Abstract
The administration of live attenuated Salmonella strains has proven to be an effective way to generate protective immunity against Salmonella infection in humans and mice. Studies in the mouse model have shown that protection requires Salmonella-specific Th1 cells, however the timing and stimulatory requirements for generating optimal Th1 responses have not been carefully examined. We used antibiotic interruption of vaccination with live attenuated Salmonella to examine the requirements for Salmonella-specific Th1 development and protective immunity. Optimal development of protective immunity to Salmonella infection required at least one week of exposure to the live attenuated Salmonella strain. In contrast, optimal development of Salmonella-specific Th1 cells required two weeks of in vivo colonization. Thus, sustained in vivo stimulation with a live vaccine strain is essential for the development of robust Salmonella-specific Th1 cells.
Introduction
Salmonella enterica serovar Typhi (hereafter referred to as S. Typhi), the causative agent of human typhoid, is responsible for significant morbidity and mortality throughout the world, particularly in developing countries. The most recent estimate is that S. Typhi annually infects 21.7 million people and causes over 200,000 fatalities [1]. Salmonella infections are typically transmitted by the fecal-oral route via contaminated food or water, and typhoid outbreaks concentrate in geographical areas lacking adequate sanitation or access to clean drinking water [2]. For infected individuals with access to medical treatment, antimicrobial resistance is a growing concern [3] and reliance on antibiotic treatment alone is not a realistic long-term strategy to combat typhoid. The most obvious solution to endemic typhoid in developing nations would be global implementation of infrastructure for improved sanitation and water treatment, however this would impose a huge financial burden on already impoverished nations [4]. Therefore, the development of an effective, safe, and inexpensive typhoid vaccine that can be administered in endemic areas is urgently required [5-7].
Two typhoid vaccines are currently available in the US and are widely used by foreign travelers to endemic areas [6, 7]. The first of these is a parenteral subunit vaccine consisting of the Vi capsular polysaccharide of S. Typhi [8], and the second is a live attenuated oral vaccine, which has been shown to be a safe and effective way to induce strong protective immunity in immunocompetent individuals [6, 9, 10]. Much of our knowledge about the mechanism of protective immunity to Salmonella infection comes from the development and study of animal models [10, 11]. In particular, laboratories have studied acquired protective immunity to Salmonella after vaccination of mice with attenuated strains of S. enterica serovar Typhimurium (hereafter referred to as S. Typhimurium) [12]. Immunization of susceptible mice with these live vaccine strains confers solid protective immunity to challenge with virulent Salmonella, and this protection is known to require both cellular and humoral immunity [13-18]. Many of these studies have emphasized an essential role for Salmonella-specific CD4+ Th1 cells in the acquisition of protective immunity by vaccination [19]. Importantly, these Salmonella-specific Th1 cells secrete the cytokine IFN-γ, which is capable of activating infected macrophages to kill intracellular bacteria [20].
Although Th1 cells are essential for immunity to Salmonella infection, the timing and development of Salmonella-specific Th1 cells has not been carefully examined in vivo. Studies of T cell memory development in other disease models suggest that sustained access to antigen can be an important variable for the optimal maturation and development of long-term memory responses in some situations, but not in others [21-24]. For example, in a mouse model of systemic Listeria monocytogenes infection, CD8 T cells require a very short period of antigen exposure in order to differentiate and develop into memory cells [21]. However, in another model, sustained engagement of the T cell receptor (TCR) by cognate ligand was required throughout the expansion phase for the development of CD4 memory cells [24]. It is not yet clear whether this requirement for sustained TCR stimulation represents an intrinsic difference between CD4 and CD8 T cell effector/memory development or whether a period of prolonged antigenic stimulation might be specifically required for memory T cell development in some disease models, but not in others.
As CD4 Th1 cells are essential for the induction of acquired immunity following vaccination of mice with attenuated S. Typhimurium, we sought to determine the length of time that mice would need to be colonized with a live vaccine strain in order for optimal Th1 development and vaccine-mediated protection to be induced. Our data demonstrate that the induction of robust protective immunity requires at least one week of antigenic exposure while maximal development of Th1 responses takes considerably longer.
Materials and Methods
Mouse strains
C57BL/6 mice were purchased from the National Cancer Institute (Frederick, MD) and used at 6-12 weeks of age. CD90.1 RAG-deficient SM1 TCR transgenic mice have been described previously [25, 26], and were bred in our facility. All mice were housed in specific pathogen-free conditions and cared for in accordance with Research Animal Resources (RAR) practices at the University of Minnesota.
Salmonella infection, vaccination, and antibiotic treatment
S. Typhimurium strain BRD509 (AroA−D−) was grown overnight in Luria-Bertani broth without shaking and diluted in PBS after determining bacterial concentrations using a spectrophotometer. Mice were vaccinated orally by gavage with 5×109 bacteria (live or heat-killed), immediately following administration of 100ul of a 5% NaHCO3 solution. In all infection experiments, the actual bacterial dose was confirmed by plating serial dilutions onto MacConkey’s agar plates and incubating overnight at 37°C. Some mice were treated with Enrofloxacin (Baytril) at 2mg/ml in their drinking water, beginning on various days post-infection. Five days after antibiotic withdrawal (six weeks after primary infection), mice were infected with 5×107 SL1344 and monitored daily for survival. When moribund (displaying no movement when gently prodded with an index finger), mice were euthanized by cervical dislocation as stipulated by our animal care protocol. In some experiments, mice were immunized intravenously with heat-killed S. Typhimurium (approximately 108 bacteria).
