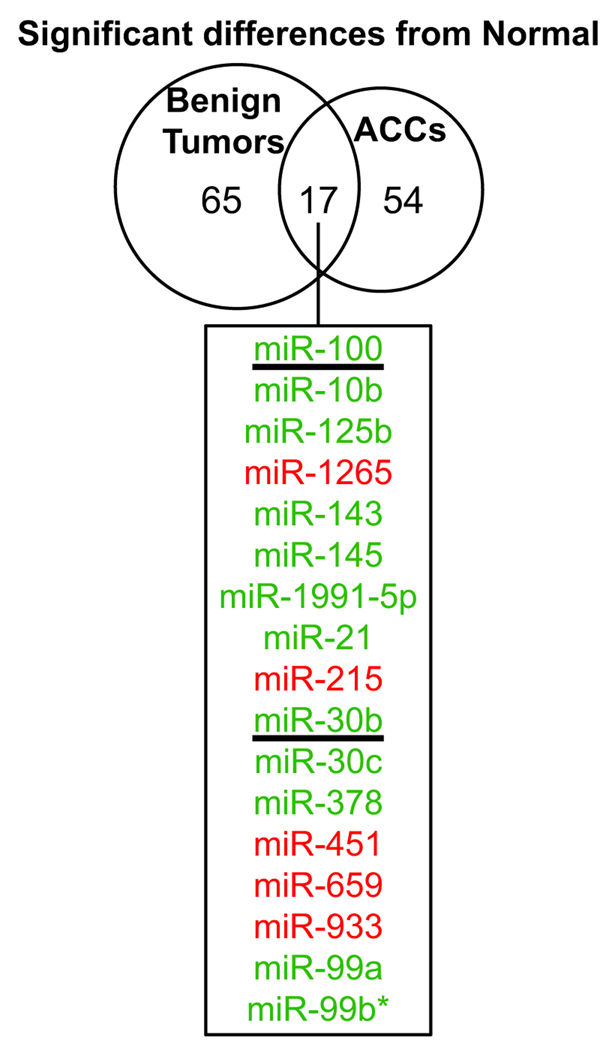
We first analyzed the difference between tumors and normal adrenocortical tissue. Differentially expressed miRNAs were defined as those that had an adjusted p-value of less than 0.01 as described in the Materials and Methods. There were a similar number of differentially expressed human miRNAs in both benign and malignant tumors as compared to normal (82 and 71, respectively) (Fig. 1). Interestingly, among these, only 17 miRNAs were differentially expressed in both comparisons suggesting that benign and malignant tumors have relatively distinct patterns of miRNA dysregulation. For the majority of the 17 common miRNAs, the fold change in expression was more dramatic in ACC. For example, in comparison to normal tissue, miR-100 was downregulated 1.5-fold in benign tumors whereas in malignant tumors it was 2.6-fold lower. It is possible that these common miRNAs are involved in neoplastic proliferation in the adrenal cortex. In fact, miR-100 was recently shown to regulate Polo-like kinase 1 (Plk1), a critical regulator of mitosis 15.
Figure 1. The miRNAs significantly up- or down-regulated in malignant and/or benign adrenocortical tumors as compared to normal adrenocortical tissue.
Microarray analysis compared tumors (benign or malignant) to normal adrenocortical tissue. Differentially expressed genes were defined as those that had a p-value <0.01 (Ho: there is no difference between expression in tumor and normal). 17 miRNAs were misexpressed in both benign and malignant tumors (listed in the box, green and red indicate lower and higher expression in tumors, respectively). The underlined miRNAs were chosen to be validated by real-time quantitative RT-PCR.
Unsupervised cluster analysis of the most differentially expressed miRNAs
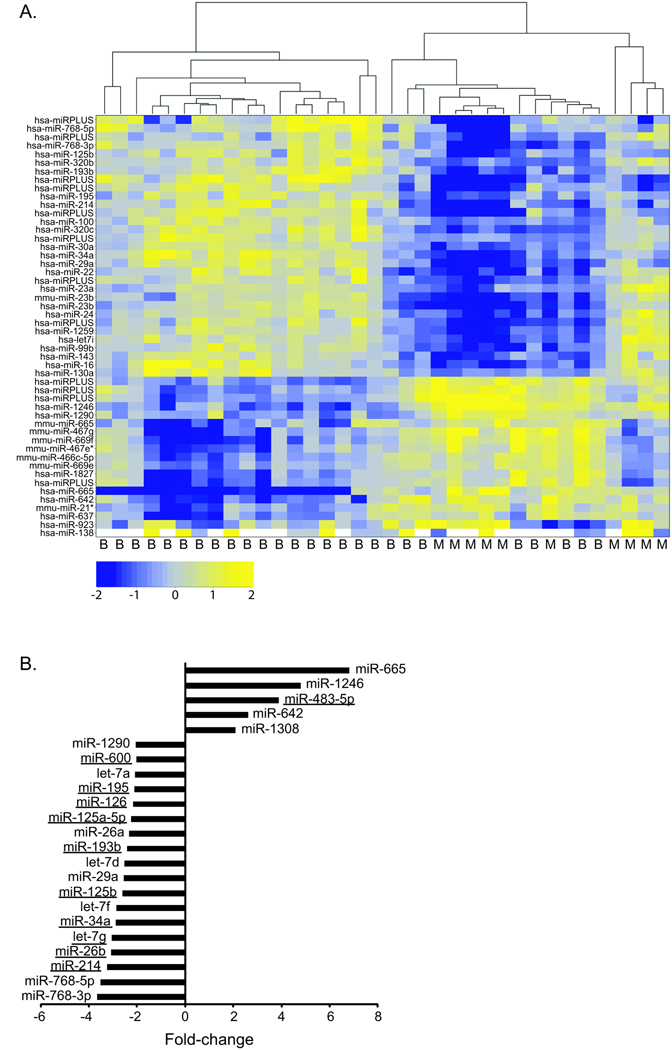
Since a major interest of this study was to identify a miRNA(s) that can distinguish ACCs from benign tumors, we next compared the miRNA expression differences between these classes. As mentioned above, the microarray design compared each tumor sample to a common reference (pooled normal), thereby allowing for direct assessment of the miRNA expression differences between benign and malignant adrenocortical tumors. Unsupervised clustering was performed on the top 50 most variable miRNAs. The heatmap showed some structure, with the malignant samples clustering separately from the majority of the benign samples (Fig. 2A). However, we found that the miRNA expression profiles for a few of the benign samples were more similar to malignant. It is possible that for this subset of tumor samples that these tumors have more potential for malignant transformation.
