Abstract
Huntington disease (HD) is an inherited and incurable neurodegenerative disorder caused by an abnormal polyglutamine (polyQ) expansion in huntingtin (HTT). PolyQ length determines disease onset and severity with a longer expansion causing earlier onset. The mechanisms of mutant HTT-mediated neurotoxicity remain unclear; however, mitochondrial dysfunction is a key event in HD pathogenesis1,2. Here we tested whether mutant HTT impairs the mitochondrial fission/fusion balance and thereby causes neuronal injury. We show that mutant HTT triggers mitochondrial fragmentation in neurons and fibroblasts of HD individuals in vitro and HD mice in vivo before the presence of neurological deficits and HTT aggregates. Interestingly, mutant HTT abnormally interacts with the mitochondrial fission GTPase dynamin-related protein 1 (DRP1) in HD mice and individuals which in turn stimulates its enzymatic activity. Importantly, mutant HTT-mediated mitochondrial fragmentation, defects in anterograde and retrograde mitochondrial transport, and neuronal cell death are all rescued by reducing DRP1 GTPase activity with the dominant-negative DRP1K38A mutant. Thus, DRP1 might represent a new therapeutic target to combat neurodegeneration in HD.
Cycles of mitochondrial fission and fusion are necessary for neuronal function and there is strong evidence that an imbalance in these opposing processes initiates neurodegeneration3,4. Large GTPases of the dynamin family mediate mitochondrial fission and fusion with DRP1 regulating mitochondrial fission and mitofusin (MFN) regulating mitochondrial fusion5,6. Mitochondrial defects and neuronal cell death caused by mutant HTT are events that could be explained by an altered mitochondrial fission/fusion equilibrium1.
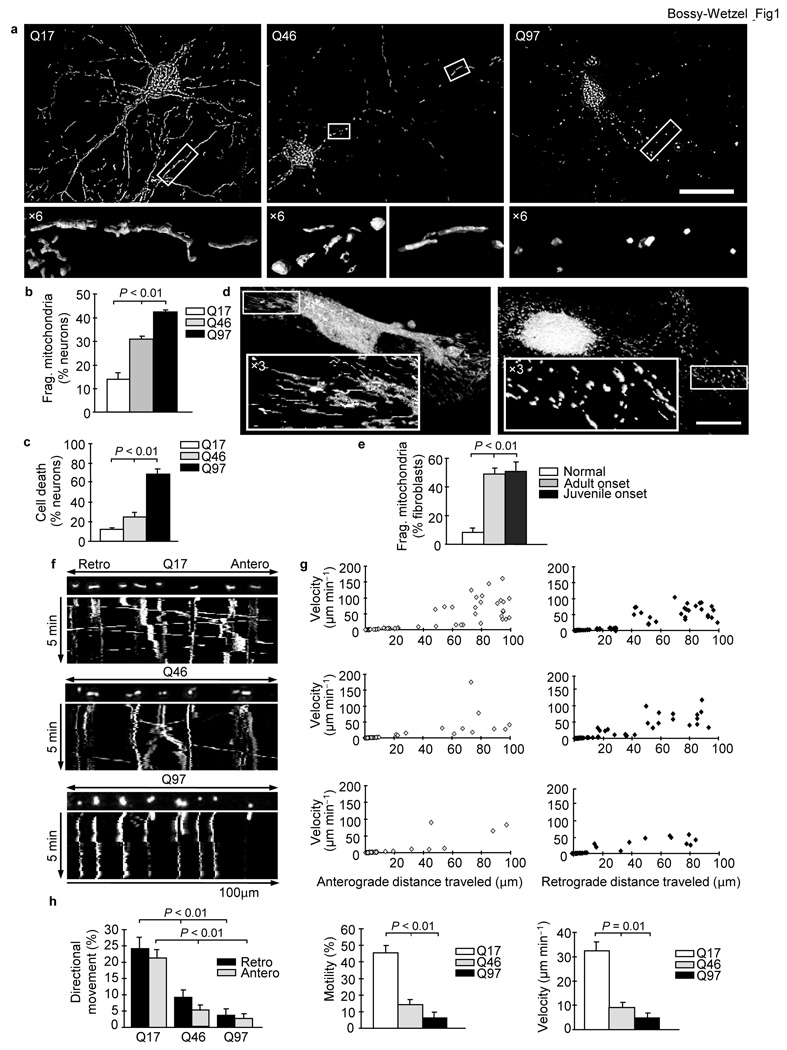
To explore this possibility, we tested whether mutant HTT alters the mitochondrial morphology in rat cortical neurons or human HD fibroblasts. Neurons with exogenous expression of wild-type HTT exon1-Q17 exhibited filamentous mitochondrial morphology, typical of healthy neurons (Fig. 1a, left), with an average length of 3,200 nm. In contrast, neurons expressing mutant HTT exon1-Q46 exhibited round and elongated mitochondria (Fig. 1a, center), suggesting the occurrence of mitochondrial fragmentation. Neurons expressing mutant HTT exon1-Q97 revealed more profound mitochondrial fragmentation (Fig. 1a, right), with an average of mitochondrial length of 800 nm. Further quantitative analysis indicated an increase in the percentage of neurons with fragmented mitochondria in neuronal populations expressing polyQ’s linked to disease (Q46 and Q97), with the Q97 populations exhibiting the most fragmentation (Fig. 1b). Mitochondrial numbers decreased (Supplementary Fig. 1) and autophagy increased (Supplementary Fig. 2) in neurons expressing mutant HTT. Using time-lapse imaging, we observed an increased ratio of fission over fission plus fusion events in neurons expressing mutant HTT, suggesting an imbalance in mitochondrial dynamics (Supplementary Fig. 3). Finally, mitochondrial fragmentation correlated with neuronal cell death (Fig. 1c). Mutant HTT exon1 expression alone did not cause mitochondrial fragmentation in HeLa cells, but only increased the vulnerability to hydrogen peroxide-induced fragmentation7.