Bacterial colonization in vivo
On days two, three, four, and seven, spleens and mesenteric lymph nodes (MLNs) from vaccinated mice were removed and homogenized in Eagle’s Hanks amino acids (EHAA; Gibco) containing 2% FBS. Serial dilutions were plated on MacConkey’s agar plates and incubated overnight at 37°C, and bacterial counts were calculated for each organ.
Salmonella-specific antibody responses
Blood was recovered retro-orbitally from infected mice and sera collected after centrifugation. High protein binding plates (Costar Inc., Corning, NY) were coated overnight with heat-killed S. Typhimurium (HKST) diluted in 0.1M NaHCO3. After incubation in 10% FBS/PBS for one hour at 37°C, plates were washed in PBS/0.5% Tween20 and serum samples added in serial dilutions. After a two-hour incubation at 37°C, plates were washed before biotin-conjugated antibody specific for the desired antibody isotype was added to each well (BD Bioscience and eBioscience). After further incubation at 37°C, plates were washed and incubated with ExtrAvidin-Peroxidase (Sigma-Aldrich) diluted in 10% FCS/PBS. Plates were washed and a 3,3′, 5,5′ tetramethylbenzidine (TMB) substrate (BD OptEIA TMB Substrate Reagent Set, BD Biosciences) was used to develop plates. After color-change was observed, 50μl of 2N H2SO4 was added to plates to stop the reaction and plates were analyzed using an ELISA plate reader (SpectraMax M2, Molecular Devices).
Adoptive transfer of SM1 T cells
Spleen and lymph nodes (cervical, axillary, brachial, inguinal, and mesenteric) of RAG-deficient, CD90.1 congenic, SM1 TCR transgenic mice were harvested. After generating a single-cell suspension, the percentage of SM1 cells was determined using a small aliquot of this suspension and Abs to CD4+Vβ2+ SM1 cells, and the total number of SM1 cells was calculated. SM1 cells were then incubated with CFSE at 37°C for eight minutes, with shaking every 2-3 minutes. Cells were washed twice in cold HBSS before adjusting the concentration and injecting 1-2 × 106 SM1 T cells into the lateral tail vein of recipient C57BL/6 mice.
Flow cytometry
A single-cell suspension was generated from harvested mouse MLNs and spleens. Samples were incubated on ice in the dark for 30 minutes in FACS staining buffer (Hank’s Balanced Salt Solution containing 2% FBS and 0.1% sodium azide) containing primary Abs. FITC-, PE-, PE-Cy7-, eFluor450-, or APC-conjugated Abs specific for CD4, CD11a, CD90.1, Vβ2, IFNγ, and TNFα were purchased from eBiosciences and BD Biosciences. After staining, cells were analyzed by flow cytometry using a FACS Canto, and data were analyzed using FlowJo software (Tree Star).
Tracking SM1 cells in vivo
After i.v. injection of 5ug of flagellin peptide 427-441 (VQNRFNSAITNLGNT) [27] or oral infection with live Salmonella, spleens and MLNs were harvested on days 3 and 10 into EHAA medium containing 2% FBS. Cells were stained, exactly as described above. The percentage of SM1 cells per organ was determined, as well as the activation and expansion of SM1 cells, using the cell surface marker CD11a and CFSE dye dilution, respectively, using flow cytometry.
Detection of in vivo cytokine production
Mice were vaccinated with BRD509 and some were treated with antibiotics beginning on various days following infection, as described above. Six weeks after vaccination, and five days after antibiotic withdrawal, mice were injected i.v. with 108 heat-killed BRD509 to activate Salmonella-specific T cells [28]. Six hours later, spleens and mesenteric lymph nodes were harvested into EHAA containing 2% FBS, and a single-cell suspension was generated. Following rapid surface staining on ice, cells were fixed with formaldehyde, permeabilized using saponin (Sigma-Aldrich), and stained intracellularly using cytokine-specific Abs.
Statistical analysis
Statistical analysis was performed using Prism 5 for MAC OS X (GraphPad Software, Inc., San Diego, CA). Groups of normally distributed data were compared using an unpaired two-tailed t test. Survival data was compared using a Log-rank (Mantel-Cox) test. Differences were considered significant (*) with a p value of 0.01 to 0.05, very significant (**) with a p value of 0.001 to 0.01, and extremely significant (***) with a p value of <0.001.
Results
Elimination of live vaccine strain colonization using antibiotic treatment
Fluoroquinolones, such as ciprofloxacin, are commonly used to treat typhoid in endemic areas [29]. Enrofloxacin is a veterinary fluoroquinolone derivative known to act on the type-II topoisomerase DNA gyrase of Gram-negative bacteria, thereby inhibiting DNA synthesis [30]. Our laboratory previously reported that antibiotic treatment of susceptible mice with enrofloxacin rapidly halted the growth of virulent bacteria from spleen and mesenteric lymph nodes (MLNs) [31]. Antibiotic treatment of mice immunized with a live vaccine strain of S. Typhimurium also hindered the development of protective immunity to rechallenge with a virulent strain [31]. These findings prompted us to examine the clearance of this vaccine strain in more detail, and we hypothesized that early treatment of vaccinated mice with antibiotics rapidly eliminates attenuated bacteria, and limits the available antigen to induce protective immunity.