Figure 2. Identification of differentially expressed miRNAs using microarray analysis.
A. Unsupervised clustering was performed on the 50 most variable miRNAs. The most variable miRNAs were defined by the greatest absolute deviation from the mean across all samples. The Pearson correlation coefficient was used as the similarity metric in this analysis. The heatmaps show the clustering between clinical samples (columns) and the intensity of miRNA expression (log2 (tumor/normal), rows). The tumor diagnosis, either benign (B) or malignant (M), was indicated below. The microarrays included probes for mouse (mmu), human (hsa), and unannotated (miRPLUS) miRNAs (Exiqon LNA miRNA array v. 11.0). B. The top miRNAs significantly up- or down-regulated in ACC as compared to benign adrenocortical tumors were identified as those that were statistically significantly different (p<0.01, Ho: there is no difference between the expression in benign and malignant tumors) and had an expression difference of at least two-fold (up or down) between the two tumor classes were classified as the most differentially expressed. 23 miRNAs fit this criteria and the fold change for each is plotted (negative values indicate decreased expression in malignant and positive values indicate increased expression in malignant). The underlined miRNAs were chosen to be validated by real-time quantitative RT-PCR.
Validation of differentially expressed miRNAs
We next focused on the human miRNAs that had the largest expression difference between benign and malignant tumors. Using a stringent significance criteria of a 2-fold or greater difference in expression level and an adjusted p-value of less than 0.01, we found that 23 miRNAs were differentially expressed. Of these 23, 5 miRNAs had higher expression and 18 miRNAs had lower expression in ACC (Fig. 2B). 13 of these top differentially expressed miRNAs were chosen for further study (Figs. 1 and 2B, underlined). Aside from being the most differentially expressed miRNAs, these 13 were selected because they possess other interesting features. For example, miRs −26b, −214, −195, −125b, 34a, −100, and let-7g lie within genomic regions that have been reported to be frequently lost in ACC 16–20. MiR-483-5p falls within a gene that has been found to be highly expressed in many ACCs, IGF2. Additionally, misexpression of many of the chosen miRNAs have been associated with different types of cancers (reviewed in 21).
The 13 miRNAs were validated in the same samples used for the microarrays (10/10 malignant and 24/26 benign tumors; two benign samples had insufficient RNA and were excluded from validation) by real-time quantitative RT-PCR. The unnormalized PCR data for 12 of the miRNAs tested showed good correlation with the microarray data (p<0.0001) (data not shown). Data was not obtained for miR-600 because it was not amplified by PCR.
Two of our malignant tumor samples had evidence of necrosis and consistently had lower miRNA expression (higher raw Ct) across all of the tested miRNAs. We initially chose to include these types of tumors in our microarray studies since they are representative of the tumor tissue heterogeneity encountered in ACCs. In fact, in both benign and malignant samples we observed that a variable amount of fibrosis and tumor stroma were present in up to 20% of the tissue section. Given this tumor heterogeneity it was important to find a good strategy for controlling the biological variations between samples. “Invariant” small RNAs are often used as endogenous controls for miRNA quantitative RT-PCR data normalization. We initially tested a panel of six possible normalizers; three small RNAs (RNU48, RNU6, and RNU6b) and three miRNAs that according to the microarray data were least variably expressed across all samples (miRs −1285, −34c-5p, and −542-3p). We picked RNU48 as the best control since it had the lowest standard deviation across all of the samples (raw Ct) and was the most stably expressed overall and across the sample subgroups (NormFinder 14).