Figure 1.
Mutant HTT triggers mitochondrial fragmentation, decrease in anterograde and retrograde transport, and neuronal cell death, which depends on polyQ length. (a) Fluorescence micrographs and ×6 zoom of boxed regions of neurons expressing HTT exon1-Q17, -Q46, or -Q97 and DsRed2-Mito. Scale bar, 50 µm. (b) Mitochondrial fragmentation of neurons expressing HTT exon1-Q17, -Q46, or -Q97 and DsRed2-Mito. (c) Cell death of neurons expressing HTT exon1-Q17, -Q46, or -Q97. (d) Fluorescence micrographs and ×3 zoom of boxed regions of MitoTracker Red stained human fibroblasts from an unaffected (left) or an adult onset HD individual (right). Scale bar, 25 µm. (e) Mitochondrial fragmentation in fibroblasts from an unaffected or HD individuals. (f) Kymographs of mitochondrial transport in neurons expressing HTT exon1-Q17, -Q46, or -Q97 and DsRed2-Mito. See also Movies S1, S2, S3. (g) Scatter plots of mitochondrial velocity in retrograde or anterograde direction as a function of distance traveled in 5 minutes in neurons (n = 10) expressing HTT exon1-Q17, -Q46, or -Q97 and DsRed2-Mito. (h) Anterograde and retrograde movement, motility, and mean velocity of mitochondria in the same neurons analyzed in (g). Data are mean ± s.e.m. of triplicate samples of representative experiments. Results are representative of three or more independent experiments. Statistics: one-way ANOVA test.
Next, we used fibroblasts from HD individuals to investigate whether endogenous full-length mutant HTT can also alter mitochondrial morphology. Mitochondria in fibroblasts of an unaffected individual exhibited the expected tubular morphology. In contrast, mitochondria from a HD person revealed shorter and rounder morphology, suggesting mitochondrial fragmentation (Fig. 1d). Quantitative analysis demonstrated an increased percentage of fibroblasts with fragmented mitochondria for juvenile and adult onset HD (Fig. 1e). Thus, endogenous full-length mutant HTT causes mitochondrial fragmentation in human HD fibroblasts.
Mitochondrial fragmentation induced by nitric oxide (NO) arrests mitochondrial transport8–10. Furthermore, defects in transport of BDNF vesicles and mitochondria have been linked to HD11,12. To test whether mutant HTT-induced mitochondrial fragmentation stalls mitochondrial transport, we tracked mitochondrial movement. Neurons expressing HTT exon1-Q17 had high anterograde and retrograde mitochondrial transport and velocity (Fig. 1f, g top and Supplementary Movie 1). In contrast, neurons expressing HTT exon1-Q46 demonstrated a decrease in both anterograde and retrograde mitochondrial transport and velocity (Fig. 1f, g center and Supplementary Movie 2). The arrest in directional mitochondrial movement and velocity was even more pronounced in neurons expressing HTT exon1-Q97 (Fig. 1f, g bottom and Supplementary Movie 3). Quantitative analysis revealed a polyQ-length dependent loss of bidirectional transport, mobility, and velocity of mitochondria (Fig. 1h). Thus, mutant HTT-induced mitochondrial fragmentation correlates with an arrest in transport, which might negatively impact energy distribution at sites of high demand, such as synapses.