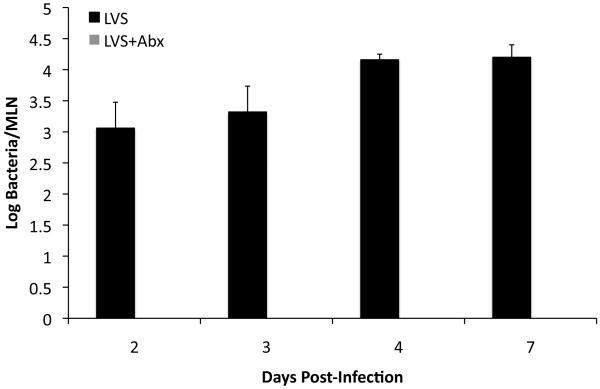
Initially, we examined whether antibiotic treatment actually led to the efficient removal of live attenuated Salmonella, using a strain that grows slowly in vivo due to an inability to generate essential metabolites. C57BL/6 mice were immunized orally with an AroA−D− live vaccine strain (LVS) of S. Typhimurium and treated with enrofloxacin in their drinking water beginning two days post-immunization. After 24 hours, bacteria were undetectable in the MLNs of antibiotic-treated mice but were still present in the MLNs of mice not treated with antibiotics (Fig. 1). Therefore, enrofloxacin rapidly eliminates a vaccine strain of Salmonella in vivo and can be used as a simple intervention to modulate the period of antigen exposure during live vaccination.
Figure 1. Treatment with enrofloxacin rapidly eliminates LVS Salmonella from mouse MLNs.
C57BL/6 mice were orally infected with 5 × 109 attenuated S. Typhimurium (BRD509). Some mice were treated with enrofloxacin in their drinking water beginning 2 dpi. MLNs were harvested from infected mice at various time points after infection, and bacterial loads were determined by plating organ homogenates on MacConkey’s agar. Data show mean bacterial load +/− SD for three to five mice per time point.
Seven days of live vaccine colonization is required for the development of robust protective immunity to Salmonella
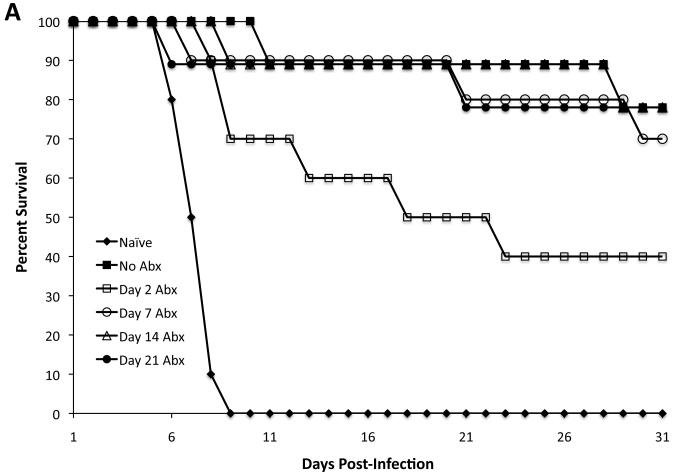
Live vaccine strains of Salmonella typically colonize the host for 3-5 weeks after immunization and induce protective immunity against secondary Salmonella infection. We used antibiotic treatment to define the minimum period of live vaccine colonization required to develop protective immunity. Groups of mice were vaccinated orally with LVS S. Typhimurium and administered antibiotics starting 2, 7, 14, or 21 days later. All antibiotic-treated mice continued treatment until 37 days post-infection followed by five days of untreated drinking water [31]. At 42 days post-vaccination, each group of mice was challenged with a virulent strain of S. Typhimurium and protection against infection evaluated. Mice that received antibiotics beginning 7, 14, and 21 days post-immunization displayed marked resistance to Salmonella infection when compared to naïve mice (Fig. 2A). However, mice that received antibiotics starting 2 days after live vaccination displayed a significantly reduced ability to resist subsequent Salmonella infection (Fig. 2A). Therefore, the development of protective immunity to Salmonella infection requires between 2 and 7 days of colonization with a live vaccine strain.
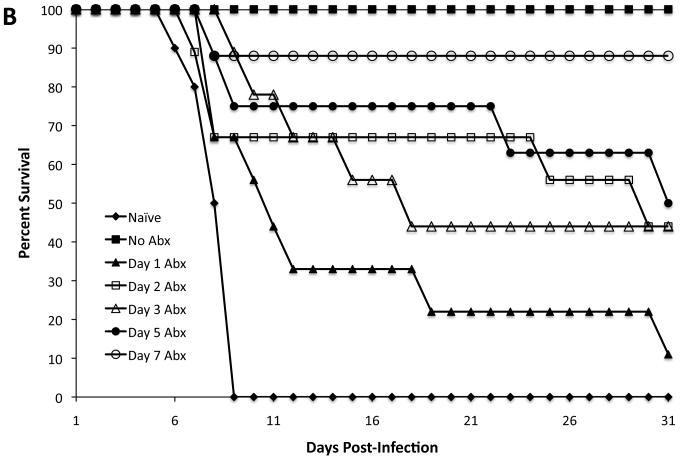
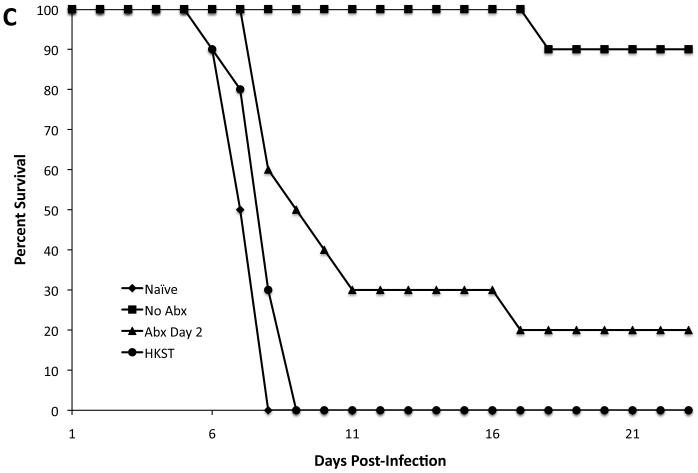
Figure 2. One week of exposure to bacterial antigens leads to protective immunity.