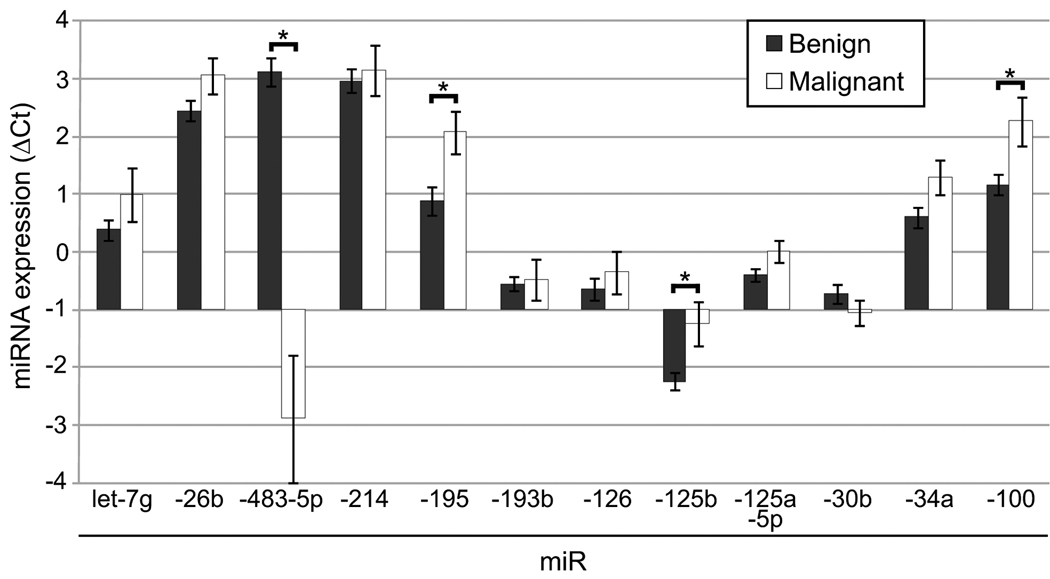
The mean expression data normalized to RNU48 (ΔCt) for benign tumors (n=24) and ACC (n=10) for the 12 validation miRNAs is shown in Fig. 3. For the most part, the overall trend observed by microarray analysis was recapitulated (higher expression of miR-483-5p (lower ΔCt) and lower expression of most of the other miRs (higher ΔCt)) in ACC as compared to benign tumors. Four of the miRNAs, miRs −483-5p, −195, −125b, and −100, had a statistically significant difference between the tumor types (Mann Whitney U test, p<0.05).
Figure 3. Real-time quantitative RT-PCR validation.
Real-time quantitative RT-PCR was used to assay the expression of various miRNAs in tumor samples. The expression of each miRNA is expressed as the ΔCt (Ct miR of interest – Ct RNU48). The mean expression was calculated for benign tumors (n=24, except miRs-483-5p, 193b, and −125-5p where n=23) and ACCs (n=10) and is plotted (± standard error of the mean). An increase in ΔCt indicated lower expression whereas a decrease in ΔCt indicated higher expression (compare benign to malignant). A statistically significant difference between benign and malignant was indicated with an asterik (*) (p<0.05, Mann Whitney U test).
Identification of a diagnostic miRNA for ACC
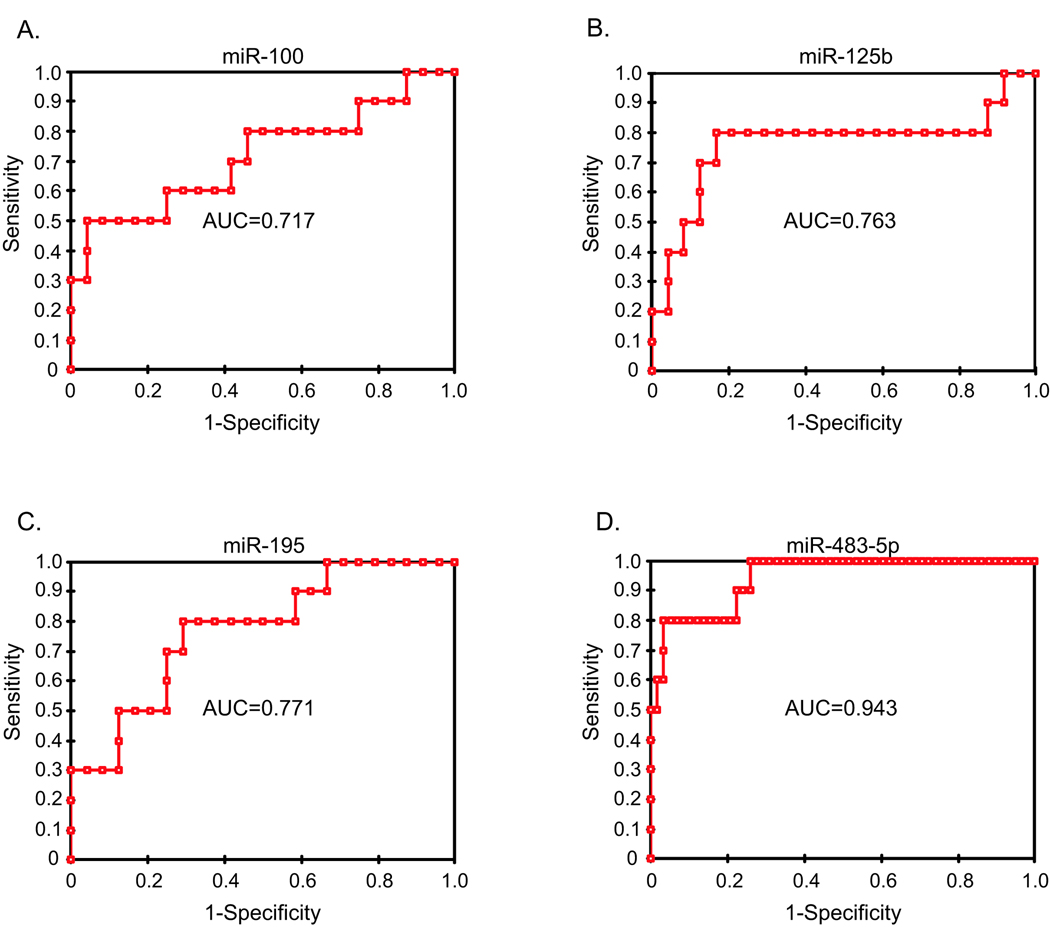
We hypothesized that one or more of the four differentially expressed miRNAs could be used to accurately classify benign and malignant tumors. To determine the diagnostic accuracy of these miRNAs, the area under the receiver operator characteristic curve (AUC) was calculated (Fig. 4A–D). MiR-483-5p had the highest AUC (0.95) indicating that the expression of this miRNA could accurately distinguish between benign tumors and ACCs (Fig. 4D). Combining miR-483-5p with the other miRNAs did not improve accuracy (data not shown). Furthermore, classification based miR-483-5p expression alone in these same tumor samples (10 primary malignant and 23 benign tumors) resulted in 8 of 10 malignant samples being classified as malignant (80% sensitivity) and 24 of 24 truly benign samples being classified as benign (100% specificity). This results in a positive predictive value of 100% and a negative predictive value of 92%.