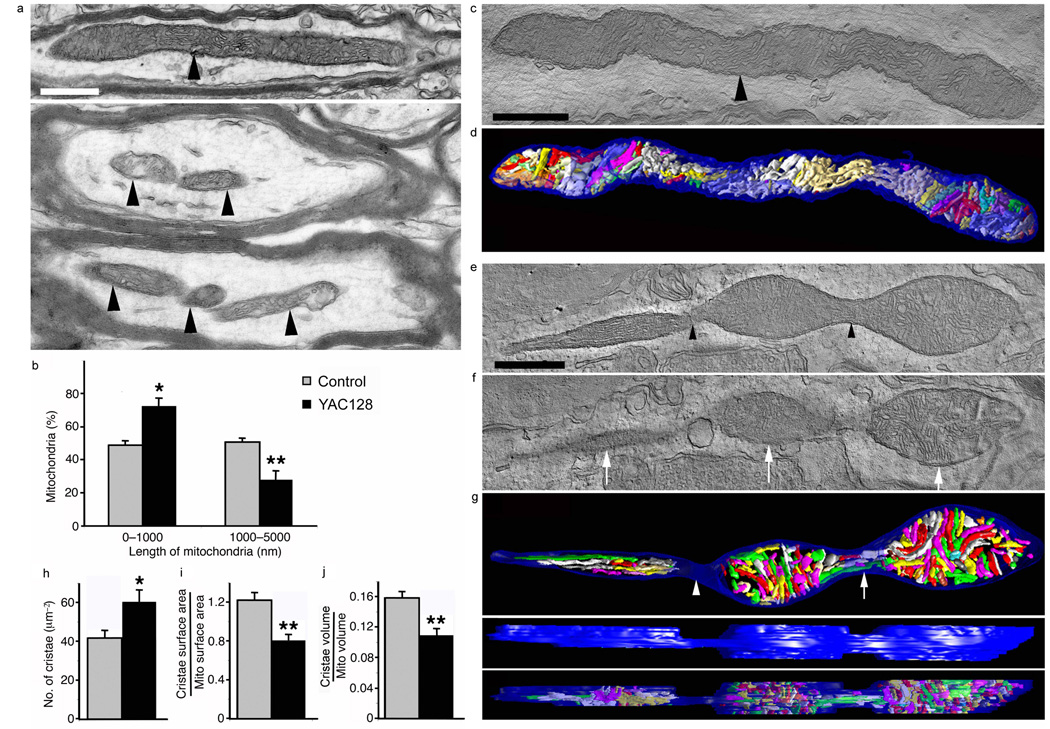
To investigate whether mitochondrial fragmentation initiated by the N-terminal HTT fragment and full-length endogenous HTT in vitro also occurs in vivo, we employed mice that express the human full-length HTT with 18Q repeats (YAC18 mice) or with 128Q repeats (YAC128 mice), driven by the endogenous promoter13. To test whether fragmentation occurs in mutant HD YAC128 mice before disease onset and in the absence of HTT aggregates, we used electron microscopy (EM). Mitochondria in the striatum of control mice exhibited elongated morphology in neuronal processes (Fig. 2a top). In YAC128 mice, we found fewer filamentous mitochondria and more small mitochondria with close proximity to each other (Fig. 2a bottom). There was a significant increase (P < 0.05) in short mitochondria (< 1,000 nm) and a decrease (P < 0.01) in long mitochondria (> 1,000 nm) (Fig. 2b). These results suggest that mitochondrial fragmentation occurs in vivo in HD mice prior to neurological deficits, neuronal cell death, and HTT aggregate formation14.
Figure 2.
Mutant HTT increases the number of small mitochondria and cristae, but decreases cristae surface area and volume in the striatum of six-month old YAC128 mice. (a) EM of control (top) and YAC128 (bottom) brain showing an elongated neuronal mitochondrion (arrowhead) and several short mitochondria (arrowheads), respectively. Scale bar, 500 nm. (b) Percentage of mitochondria of short length (0–1,000 nm) and long length (1,000–5,000 nm). (c) EM of a control mitochondrion (arrowhead) nearly 4 µm long. (d) Surface-rendered volume showing the normal structure of its 84 cristae. (e) EM of a mitochondrion fissioning into three parts (arrowheads). (f) A slice of the volume shows the separation of the three mitochondrial bodies (arrows). (g) Top view showing all 223 cristae. There are no cristae in the constriction (arrowhead) between the left and middle bodies, but a few cristae appear in the constriction between the middle and right bodies (arrow). Side view of the outer membrane showing the width of the constrictions. Side view demonstrating cristae fragmentation by how few cristae extend from top to bottom of the volume. (h) Number of cristae per mitochondrial cross-sectional area. (i, j) Cristae surface area and volume. Data are mean ± s.e.m. Statistics: Student’s t-test, P < 0.05*, P < 0.01**.