C57BL/6 mice were orally infected with 5 × 109 live or heat-killed (HKST) attenuated S. Typhimurium (BRD509). Some mice were treated with enrofloxacin in their drinking water beginning at various time points after infection. Five days after antibiotic withdrawal (six weeks post-infection), mice were rechallenged with 5 × 107 virulent S. Typhimurium. Data show the percentage of survival of infected mice. (A) There was a statistically significant difference between No Abx and Day 2 Abx (*p=0.0369). There was no statistically significant difference between No Abx and Day 7 Abx or No Abx and Day 14 Abx. (B) There was a statistically significant difference between No Abx and Day 1 Abx (***p=0.0001), No Abx and Day 2 Abx (*p=0.0101), No Abx and Day 3 Abx (*p=0.0101), and No Abx and Day 5 Abx (*p=0.0175). There was no statistically significant difference between No Abx and Day 7 Abx (p=0.2888). (C) There was no statistically significant difference between naïve and HKST (p=0.0620).
In a second series of experiments, we further defined this narrow time period by intervening with antibiotic treatment at 1, 2, 3, 5, or 7 days following live vaccination. Groups of mice were orally immunized with LVS S. Typhimurium, treated with antibiotics, challenged with virulent S. Typhimurium, and monitored for the ability to resist infection. Vaccinated mice that were treated with antibiotics as early as day 1, 2, 3, or 5 displayed markedly reduced ability to resist Salmonella infection (Fig. 2B). In contrast, the vast majority of mice that were antibiotic treated beginning 7 days post vaccination were able to resist Salmonella infection (Fig. 2B). Together, these data demonstrate that mice require colonization for around 7 days with a live vaccine strain in order to develop robust protective immunity to Salmonella infection.
As an additional control, we compared protective immunity following vaccination with live S. Typhimurium versus heat-killed S. Typhimurium (HKST). However, mice immunized with HKST were not protected against rechallenge with virulent S. Typhimurium, and displayed similar survival times as naïve control mice (Fig. 2C).
Antibiotic treatment does not affect initial CD4 T cell activation and expansion
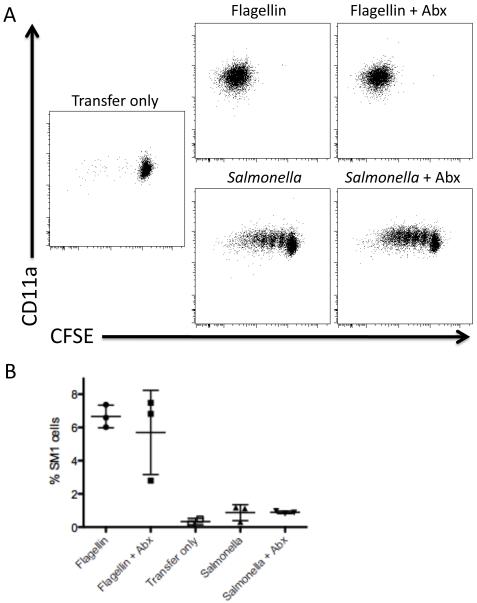
These results demonstrated that antibiotic treatment during vaccination has a profound detrimental effect on the development of protective immunity. However, rather than simply reducing access to antigen, it seemed possible that antibiotics could have a direct effect on CD4 T cell activation or expansion. In order to examine this issue, we monitored the effect of antibiotic treatment on Salmonella-specific CD4 T cell expansion following peptide immunization or administration of live bacteria. C57BL/6 mice were adoptively transferred with Salmonella-specific SM1 CD4 T cells and immunized with the relevant cognate antigen (flagellin peptide 427-441) or vaccinated with Salmonella [25, 27] while also being treated with antibiotics. SM1 T cells were activated to undergo cell division and clonally expand in mice treated with antibiotics (Fig. 3A), indicating that antibiotic treatment alone does not hinder the ability of CD4 T cells to be activated or expand in vivo. Similar results were also obtained when SM1 T cell responses to live bacteria were examined (Fig. 3A). Statistical analysis showed that clonal expansion in groups of mice treated with antibiotics versus untreated mice was not statistically significant (Fig. 3B). Thus, antibiotic treatment does not appear to directly inhibit the ability of CD4 T cell to respond to antigen or live bacteria.
Figure 3. CD4 T cells become activated and expand in the presence of antibiotics.
C57BL/6 mice were adoptively transferred with 1 × 106 CFSE-stained CD90.1+ SM1 T cells. Some of the mice were administered antibiotics in their drinking water. The following day, mice were injected i.v. with 5ug flagellin peptide 427-441 or were vaccinated orally with 5 × 109 attenuated S. Typhimurium (BRD509). Three days after injection, MLNs were harvested, and SM1 T cells were identified by flow cytometry staining for CD4 and CD90.1. Plots show (A) CD11a surface staining, CFSE dye dilution, and (B) the percentage of SM1 T cells from mice that were flagellin-injected +/− antibiotics and vaccinated +/− antibiotics. Differences between these groups (Flagellin versus Flagellin + Abx; and Salmonella versus Salmonella + Abx) are not statistically significant.