Figure 4. MiR-483-5p expression can distinguish benign from malignant tumors.
The receiver operating characteristic curve (ROC) was plotted based on the real-time PCR expression profiling of miRs −100 (A), −125b (B), −195 (C), and −483-5p (D) (normalized to RNU48) for 34 samples (10 primary ACCs and 24 benign adrenocortical tumors). The area under the curve (AUC) was listed on the graph. With an AUC of 0.95, miR-483-5p had the greatest diagnostic accuracy (a perfect diagnostic marker without any false- negative or false-positives would have an AUC of 1).
Increased miR-483-5p expression is a marker of malignant phenotype in adrenocortical tumors
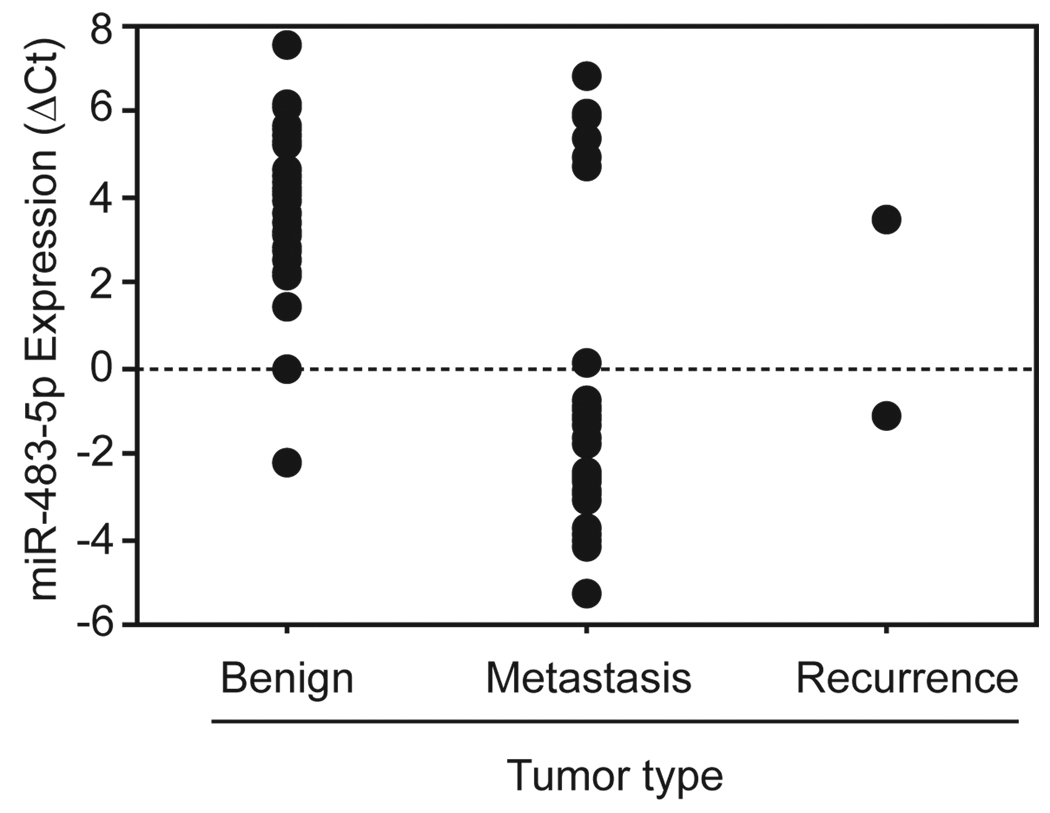
Our data suggests that overexpression of miR-483-5p occurs frequently and consistently in primary ACCs. We speculated that high expression of miR-483-5p may be a persistent and general characteristic of adrenocortical malignancy. To address this hypothesis we measured the expression of miR-483-5p in an independent cohort of benign tumors (n=35), locoregional ACC recurrences (n=2), and ACC metastases (n=29) by real-time quantitative RT-PCR. We found that the majority of malignancies had higher expression of miR-483-5p (lower ΔCt) relative to the benign group and that this difference was statistically significant (p<0.0001, Mann Whitney U test) (Fig. 5). We found that a small subset (7 out of 31) of the malignant samples had miR-483-5p expression that more closely resembled that of benign tumors. These samples came from various sites of metastases in four different ACC patients (these were the only samples from these patients included in this analysis). Interestingly, these patients’ tumors responded well to chemotherapy and they had the best overall response among the group tested.