To explore whether the mutant HTT-induced mitochondrial fragmentation produces ultrastructural changes in mitochondria, we performed EM tomography. The 3D structure of a typical mitochondrion from control mice exhibits an elongated form and well-preserved outer and inner membrane structures, including tubular and lamellar cristae, commonly observed in striatal mitochondria (Fig. 2c, d and Supplementary Movie 4)15. In contrast, the 3D structure of the YAC128 mitochondrion exhibits the process of fragmentation into three parts (Fig. 2e–g and Supplementary Movies 5). The outer membrane shows constrictions between the three bodies (Fig. 2e–g). The segregation of cristae during the fragmentation process was complete between left and middle bodies, but incomplete between middle and right bodies. YAC128 mitochondria had greater cristae density (Fig. 2h). Potential explanations for the cristae increase are that mutant HTT evokes cristae biogenesis or that cristae fragmentation occurs in addition to mitochondrial fragmentation. To distinguish between these two possibilities, we measured the ratio of total cristae membrane surface area to outer mitochondrial membrane surface area and the ratio of total cristae volume to mitochondrial volume (Fig. 2i, j). The YAC128 mitochondria had a mean cristae surface area ratio of 0.8 compared to 1.2 in controls (Fig. 2i). Similarly distinct, the YAC128 mitochondria had a mean cristae volume ratio of 0.11 compared to 0.16 in controls (Fig. 2j). In sum, YAC128 mitochondria have almost 50% more cristae, yet lower cristae surface area and volume ratios. These data suggest that mutant HTT does not cause cristae biogenesis, but rather cristae fragmentation. These data agree with the inhibition of PGC1-α-mediated mitochondrial biogenesis by mutant HTT16. Thus, mitochondrial deficits in HD may originate both from inappropriate and excessive mitochondrial fragmentation and impaired PGC-1α-mediated de novo biogenesis1.
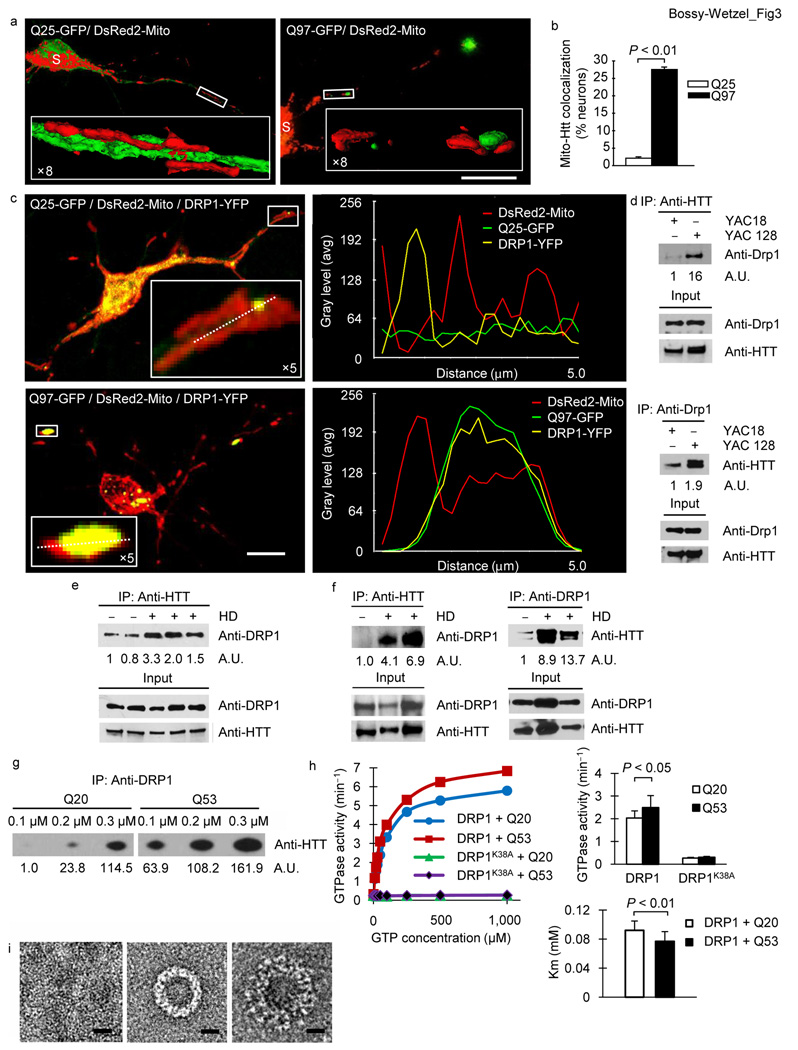
Mitochondrial fragmentation can be caused by stimulation of fission and/or inhibition of fusion. A potential mechanism by which mutant HTT may cause mitochondrial fragmentation could be via DRP1 S-nitrosylation 8,9,17–19. However, we find that DRP1 S-nitrosylation does not increase DRP1 GTPase activity20, leading us to conclude that DRP1 S-nitrosylation is not the basis for mutant HTT-mediated mitochondrial fragmentation. Alternatively, mutant HTT might trigger mitochondrial fragmentation by abnormal protein interactions21,22. The normal function of HTT is that of a scaffold molecule regulating physiological processes via selective protein interactions. There is evidence that altered binding of mutant HTT with this group of target proteins participates in HD pathogenesis22. Mutant HTT binds to mitochondria where it can accumulate in large oligomers12,23,24. A similar subcellular distribution pattern has been documented for DRP1, which forms oligomers at mitochondrial division sites25. While wild-type HTT exon1-Q25 exhibited largely cytoplasmic distribution, mutant HTT exon1-Q97 revealed a significant increase (P < 0.01) in mitochondrial co-localization (Fig. 3a, b). Based on these observations and supporting literature2,21,23,24,26, we speculated that mutant HTT might abnormally bind to DRP1, thereby inappropriately shifting the mitochondrial fission/fusion balance.