Two weeks of live vaccine colonization is required for optimal development of Salmonella-specific Th1 responses
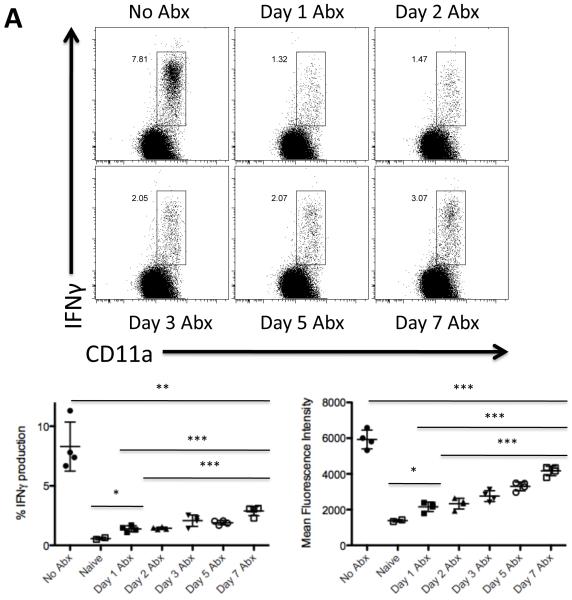
Endogenous Salmonella-specific CD4 T cell responses can be visualized directly ex vivo following brief stimulation in vivo with heat-killed Salmonella Typhimurium (HKST) [28, 32]. In order to examine the requirement for vaccine strain colonization on Th1 development, we monitored the development of Salmonella-specific IFN-γ-producing CD4 T cells in parallel to the protection experiments outlined above. Mice that received a live vaccine strain of Salmonella without antibiotic treatment generated an expanded population of CD11aHi cells that were able to produce IFN-γ following restimulation with HKST (Fig. 4A, No Abx). In marked contrast, vaccinated mice that were treated with antibiotics beginning one day later had a severely impaired Salmonella-specific Th1 response (Fig. 4A, Day 1). A weak Salmonella-specific Th1 response was also detected in mice colonized for 2-5 days before receiving antibiotic treatment (Fig. 4A, Day 2, 3, and 5). However, a large increase in the Th1 response was noted in groups of mice exposed to live Salmonella for 7 days before antibiotic treatment started (Fig. 4A, Day 7), although this response remained substantially lower than the maximal response detected without any antibiotic treatment (Fig. 4A, No Abx). Furthermore, in addition to the lower percentage of IFN-γ-producing cells in antibiotic-treated mice, CD4 Th1 cells from these groups also produced a lower amount of IFN-γ on a per cell basis (Fig. 4A).
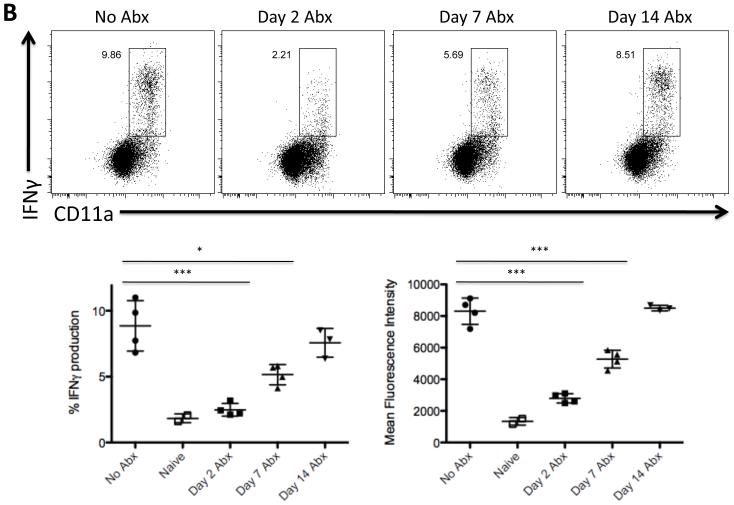
Figure 4. Exposure to bacterial antigens correlates with development of Salmonella-specific Th1 development.
C57BL/6 mice were orally infected with 5 × 109 attenuated S. Typhimurium (BRD509). Some mice were treated with enrofloxacin in their drinking water beginning at various time points after infection. Five days after antibiotic withdrawal (six weeks post-infection), CD4 Th1 cell responses in the spleen were examined by restimulation with Salmonella lysate and direct ex vivo detection of intracellular cytokine production. FACS plots show IFNγ production by gated CD4 T cells in response to 6 h of stimulation with Salmonella lysate. Graphs show mean percentage of IFNγ+ CD4 T cells +/− SD (left) and mean fluorescence intensity +/− SD of IFNγ-producing CD4 T cells (right) from four mice per group. Differences between the following groups are statistically significant: (A) %IFNγ production: No Abx and Day 7 Abx (**p=0.0021), Day 1 Abx and Day 7 Abx (***p=0.0007), Day 2 Abx and Day 7 Abx (***p=0.0004), Naïve and Day 1 Abx (*p=0.0141); Mean Fluorescence Intensity: No Abx and Day 7 Abx (***p=0.0009), Day 1 Abx and Day 7 Abx (***p<0.0001), Day 2 Abx and Day 7 Abx (***p<0.0001), Naïve and Day 1 Abx (*p=0.0174); and (B) %IFNγ production: No Abx and Day 2 Abx (***p=0.0007), No Abx and Day 7 Abx (*p=0.0115); Mean Fluorescence Intensity: No Abx and Day 2 Abx (***p<0.0001), No Abx and Day 7 Abx (***p=0.0009).