Figure 5. High miR-483-5p expression is associated with adrenocortical malignancy.
Real-time quantitative RT-PCR was used to assay the expression of miR-483-5p in tumor samples, either benign (n=35) or malignant (ACC recurrence, n=2; ACC metastasis, n=29). Each individual point on the plot denotes a patient sample. The y-axis represents the expression of miR-483-5p which was expressed as the ΔCt (Ct miR-483-5p – Ct RNU48). A lower ΔCt (compare benign to malignant) indicated higher miRNA expression. There was a statistically significant difference between the benign and malignant groups (p<0.0001, Mann Whitney U test).
We found it striking that, based on miR-483-5p expression, the malignant samples formed two mutually exclusive groups; the majority highly expressed miR-483-5p (low ΔCt) and, as mentioned above, a small subset had lower expression of this miRNA (high ΔCt) (Fig. 5). As discussed previously, the miR-483 locus maps to the epigenetically regulated IGF2 gene. Therefore the dichotomous expression pattern we observed may be reflective of the transcriptional status of the imprinted locus being either ‘on’ or ‘off’ 22. If this is the case, it also suggests that these tumors are rather homogenous, at least in terms of gene expression from this particular region.
Increased miR-483 is correlated with high IGF2 expression in adrenocortical tumors
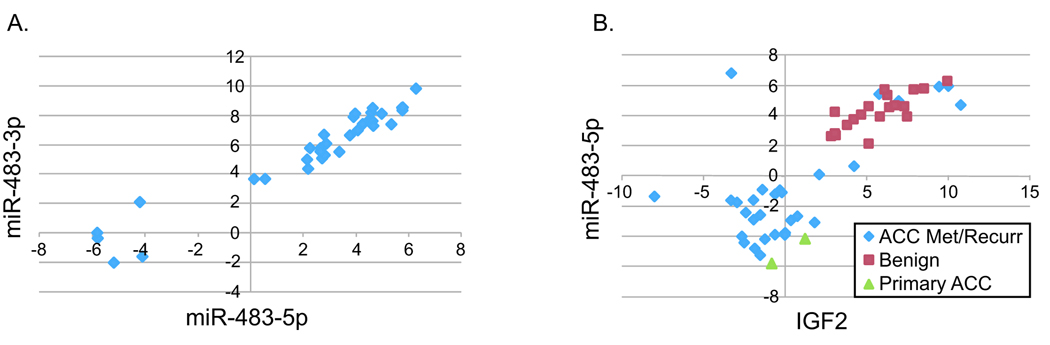
MiR-483 is expressed from intron two of the IGF2 gene. Both the 5’ and 3’ arm of this precursor can express a mature miRNA (either miR-483-5p or miR-483-3p, respectively). Although our miRNA microarray did not identify miR-483-3p as differentially expressed between malignant and benign tumors, we decided to analyze the expression of this miRNA directly by real-time quantitative RT-PCR. Surprisingly, in a manner identical to miR-483-5p, miR-483-3p is highly expressed in malignant samples. In fact the expression of these two miRNAs are highly correlated (Fig. 6A, r=0.965).
Figure 6. The expression of miR-483-3p, miR-483-5p, and IGF2 mRNA are highly correlated.
A. Real-time quantitative RT-PCR was used to assay the expression of miR-483-3p in tumor samples, either benign (n=24) or malignant (primary, n=10). MiR-483-3p expression, expressed as the ΔCt (Ct miR-483-3p – Ct RNU48), is plotted against the ΔCt of miR-483-5p for each patient sample. A strong positive correlation was detected (r=0.965, Pearson correlation). B. Real-time quantitative RT-PCR was used to assay the expression of IGF2 mRNA in tumor samples, either benign (n=19) or malignant (primary, n=2; ACC metastasis, n=27; ACC recurrence, n=3). IGF2 expression, as the ΔCt (Ct IGF2 – Ct GAPDH), is plotted against the ΔCt of miR-483-5p (Ct miR-483-5p – Ct RNU48) for each patient sample. A significant positive correlation was detected (r=0.799, Pearson correlation).
Given that IGF2 has been identified in many studies as commonly overexpressed in ACC 7, 23–29, we speculated that the high expression of miR-483 detected in malignant tumors is a byproduct of overexpression of the IGF2 mRNA. To address this, IGF2 expression was measured in the same patient samples by real-time quantitative RT-PCR. Comparison of IGF2 mRNA and miR-483 expression showed a significant positive correlation (Fig. 6B, r=0.799) suggesting that they are coexpressed from this locus.
Discussion
In this study we performed global miRNA profiling on benign and malignant adrenocortical tumors and identified four human miRNAs, miR −100, −125b, −195, and −483-5p, that had a significant difference in expression between the two tumor classes. Recently, Soon et al. published the results of a similar microarray miRNA profiling in benign and malignant adrenocortical tumors 30. This study identified miRNAs that were differentially expressed between tumor subtypes and found that two miRNAs, miRs −195 and −483-5p, could predict ACC prognosis. Our work supports this study’s observation that miR-195 is significantly downregulated in ACC compared to benign adrenocortical tumors. In addition, by using a large sample size and expanding the analysis to ACC recurrences and metastases, we were able to extend the observations of Soon et al. and convincingly show that there is a significant increase in miR-483-5p expression in ACC (Figs. 3 and 5) and that this is likely a byproduct of IGF2 misexpression (Fig. 6B).