Figure 3.
Mutant HTT interacts with DRP1 in HD mice and human and alters DRP1 structure and function. (a) Fluorescence micrographs of neurons expressing HTT exon1 and DsRed2-Mito. Scale bar, 50 µm. (b) Co-localization of mitochondria and HTT in neurons co-expressing HTT exon1 and DsRed2-Mito. (c) Fluorescence micrographs with line scan of co-localization of HTT, DRP1, and mitochondria in neurons co-expressing DsRed2-Mito, DRP1-YFP, and HTT exon1. Scale bar, 10 µm. (d) Co-immune precipitations of brain mitochondrial lysates from 1.5 or 2 month old YAC18 and YAC128 mice followed with HTT or Drp1 antibodies. The intensities of the signals are presented as arbitrary units (A.U.) and are normalized to input signals. (e) Co-immune precipitations of human lymphoblast lysates from unaffected or HD individuals with HTT antibodies. (f) Co-immune precipitations of human postmortem brain tissue lysates from unaffected or HD individuals with DRP1 or HTT antibodies. (g) Co-immune precipitations of bacterial DRP1 protein and bacterial HTT exon1-Q20-GST or -Q53-GST protein with DRP1 antibodies. (h) Steady-state kinetics of DRP1 GTPase activity (left), bar graph of GTPase activity at 0.05 mM GTP and apparent Michaelis-Menten constant (Km) (right) in the presence of wild-type or mutant HTT exon1 protein and MOM liposomes. Data are mean ± s.d. from three independent measurements. Statistics: Student’s t-test. (i) Negative stain EM images of baculovirus DRP1 in the absence of nucleotides (left), the presence of GTP (center), and the presence of GTP and HTT exon1-Q53 -GST protein (right). Scale bars, 10 nm.
To test whether mutant HTT interacts with DRP1 we first explored whether HTT and DRP1 co-localize on mitochondria in neurons. We found an overlap of mutant HTT and DRP1 fluorescent signals on mitochondria, suggesting that they may form a physical complex (Fig. 3c and Supplementary Fig. 4). To further investigate whether mutant HTT indeed binds Drp1, we isolated brain lysates from pre-symptomatic YAC18 and YAC128 and performed co-immune precipitations (Fig. 3d). While wild-type HTT also binds Drp1, we observed an increase in the mutant HTT-Drp1 interaction. (Fig. 3d). Reverse immune precipitations confirmed the results (Fig. 3d). Furthermore, the increased interaction was Drp1 specific and did not occur for Mfn2 (Supplementary Fig. 5 and Supplementary Fig. 6).
To test whether the mutant HTT-Drp1 complex also exists in human HD and occurs with endogenous HTT, we analyzed lymphoblasts and brain tissue of HD individuals. Similar to our results with HD mice, mutant HTT co-precipitated with DRP1 more strongly (Fig. 3e, f). In sum, the increased mutant HTT-DRP1 interaction is of relevance to human HD and occurs with endogenous full-length HTT.
Because mutant HTT interacts with DRP1 in vivo, we tested whether mutant HTT binds DRP1 directly and with greater affinity than wild-type HTT in vitro. Using co-immune precipitations with purified proteins, we detected that wild-type HTT exon1-Q20 interacted either not at all or only weakly with DRP1. By contrast, mutant HTT was able to bind DRP1, suggesting that mutant HTT exon1-Q53 exhibits greater affinity to DRP1 than wild-type HTT (Fig. 3g).
Next, it was important to test whether this interaction would alter DRP1’s enzymatic activity and structure. Using liposomes that mimic the mitochondrial outer membrane (MOM) we observed a significant increase in the enzymatic activity (P < 0.05) and a significant decrease in the apparent Km (P < 0.01) with mutant HTT (Fig. 3h and Supplementary table 1). This finding was not due to a contamination with another GTPase, because our DRP1 protein was pure (Supplementary Fig. 7) and replacement of wild-type DRP1 with the enzymatically inactive DRP1K38A mutant revealed no activity (Fig. 3h and Supplementary table1). Using EM negative stain we found that DRP1 forms ring- and spiral-like oligomers, similar to its yeast homolog (Fig. 3i and Supplementary Fig. 8)27,28. The DRP1 ring-like oligomer remained largely unaltered with wild-type HTT. However, when mutant HTT was added, we observed an additional layer of density around the density corresponding to the DRP1 ring-like oligomer (Fig. 3i). In sum, our in vitro results suggest that mutant HTT exhibits enhanced affinity to DRP1, which in turn alters its structure and stimulates its enzymatic activity. Although our data suggest a primary and direct activation of DRP1 by mutant HTT, we cannot also exclude an inhibition of mitochondrial fusion.