The clear recovery of Th1 responses in mice that were colonized with a vaccine strain for 7 days, correlated with the ability of these mice to resist Salmonella infection (Fig. 3). However, as noted above this Th1 response was still lower than that detected in vaccinated mice that did not receive any antibiotics. Therefore, we examined whether further delaying treatment would additionally amplify the Salmonella-specific Th1 response. Indeed, maximal Th1 effector function was observed if antibiotic treatment was delayed for 14 days after vaccination, as IFN-γ-producing CD4 T cells from this group were equivalent in percentage and MFI to those detected from the untreated but vaccinated mice (Fig. 4B). Therefore, although a single week of live vaccine colonization was sufficient to develop effective protective immunity against Salmonella infection, up to 2 weeks of exposure was required for optimal Salmonella-specific Th1 development.
Effect of antibiotic treatment on Salmonella-specific antibody
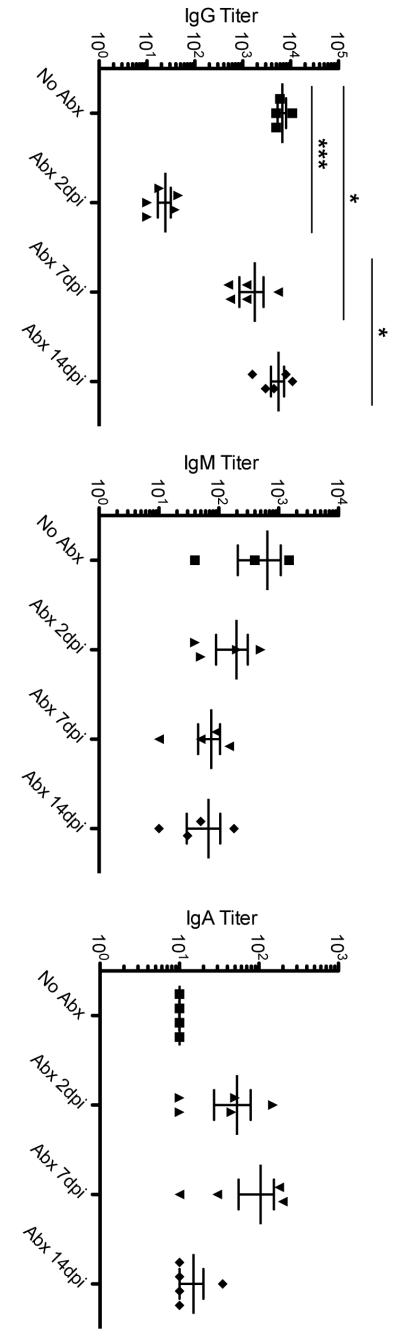
Our data correlate the development of Salmonella-specific Th1 cells with the generation of protective immunity against infection but suggest that robust protection can still occur while Th1 responses are sub-optimal. We examined whether antibody responses occurred in vaccinated mice administered antibiotics, since this response might complement a weak Th1 response and thus allow protective immunity to develop. As expected, immunized mice developed Salmonella-specific IgM, IgG, and IgA responses even after antibiotic treatment (Fig. 5). However, mice that were treated beginning two days post-infection developed very weak Salmonella-specific IgG responses, which correlated with the absence of protective immunity exhibited by these mice. These data suggest that IgG most likely plays an additional protective role when Th1 responses are sub-optimal.
Figure 5. Development of Salmonella-specific serum antibody responses in antibiotic-treated mice.
C57BL/6 mice were orally infected with 5 × 109 attenuated S. Typhimurium (BRD509) and some mice treated with antibiotics beginning at various time points after infection. Five days after antibiotic withdrawal (six weeks post-infection), blood was collected and Salmonella-specific antibody responses were examined using isotype-specific antibody ELISAs. The limit of detection is a titer of 10. (A) Differences between the following groups are statistically significant: No Abx and Day 2 Abx (***p=0.0009), No Abx and Day 7 Abx (*p=0.0190), and Day 2 Abx and Day 14 Abx. (B and C) Differences between these groups are not statistically significant.
Discussion
Vaccination is currently bestthe best option for combating infectious diseases [5-7], and live attenuated vaccine strains have proven to be effective at inducing strong protective immunity against several pathogens [6, 9, 10]. Although live attenuated vaccines are widely used they are not always approved for all age groups and multiple doses of live attenuated typhoid vaccine are sometimes required due to poor immunogenicity [5]. Human typhoid remains a considerable problem in many developing countries and development of an improved live attenuated vaccine will require greater understanding of how Salmonella strains induce a protective immune response.
Our laboratory recently used antibiotic treatment of virulent Salmonella strains to examine the development of immunity to Salmonella [31]. In completing these studies we noticed that antibiotic treatment during immunization with a live vaccine strain severely reduced the development of protective immunity. This was somewhat surprising, given previous work where early interruption of Listeria vaccination did not significantly reduce CD8 T cell development [21]. Our current experiments have examined this process in more detail and demonstrate that prolonged colonization with a live vaccine strain of Salmonella is required to develop robust immunity to challenge infection. The most likely explanation for this finding is that early antibiotic treatment hinders the development of Th1 cells by rapidly eliminating a source of bacterial antigens. Indeed, our TCR transgenic adoptive transfer experiments show that antibiotics do not directly affect the ability of CD4 T cells to proliferate and become activated in vivo. Thus, our data are most consistent with a model where optimal development of protective Salmonella-specific Th1 cells requires between 1-2 weeks of antigen exposure in order for prolonged stimulation of responding CD4 T cells.