High expression of miR-483-5p is a distinguishing feature of malignant adrenocortical tumors
Preoperative diagnosis of adrenocortical tumors is not always straightforward. Lesions of intermediate size with some suspicious features are often removed to rule out the possibility of malignancy. In these cases, we instead envision using molecular diagnostics on biopsy samples to rule out the possibility of malignancy and avoid unnecessary operations. miRNA markers are ideal in this type of testing since miRNAs seem to be more stable and less prone to degradation then mRNAs and measuring miRNAs requires little starting RNA. Our work found that using miR-483-5p expression has a high negative predictive value of 92% suggesting that the majority of patients with negative test results would be correctly diagnosed. Future studies will be necessary to extend this finding to biopsy samples or as an adjunct to routine histopathology.
Our work indicates that high expression of miR-483-5p is a common occurrence in primary and metastatic ACCs. The miR-483 gene locus has been mapped to intron two of IGF2, one of the most commonly overexpressed genes in ACC 7, 23–29. We hypothesized that the high expression of miR-483-5p observed in ACC is an indirect consequence of IGF2 overexpression. In support of this, miR-483-5p seems to be expressed concordantly with IGF2 mRNA (Fig. 6). Recent studies have found similar positive correlation between miR-483 and IGF2 mRNA levels in Wilms’ tumor, hepatocarcinoma, colorectal cancer, and malignant pheochromocytoma samples 31, 32. Taken together these data suggest that IGF2 and miR-483 are frequently coregulated. Based on this, we propose that miR-483 expression could serve as a useful indicator of IGF2 expression. Clinical trials for ACC patients are evaluating therapies targeting the IGF signaling pathway because of promising preclinical studies that showed a significant antineoplastic effect from inhibiting IGF signaling 33, 34. Assessing the miR-483 expression level from tumor samples or possibly even in serum 35 may serve to quickly identify those patients that would most benefit from this type of therapy.
The role of miRNA misregulation in adrenocortical malignancy
Increasingly studies are finding that altered expression of specific miRNAs can contribute to the initiation and progression of cancer (reviewed in 36). Our work provides the miRNA expression phenotype of ACC, a powerful tool for beginning to better understand the pathological mechanisms involved in the development of this disease.
Aside from being an accurate marker of adrenocortical malignancy, miR-483 is also an appealing therapeutic target for treating both primary and metastatic ACC lesions. Our observation that high expression of miR-483-5p and miR-483-3p occur frequently in malignant adrenocortical tumors coupled with previous work suggesting that higher miR-483-5p is indicative of worse disease outcome 30, leads us to hypothesize that miR-483 expression may promote adrenocortical carcinogenesis. In support of this, a recent study found that miR-483-3p can function as an anti-apoptotic oncogene in cancer cell lines (HEPG2, liver carcinoma and HCT116, colorectal carcinoma) 31. Future functional studies are necessary to assess if miR-483-3p plays a similar role in ACC and the exact role of miR-483-5p in cancer cell biology.
Our targeted validation found that, in addition to high expression of miR-483-5p, miRs −195, was significantly downregulated in malignant adrenocortical tumors as compared to benign tumors (Fig. 3). As mentioned above, this is the second report of low miR-195 expression in ACC 30. In addition, miR-195 expression has been shown to be reduced in hepatocellular carcinoma (HCC). In fact, in HCC, this miRNA seems to play an important functional role in regulating cell proliferation as ectopic expression of miR-195 reduces tumorigenicity and regulates cell cycle progression in HCC cells 37.
This study also found that miR-100 is significantly downregulated in malignant adrenocortical tumors as compared to benign. Decreased miR-100 expression has also been observed in a type of benign adrenocortical tumor, primary pigmented nodular adreoncortical disease (PPNAD, 38), and in childhood adrenocortical tumors 39. Interestingly the latter study used adrenocortical tumor cell lines and primary culture to demonstrate that miR-100 can regulate the IGF-1R and mTOR signaling cascades and that a rapamycin analogue could inhibit cell growth 39.
In summary, this study identified four miRNAs that are misexpressed in ACCs. MiR-483-5p was more highly expressed in malignant than benign tumors and our work indicates that expression of this miRNA can accurately distinguish between tumor types (Fig. 4D). Furthermore, as discussed above, the misexpressed miRNAs have interesting connections to oncogenesis. We anticipate that future functional studies in adrenocortical cell lines will be the key to better understanding not only the specific role(s) that these miRNAs play in adrenocortical carcinogenesis but, more broadly, the molecular pathological mechanisms involved in development of malignant tumors of the adrenal cortex.
Acknowledgments
We thank S. Steinberg for help with statistical analysis.
Funding: This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Conflict of Interest: There are no financial disclosures.