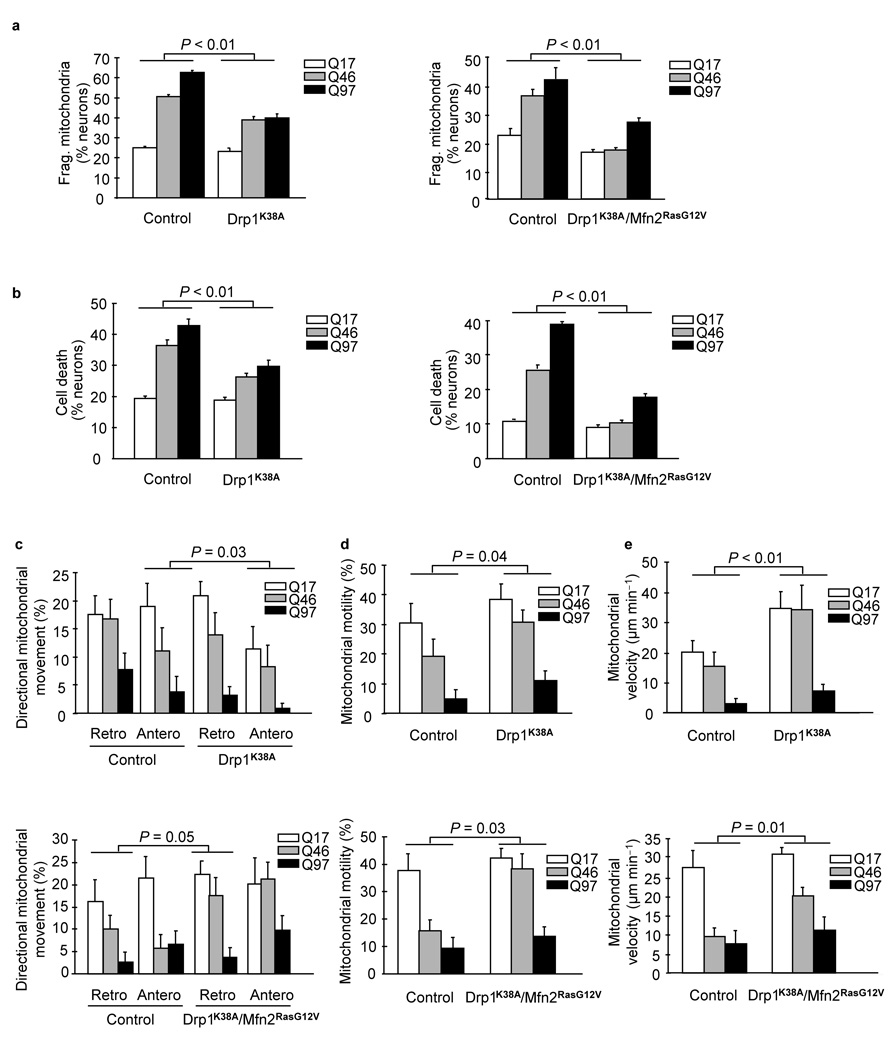
Our data show that mutant HTT triggers mitochondrial fragmentation, which is linked to an arrest in mitochondrial transport and neuronal cell death. To test whether DRP1 and an altered fission/fusion equilibrium causes these events, we expressed either the mutant DRP1K38A alone or in combination with the constitutively active mutant MFN2RasG12V 29. Loss of DRP1 by shRNA expression altered the neuronal differentiation (Supplementary Fig. 9), similar to DRP1 null mice30,31. We found a significant reduction in mitochondrial fragmentation (P < 0.01) (Fig. 4a) and neuronal cell death (P < 0.01) (Fig. 4b) in neurons co-expressing mutant HTT with either DRP1K38A alone or in combination with MFN2RasG12V. Last, transport, motility, and velocity of mitochondria were all improved by re-stablishing the mitochondrial fission and fusion balance (Fig. 4c–e and Supplementary Fig. 10). In a C. elegans model of HD, DRP1 knockdown increased worm motility, however the authors did not examine mitochondrial morphology or motility7. In sum, our data provide evidence that DRP1 is functionally implicated in mutant HTT-mediated neuronal injury and cell death.
Figure 4.
Restoring mitochondrial fusion with DRP1K38A or in combination with MFN2RasG12V rescues neurons from neuritic trafficking defects and cell death. (a) Mitochondrial fragmentation of neurons expressing either HTT exon1-Q17, -Q46, or -Q97 alone or in combination with either DRP1K38A alone, or DRP1K38A and MFN2RasG12V. (b) Cell death of neurons expressing either mutant HTT exon1-Q17, -Q46, or -Q97 and DsRed2-Mito alone or in combination with either DRP1K38A alone or DRP1K38A and MFN2RasG12V. (c) Anterograde and retrograde movement of mitochondria in neurons expressing either HTT exon1-Q17, -Q46, or -Q97 alone or in combination with DRP1K38A alone or DRP1K38A and MFN2RasG12V. (d) Motility of mitochondria in neurons expressing either HTT exon1-Q17, -Q46, or -Q97 alone or in combination with either DRP1K38A alone or both DRP1K38A and MFN2RasG12V. (e) Mean velocity of mitochondria in neurons expressing either HTT exon1-Q17, -Q46, or -Q97 alone or in combination with DRP1K38A alone or DRP1K38A and MFN2RasG12V. Results are representative of three or more independent experiments. Statistics: Two-way ANOVA with post-hoc test.