Other studies have noted that sustained antigen exposure can enhance the survival of CD4 T cells or allow greater development of effector potential [22-24]. In experiments directly examining CD4 Th1 development, we found that antibiotic interruption up to 2 weeks after vaccination could hinder the maturation of Th1 cells. The longer time period for optimal development of Th1 cells versus the development of protective immunity, may be explained by the challenge dose used in our studies, i.e. one week of vaccine stimulation allows sufficient Th1 development to protect against the challenge dose used in our experiments, while longer exposure to antigen is required for maximal Th1 development. An alternative explanation is that the significant Salmonella-specific antibody responses detected in vaccinated mice allow protective immunity to develop even with sub-optimal Th1 responses. Future experiments will compare each of these possibilities, but it seems likely that Salmonella-specific antibody contributes significantly to immunity to secondary infection in this model.
Our finding that protective immunity requires at least one week of colonization with a live vaccine strain of Salmonella and that optimal Th1 development requires two weeks, is important with respect to the design of typhoid vaccination and provides an explanation for why multiple doses of live attenuated Salmonella strains are often required to generate robust immunity. Indeed, if the attenuation of Salmonella strains simply shortens the time period of colonization then such strains would be expected to generate weak Th1 responses and provide poor immunity. Thus, focused development of highly attenuated strains that maintain prolonged in vivo colonization should be a goal of future vaccine design. In conclusion, our data suggest a surprisingly long time period of CD4 T cell stimulation for the development of effective anti-Salmonella immunity. These studies should aid in the design of effective vaccines that can be widely distributed in developing nations where the burden of typhoid is most detrimental.
Acknowledgements
This work was supported by grants from the National Institutes of Health to SJM (AI055743 and AI56172).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Crump JA, Luby SP, Mintz ED. The global burden of typhoid fever. Bull World Health Organ. 2004;82(5):346–53. [PMC free article] [PubMed] [Google Scholar]
- [2].Farthing MJ. Diarrhoea: a significant worldwide problem. Int J Antimicrob Agents. 2000;14(1):65–9. doi: 10.1016/s0924-8579(99)00149-1. [DOI] [PubMed] [Google Scholar]
- [3].Parry CM, Hien TT, Dougan G, White NJ, Farrar JJ. Typhoid fever. N Engl J Med. 2002;347(22):1770–82. doi: 10.1056/NEJMra020201. [DOI] [PubMed] [Google Scholar]
- [4].Mara DD. Water, sanitation and hygiene for the health of developing nations. Public Health. 2003;117(6):452–6. doi: 10.1016/S0033-3506(03)00143-4. [DOI] [PubMed] [Google Scholar]
- [5].Crump JA, Mintz ED. Global trends in typhoid and paratyphoid Fever. Clin Infect Dis. 2010;50(2):241–6. doi: 10.1086/649541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Guzman CA, Borsutzky S, Griot-Wenk M, Metcalfe IC, Pearman J, Collioud A, et al. Vaccines against typhoid fever. Vaccine. 2006;24(18):3804–11. doi: 10.1016/j.vaccine.2005.07.111. [DOI] [PubMed] [Google Scholar]
- [7].Whitaker JA, Franco-Paredes C, del Rio C, Edupuganti S. Rethinking typhoid fever vaccines: implications for travelers and people living in highly endemic areas. J Travel Med. 2009;16(1):46–52. doi: 10.1111/j.1708-8305.2008.00273.x. [DOI] [PubMed] [Google Scholar]
- [8].Acharya IL, Lowe CU, Thapa R, Gurubacharya VL, Shrestha MB, Cadoz M, et al. Prevention of typhoid fever in Nepal with the Vi capsular polysaccharide of Salmonella typhi. A preliminary report. N Engl J Med. 1987;317(18):1101–4. doi: 10.1056/NEJM198710293171801. [DOI] [PubMed] [Google Scholar]
- [9].Hoiseth SK, Stocker BA. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature. 1981;291(5812):238–9. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- [10].Pasetti MF, Levine MM, Sztein MB. Animal models paving the way for clinical trials of attenuated Salmonella enterica serovar Typhi live oral vaccines and live vectors. Vaccine. 2003;21(5-6):401–18. doi: 10.1016/s0264-410x(02)00472-3. [DOI] [PubMed] [Google Scholar]
- [11].Santos RL, Zhang S, Tsolis RM, Kingsley RA, Adams LG, Baumler AJ. Animal models of salmonella infections: enteritis versus typhoid fever. Micrtobes Infect. 2001;3:1335–44. doi: 10.1016/s1286-4579(01)01495-2. [DOI] [PubMed] [Google Scholar]
- [12].Mastroeni P, Chabalgoity JA, Dunstan SJ, Maskell DJ, Dougan G. Salmonella: immune responses and vaccines. Vet J. 2001;161(2):132–64. doi: 10.1053/tvjl.2000.0502. [DOI] [PubMed] [Google Scholar]
- [13].Nauciel C. Role of CD4+ T cells and T-independent mechanisms in aquired resistance to Salmonella typhimurium infection. J Immunol. 1990;145:1265–9. [PubMed] [Google Scholar]