References
- 1.Icard P, Goudet P, Charpenay C, Andreassian B, Carnaille B, Chapuis Y, et al. Adrenocortical carcinomas: surgical trends and results of a 253-patient series from the French Association of Endocrine Surgeons study group. World J Surg. 2001;25(7):891–897. doi: 10.1007/s00268-001-0047-y. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11572030. [DOI] [PubMed]
- 2.Favia G, Lumachi F, Carraro P, D'Amico DF. Adrenocortical carcinoma. Our experience. Minerva Endocrinol. 1995;20(1):95–99. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=7651289. [PubMed]
- 3.Wajchenberg BL, Albergaria Pereira MA, Medonca BB, Latronico AC, Campos Carneiro P, Alves VA, et al. Adrenocortical carcinoma: clinical and laboratory observations. Cancer. 2000;88(4):711–736. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10679640. [PubMed]
- 4.Brunt LM, Moley JF. Adrenal incidentaloma. World J Surg. 2001;25(7):905–913. doi: 10.1007/s00268-001-0029-0. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11572032. [DOI] [PubMed]
- 5.Lau SK, Weiss LM. The Weiss system for evaluating adrenocortical neoplasms: 25 years later. Hum Pathol. 2009;40(6):757–768. doi: 10.1016/j.humpath.2009.03.010. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19442788. [DOI] [PubMed]
- 6.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–531. doi: 10.1038/nrg1379. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15211354. [DOI] [PubMed]
- 7.Fernandez-Ranvier GG, Weng J, Yeh RF, Khanafshar E, Suh I, Barker C, et al. Identification of biomarkers of adrenocortical carcinoma using genomewide gene expression profiling. Arch Surg. 2008;143(9):841–846. doi: 10.1001/archsurg.143.9.841. discussion 46. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18794420. [DOI] [PubMed]
- 8.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15461798. [DOI] [PMC free article] [PubMed]
- 9.Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, et al. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30(4):e15. doi: 10.1093/nar/30.4.e15. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11842121. [DOI] [PMC free article] [PubMed]
- 10.Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31(4):265–273. doi: 10.1016/s1046-2023(03)00155-5. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=14597310. [DOI] [PubMed]
- 11.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16646809. [DOI] [PubMed]
- 12.Smyth GK. limma: Linear Models for Microarray Data. In: Robert Gentleman VJC, Wolfgang Huber, Irizarry Rafael A, Sandrine Dudoit, editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. vol. V. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- 13.Benjamini Y. Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological. 1995;57(1):289–300. Available from <Go to ISI>://A1995QE45300017. [Google Scholar]
- 14.Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization applied to bladder and colon cancer data sets. Cancer Res. 2004;64(15):5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15289330. [DOI] [PubMed]
- 15.Shi W, Alajez NM, Bastianutto C, Hui AB, Mocanu JD, Ito E, et al. Significance of Plk1 regulation by miR-100 in human nasopharyngeal cancer. Int J Cancer. 2010;126(9):2036–2048. doi: 10.1002/ijc.24880. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19739117. [DOI] [PubMed]
- 16.Stephan EA, Chung TH, Grant CS, Kim S, Von Hoff DD, Trent JM, et al. Adrenocortical carcinoma survival rates correlated to genomic copy number variants. Mol Cancer Ther. 2008;7(2):425–431. doi: 10.1158/1535-7163.MCT-07-0267. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18281524. [DOI] [PubMed]
- 17.Kjellman M, Kallioniemi OP, Karhu R, Hoog A, Farnebo LO, Auer G, et al. Genetic aberrations in adrenocortical tumors detected using comparative genomic hybridization correlate with tumor size and malignancy. Cancer Res. 1996;56(18):4219–4223. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=8797595. [PubMed]
- 18.Zhao J, Speel EJ, Muletta-Feurer S, Rutimann K, Saremaslani P, Roth J, et al. Analysis of genomic alterations in sporadic adrenocortical lesions. Gain of chromosome 17 is an early event in adrenocortical tumorigenesis. Am J Pathol. 1999;155(4):1039–1045. doi: 10.1016/S0002-9440(10)65205-4. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10514385. [DOI] [PMC free article] [PubMed]
- 19.Sidhu S, Marsh DJ, Theodosopoulos G, Philips J, Bambach CP, Campbell P, et al. Comparative genomic hybridization analysis of adrenocortical tumors. J Clin Endocrinol Metab. 2002;87(7):3467–3474. doi: 10.1210/jcem.87.7.8697. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12107267. [DOI] [PubMed]
- 20.Soon PS, Libe R, Benn DE, Gill A, Shaw J, Sywak MS, et al. Loss of heterozygosity of 17p13, with possible involvement of ACADVL and ALOX15B, in the pathogenesis of adrenocortical tumors. Ann Surg. 2008;247(1):157–164. doi: 10.1097/SLA.0b013e318153ff55. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18156936. [DOI] [PubMed]
- 21.Osaki M, Takeshita F, Ochiya T. MicroRNAs as biomarkers and therapeutic drugs in human cancer. Biomarkers. 2008;13(7):658–670. doi: 10.1080/13547500802646572. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19096960. [DOI] [PubMed]
- 22.DeChiara TM, Robertson EJ, Efstratiadis A. Parental imprinting of the mouse insulin-like growth factor II gene. Cell. 1991;64(4):849–859. doi: 10.1016/0092-8674(91)90513-x. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=1997210. [DOI] [PubMed]