Our findings provide evidence that full-length mutant HTT expressed at physiological levels is sufficient to cause mitochondrial fragmentation in HD mice and individuals prior to cell death and aggregate formation. Mutant HTT-induced mitochondrial fragmentation impairs transport of mitochondria and reduces neuronal survival as evidenced by the amelioration of these events upon prevention of mitochondrial fragmentation by DRP1K38A alone or in combination with MFN2RasG12V. Furthermore, mutant HTT abnormally interacts with DRP1 in HD mice prior to disease onset. Last, the mutant HTT-DRP1 complex is likely of pathophysiological significance because it is present in HD individuals. Thus, DRP1 might constitute a new target for HD therapy.
Methods
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturemedicine/.
Supplementary Material
Acknowledgments
We thank S. Finkbeiner (University of California San Francisco) for the HTT plasmids pGW1-HTT exon1-Q17-GFP, -Q46, and -Q97; L. Thompson (University of California Irvine) for the HTT plasmids pcDNA3.1-Q25-GFP and -Q97; U. Hartl (Max Planck Institute of Biochemistry) for the GST-HTT exon1-Q20 and -Q53 constructs; Van der Bliek (University of California Los Angeles) for the pcDNA3-DRP1K38A plasmid (NCBI accession number: NM_005690.3) baculovirus; R. Youle (US National Institute of Health) for the YFP-DRP1 plasmid; R. Slack (University of Ottawa) for the MFN2RasG12V (p82-FzoRV12pECFP-C1) plasmid; S. Strack (University of Iowa) for his pcDNA3.1β-DRP1shRNA vector. S. Lubitz, J. Johnson, V. DeAssis, C. Eldon, and B. Kincaid for technical assistance; and A. Knott for manuscript development and editing. This work is supported by NIH grants to E.B-W. (R01EY016164 and R01NS055193), a grant from the Hereditary Disease Foundation, grants to I.R., M.A.P., and M.R.H. from CIHR, and support from CHDI to M.R.H. The EM tomography work was carried out using facilities of the US National Center for Microscopy and Imaging Research, supported by NIH grant P41RR004050 awarded to M.H.E.
Footnotes
Note: Supplementary information is available on the Nature Medicine website.
Author Contributions
W.S. performed all imaging and participated in the mitochondrial fragmentation and cell death analyses. J.C. performed the GTPase assays, some of the immune precipitations and EM analysis. A.P. performed some of the neuronal cell death and immune precipitations. G.L. performed the EM stereology and generated some of the preliminary data. E.K. purified, cloned, and prepared the recombinant DRP1 protein. Y.Z. performed Western blots for the DRP1 knockdown. P.P. and J.T. participated in the EM tomography. M.A.P. and M.R.H. provided the YAC18 and YAC128 mice and advice on HTT co-immune precipitations. E.M. provided human postmortem brain samples. R.S. led the DRP1 protein purification. M.E. and G.P. led the EM tomography experiment. B.B. performed GTPase assays, prepared samples for EM, and purified the HTT protein. I.R. performed EM negative stain experiments. E.B-W. conceived the project and wrote the article. All authors participated in the data analysis and interpretation of the results.