- [14].Hess J, Ladel C, Miko D, Kaufmann SH. Salmonella typhimurium aroA- infection in gene-targeted immunodeficient mice: major role of CD4+ TCR-alpha beta cells and IFN-gamma in bacterial clearance independent of intracellular location. J Immunol. 1996;156:3321–26. [PubMed] [Google Scholar]
- [15].Ravindran R, Foley J, Stoklasek T, Glimcher LH, McSorley SJ. Expression of T-bet by CD4 T cells is essential for resistance to Salmonella infection. J Immunol. 2005;175(7):4603–10. doi: 10.4049/jimmunol.175.7.4603. [DOI] [PubMed] [Google Scholar]
- [16].McSorley SJ, Jenkins MK. Antibody is required for protection against virulent but not attenuated Salmonella enterica serovar typhimurium. Infect Immun. 2000;68(6):3344–8. doi: 10.1128/iai.68.6.3344-3348.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Mastroeni P, Simmons C, Fowler R, Hormaeche CE, Dougan G. Igh-6(−/−) (B-cell-deficient) mice fail to mount solid aquired resistance to oral challenge with virulent Salmonella enterica serovar typhimurium and show impaired Th1 T-cell responses to Salmonella antigens. Infect Immun. 2000;68:46–53. doi: 10.1128/iai.68.1.46-53.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Cunningham AF, Gaspal F, Serre K, Mohr E, Henderson IR, Scott-Tucker A, et al. Salmonella induces a switched antibody response without germinal centers that impedes the extracellular spread of infection. J Immunol. 2007;178(10):6200–7. doi: 10.4049/jimmunol.178.10.6200. [DOI] [PubMed] [Google Scholar]
- [19].Ravindran R, McSorley SJ. Tracking the dynamics of T-cell activation in response to Salmonella infection. Immunology. 2005;114(4):450–8. doi: 10.1111/j.1365-2567.2005.02140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].VanCott JL, Chatfield SN, Roberts M, Hone DM, Hohmann EL, Pascual DW, et al. Regulation of host immune responses by modification of Salmonella virulence genes. Nat Med. 1998;4:1247–52. doi: 10.1038/3227. [DOI] [PubMed] [Google Scholar]
- [21].Mercado R, Vijh S, Allen SE, Kerksiek K, Pilip IM, Pamer EG. Early programming of T cell populations responding to bacterial infection. J Immunol. 2000;165(12):6833–9. doi: 10.4049/jimmunol.165.12.6833. [DOI] [PubMed] [Google Scholar]
- [22].Bajenoff M, Wurtz O, Guerder S. Repeated antigen exposure is necessary for the differentiation, but not the initial proliferation, of naive CD4(+) T cells. J Immunol. 2002;168(4):1723–9. doi: 10.4049/jimmunol.168.4.1723. [DOI] [PubMed] [Google Scholar]
- [23].Blair DA, Lefrancois L. Increased competition for antigen during priming negatively impacts the generation of memory CD4 T cells. Proc Natl Acad Sci U S A. 2007;104(38):15045–50. doi: 10.1073/pnas.0703767104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Obst R, van Santen HM, Mathis D, Benoist C. Antigen persistence is required throughout the expansion phase of a CD4(+) T cell response. J Exp Med. 2005;201(10):1555–65. doi: 10.1084/jem.20042521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].McSorley SJ, Asch S, Costalonga M, Rieinhardt RL, Jenkins MK. Tracking Salmonella-specific CD4 T cells in vivo reveals a local mucosal response to a disseminated infection. Immunity. 2002;16:365–77. doi: 10.1016/s1074-7613(02)00289-3. [DOI] [PubMed] [Google Scholar]
- [26].Srinivasan A, Foley J, Ravindran R, McSorley SJ. Low-dose salmonella infection evades activation of flagellin-specific CD4 T cells. J Immunol. 2004;173(6):4091–9. doi: 10.4049/jimmunol.173.6.4091. [DOI] [PubMed] [Google Scholar]
- [27].McSorley SJ, Cookson BT, Jenkins MK. Characterization of CD4+ T cell responses during natural infection with Salmonella typhimurium. J Immunol. 2000;164(2):986–93. doi: 10.4049/jimmunol.164.2.986. [DOI] [PubMed] [Google Scholar]
- [28].Srinivasan A, Foley J, McSorley SJ. Massive Number of Antigen-Specific CD4 T Cells during Vaccination with Live Attenuated Salmonella Causes Interclonal Competition. J Immunol. 2004;172(11):6884–93. doi: 10.4049/jimmunol.172.11.6884. [DOI] [PubMed] [Google Scholar]
- [29].Crum NF. Current trends in typhoid Fever. Curr Gastroenterol Rep. 2003;5(4):279–86. doi: 10.1007/s11894-003-0064-0. [DOI] [PubMed] [Google Scholar]
- [30].Cattoir V, Nordmann P. Plasmid-mediated quinolone resistance in gram-negative bacterial species: an update. Curr Med Chem. 2009;16(8):1028–46. doi: 10.2174/092986709787581879. [DOI] [PubMed] [Google Scholar]
- [31].Griffin A, Baraho-Hassan D, McSorley SJ. Successful treatment of bacterial infection hinders development of acquired immunity. J Immunol. 2009;183(2):1263–70. doi: 10.4049/jimmunol.0900772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Srinivasan A, Salazar-Gonzalez RM, Jarcho M, Sandau MM, Lefrancois L, McSorley SJ. Innate immune activation of CD4 T cells in salmonella-infected mice is dependent on IL-18. J Immunol. 2007;178(10):6342–9. doi: 10.4049/jimmunol.178.10.6342. [DOI] [PubMed] [Google Scholar]