- 23.Giordano TJ, Thomas DG, Kuick R, Lizyness M, Misek DE, Smith AL, et al. Distinct transcriptional profiles of adrenocortical tumors uncovered by DNA microarray analysis. Am J Pathol. 2003;162(2):521–531. doi: 10.1016/S0002-9440(10)63846-1. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12547710. [DOI] [PMC free article] [PubMed]
- 24.de Fraipont F, El Atifi M, Cherradi N, Le Moigne G, Defaye G, Houlgatte R, et al. Gene expression profiling of human adrenocortical tumors using complementary deoxyribonucleic Acid microarrays identifies several candidate genes as markers of malignancy. J Clin Endocrinol Metab. 2005;90(3):1819–1829. doi: 10.1210/jc.2004-1075. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15613424. [DOI] [PubMed]
- 25.Velazquez-Fernandez D, Laurell C, Geli J, Hoog A, Odeberg J, Kjellman M, et al. Expression profiling of adrenocortical neoplasms suggests a molecular signature of malignancy. Surgery. 2005;138(6):1087–1094. doi: 10.1016/j.surg.2005.09.031. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16360395. [DOI] [PubMed]
- 26.Slater EP, Diehl SM, Langer P, Samans B, Ramaswamy A, Zielke A, et al. Analysis by cDNA microarrays of gene expression patterns of human adrenocortical tumors. Eur J Endocrinol. 2006;154(4):587–598. doi: 10.1530/eje.1.02116. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16556722. [DOI] [PubMed]
- 27.de Reynies A, Assie G, Rickman DS, Tissier F, Groussin L, Rene-Corail F, et al. Gene expression profiling reveals a new classification of adrenocortical tumors and identifies molecular predictors of malignancy and survival. J Clin Oncol. 2009;27(7):1108–1115. doi: 10.1200/JCO.2008.18.5678. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19139432. [DOI] [PubMed]
- 28.Giordano TJ, Kuick R, Else T, Gauger PG, Vinco M, Bauersfeld J, et al. Molecular classification and prognostication of adrenocortical tumors by transcriptome profiling. Clin Cancer Res. 2009;15(2):668–676. doi: 10.1158/1078-0432.CCR-08-1067. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19147773. [DOI] [PMC free article] [PubMed]
- 29.Soon PS, Gill AJ, Benn DE, Clarkson A, Robinson BG, McDonald KL, et al. Microarray gene expression and immunohistochemistry analyses of adrenocortical tumors identify IGF2 and Ki-67 as useful in differentiating carcinomas from adenomas. Endocr Relat Cancer. 2009;16(2):573–583. doi: 10.1677/ERC-08-0237. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19218281. [DOI] [PubMed]
- 30.Soon PS, Tacon LJ, Gill AJ, Bambach CP, Sywak MS, Campbell PR, et al. miR-195 and miR-483-5p Identified as Predictors of Poor Prognosis in Adrenocortical Cancer. Clin Cancer Res. 2009;15(24):7684–7692. doi: 10.1158/1078-0432.CCR-09-1587. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19996210. [DOI] [PubMed]
- 31.Veronese A, Lupini L, Consiglio J, Visone R, Ferracin M, Fornari F, et al. Oncogenic role of miR-483-3p at the IGF2/483 locus. Cancer Res. 70(8):3140–3149. doi: 10.1158/0008-5472.CAN-09-4456. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=20388800. [DOI] [PMC free article] [PubMed]
- 32.Meyer-Rochow GY, Jackson NE, Conaglen JV, Whittle DE, Kunnimalaiyaan M, Chen H, et al. MicroRNA profiling of benign and malignant pheochromocytomas identifies novel diagnostic and therapeutic targets. Endocr Relat Cancer. 17(3):835–846. doi: 10.1677/ERC-10-0142. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=20621999. [DOI] [PubMed]
- 33.Almeida MQ, Fragoso MC, Lotfi CF, Santos MG, Nishi MY, Costa MH, et al. Expression of insulin-like growth factor-II and its receptor in pediatric and adult adrenocortical tumors. J Clin Endocrinol Metab. 2008;93(9):3524–3531. doi: 10.1210/jc.2008-0065. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18611974. [DOI] [PubMed]
- 34.Barlaskar FM, Spalding AC, Heaton JH, Kuick R, Kim AC, Thomas DG, et al. Preclinical targeting of the type I insulin-like growth factor receptor in adrenocortical carcinoma. J Clin Endocrinol Metab. 2009;94(1):204–212. doi: 10.1210/jc.2008-1456. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18854392. [DOI] [PMC free article] [PubMed]
- 35.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18663219. [DOI] [PMC free article] [PubMed]
- 36.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19630570. [DOI] [PubMed]
- 37.Xu T, Zhu Y, Xiong Y, Ge YY, Yun JP, Zhuang SM. MicroRNA-195 suppresses tumorigenicity and regulates G1/S transition of human hepatocellular carcinoma cells. Hepatology. 2009;50(1):113–121. doi: 10.1002/hep.22919. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19441017. [DOI] [PubMed]
- 38.Iliopoulos D, Bimpaki EI, Nesterova M, Stratakis CA. MicroRNA signature of primary pigmented nodular adrenocortical disease: clinical correlations and regulation of Wnt signaling. Cancer Res. 2009;69(8):3278–3282. doi: 10.1158/0008-5472.CAN-09-0155. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19351815. [DOI] [PMC free article] [PubMed]
- 39.Doghman M, El Wakil A, Cardinaud B, Thomas E, Wang J, Zhao W, et al. Regulation of insulin-like growth factor-mammalian target of rapamycin signaling by microRNA in childhood adrenocortical tumors. Cancer Res. 70(11):4666–4675. doi: 10.1158/0008-5472.CAN-09-3970. Available from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=20484036. [DOI] [PMC free article] [PubMed]