References
- 1.Bossy-Wetzel E, Petrilli A, Knott AB. Mutant huntingtin and mitochondrial dysfunction. Trends Neurosci. 2008;31:609–616. doi: 10.1016/j.tins.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 3.Knott AB, Bossy-Wetzel E. Impairing the mitochondrial fission and fusion balance: a new mechanism of neurodegeneration. Ann N Y Acad Sci. 2008;1147:283–292. doi: 10.1196/annals.1427.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knott AB, Perkins G, Schwarzenbacher R, Bossy-Wetzel E. Mitochondrial fragmentation in neurodegeneration. Nat Rev Neurosci. 2008;9:505–518. doi: 10.1038/nrn2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen H, et al. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smirnova E, Griparic L, Shurland DL, van der Bliek AM. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang H, Lim PJ, Karbowski M, Monteiro MJ. Effects of overexpression of huntingtin proteins on mitochondrial integrity. Hum Mol Genet. 2009;18:737–752. doi: 10.1093/hmg/ddn404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barsoum MJ, et al. Nitric oxide-induced mitochondrial fission is regulated by dynamin-related GTPases in neurons. Embo J. 2006;25:3900–3911. doi: 10.1038/sj.emboj.7601253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liot G, et al. Complex II inhibition by 3-NP causes mitochondrial fragmentation and neuronal cell death via an NMDA- and ROS-dependent pathway. Cell Death Differ. 2009;16:899–909. doi: 10.1038/cdd.2009.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan H, et al. Mitochondrial fission is an upstream and required event for bax foci formation in response to nitric oxide in cortical neurons. Cell Death Differ. 2007;14:462–471. doi: 10.1038/sj.cdd.4402046. [DOI] [PubMed] [Google Scholar]
- 11.Gauthier LR, et al. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell. 2004;118:127–138. doi: 10.1016/j.cell.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 12.Orr AL, et al. N-terminal mutant huntingtin associates with mitochondria and impairs mitochondrial trafficking. J Neurosci. 2008;28:2783–2792. doi: 10.1523/JNEUROSCI.0106-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hodgson JG, et al. Human huntingtin derived from YAC transgenes compensates for loss of murine huntingtin by rescue of the embryonic lethal phenotype. Hum Mol Genet. 1996;5:1875–1885. doi: 10.1093/hmg/5.12.1875. [DOI] [PubMed] [Google Scholar]
- 14.Slow EJ, et al. Selective striatal neuronal loss in a YAC128 mouse model of Huntington disease. Hum Mol Genet. 2003;12:1555–1567. doi: 10.1093/hmg/ddg169. [DOI] [PubMed] [Google Scholar]
- 15.Perkins GA, et al. Electron tomography of mitochondria after the arrest of protein import associated with Tom19 depletion. Eur J Cell Biol. 2001;80:139–150. doi: 10.1078/0171-9335-00149. [DOI] [PubMed] [Google Scholar]
- 16.Cui L, et al. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 17.Fan J, Cowan CM, Zhang LY, Hayden MR, Raymond LA. Interaction of postsynaptic density protein-95 with NMDA receptors influences excitotoxicity in the yeast artificial chromosome mouse model of Huntington's disease. J Neurosci. 2009;29:10928–10938. doi: 10.1523/JNEUROSCI.2491-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graham RK, et al. Differential susceptibility to excitotoxic stress in YAC128 mouse models of Huntington disease between initiation and progression of disease. J Neurosci. 2009;29:2193–2204. doi: 10.1523/JNEUROSCI.5473-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho DH, et al. S-nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Science. 2009;324:102–105. doi: 10.1126/science.1171091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bossy B, et al. S-Nitrosylation of DRP1 does not affect enzymatic activity and is not specific to Alzheimer's disease. J Alzheimers Dis. 20(Suppl 2):S513–S526. doi: 10.3233/JAD-2010-100552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaltenbach LS, et al. Huntingtin interacting proteins are genetic modifiers of neurodegeneration. PLoS Genet. 2007;3:e82. doi: 10.1371/journal.pgen.0030082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li SH, Li XJ. Huntingtin-protein interactions and the pathogenesis of Huntington's disease. Trends Genet. 2004;20:146–154. doi: 10.1016/j.tig.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 23.Choo YS, Johnson GV, MacDonald M, Detloff PJ, Lesort M. Mutant huntingtin directly increases susceptibility of mitochondria to the calcium-induced permeability transition and cytochrome c release. Hum Mol Genet. 2004;13:1407–1420. doi: 10.1093/hmg/ddh162. [DOI] [PubMed] [Google Scholar]
- 24.Panov AV, et al. Early mitochondrial calcium defects in Huntington's disease are a direct effect of polyglutamines. Nat Neurosci. 2002;5:731–736. doi: 10.1038/nn884. [DOI] [PubMed] [Google Scholar]
- 25.Frank S, et al. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev Cell. 2001;1:515–525. doi: 10.1016/s1534-5807(01)00055-7. [DOI] [PubMed] [Google Scholar]
- 26.Trushina E, et al. Mutant huntingtin impairs axonal trafficking in mammalian neurons in vivo and in vitro. Mol Cell Biol. 2004;24:8195–8209. doi: 10.1128/MCB.24.18.8195-8209.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mears JA, Hinshaw JE. Visualization of dynamins. Methods Cell Biol. 2008;88:237–256. doi: 10.1016/S0091-679X(08)00413-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naylor K, et al. Mdv1 interacts with assembled dnm1 to promote mitochondrial division. J Biol Chem. 2006;281:2177–2183. doi: 10.1074/jbc.M507943200. [DOI] [PubMed] [Google Scholar]
- 29.Neuspiel M, Zunino R, Gangaraju S, Rippstein P, McBride H. Activated mitofusin 2 signals mitochondrial fusion, interferes with Bax activation, and reduces susceptibility to radical induced depolarization. J Biol Chem. 2005;280:25060–25070. doi: 10.1074/jbc.M501599200. [DOI] [PubMed] [Google Scholar]
- 30.Jagasia R, Grote P, Westermann B, Conradt B. DRP-1-mediated mitochondrial fragmentation during EGL-1-induced cell death in C. elegans. Nature. 2005;433:754–760. doi: 10.1038/nature03316. [DOI] [PubMed] [Google Scholar]
- 31.Wakabayashi J, et al. The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. J Cell Biol. 2009;186:805–816. doi: 10.1083/jcb.200903065. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.