Abstract
Genetic alterations can determine the natural history of cancer and its treatment response. With further advances in DNA sequencing technology, multiple novel genetic alterations will be discovered which could be exploited as prognostic, predictive and pharmacodynamic biomarkers in the development and use of cancer therapeutics. As such, the importance in clinical practice of efficient and robust somatic mutation testing in solid tumours cannot be overemphasized in the current era of personalized medicine. However, significant challenges remain regarding the testing of genetic biomarkers in clinical practice. Reliance on archived formalin fixed, paraffin embedded tumour, obtained from diagnostic biopsies, for testing somatic genetic alterations could restrict the scientific community in asking relevant questions about a patient’s cancer biology. Problems inherent with using formalin fixed, archival tissue are well recognized and difficult to resolve. It could be argued that to achieve rapid and efficient incorporation of genetic biomarkers into clinical practice, somatic mutation testing in cancer patients should be simpler, less invasive using a readily available clinical sample, whilst maintaining robustness and reproducibility. In this regard, use of circulating free DNA (cfDNA) from plasma or serum as an alternative and/or additional source of DNA to test cancer specific genetic alterations is an attractive proposition. In light of encouraging results from recent studies, this mini review will discuss the current role and future potential of somatic mutation testing from circulating or cell free DNA derived from the blood of patients with solid tumours.
Keywords: Mutation testing, Circulating free DNA, Genetic biomarkers, Cancer
Introduction
Genetic alterations that activate proto-oncogenes or inactivate tumour suppressor genes are key steps in the progression of normal tissue to malignancy (Kopnin 2000). In familial cancer syndromes, germline mutations predispose to malignant transformation and often result in earlier onset disease (Gatalica and Torlakovic 2008; Sluiter and van Rensburg 2011). In contrast, in more common sporadic cancers, the tumour usually originates from clonal expansion of a transformed cell through the accumulation of serial somatic mutations (Stratton et al. 2009). Currently, approximately 400 cancer related genes have been identified (http://www.sanger.ac.uk/genetics/CGP/Census). With recent advances in DNA sequencing technology, it is envisaged that multiple novel somatic genetic alterations will be discovered. These novel genetic variants may provide important biomarker information to aid prediction of the natural history of cancer or treatment response. In an era of targeted therapeutics, it is inevitable that testing the molecular characteristics of patients’ tumours will be necessary before recruitment into clinical trials to achieve the ultimate goal of personalized medicine. Already, KRAS mutation status is routinely determined in tumours of patients with metastatic colorectal cancer and only those patients with wild type KRAS are offered cetuximab, a monoclonal antibody to EGFR receptor. As most biomarkers predictive for treatment response are genetic biomarkers, the importance of efficient and standardized testing for somatic mutations is vital. However, currently significant challenges remain in testing genetic biomarkers in clinical practice.
Firstly, tumour tissue is not always available for genotyping. For example, diagnosis in a significant number of patients with lung cancer is based purely on cytology and as a result there will be insufficient material available for comprehensive molecular profiling (Sequist et al. 2009). This imposes immense difficulties if patients need to be selected or stratified by mutation status before entering into clinical trials or for routine clinical treatment with new biological targeted agents. Secondly, even if biopsy material is available, the quality and quantity of tumour tissue is often variable. Archived formalin fixed, paraffin embedded (FFPE) tumour acquired for diagnosis usually contains a small amount of tumour mixed with stroma, moreover and DNA is usually degraded by formalin fixation (Plesec and Hunt 2009). Thirdly, logistical problems around retrieving patients’ archival FFPE tumour blocks are also substantial. Within large clinical trials, samples have to be retrieved from many study sites and across different countries which is costly and often slow. As an illustration of this problem, four large studies of erlotinib and gefitinib in non-small cell lung cancer (NSCLC) have reported molecular analysis but data was derived from <30% of the patients enrolled (Riely et al. 2006). To achieve rapid and efficient incorporation of genetic biomarkers into clinical practice, somatic mutation testing in cancer patients should be simpler, less invasive from a readily available clinical sample with maintained robustness and reproducibility. In this regard, the use of circulating free DNA (cfDNA) from plasma or serum as an alternative source of DNA to test cancer specific genetic alterations rather than being reliant on archival tumour biopsy is an attractive proposition. In light of encouraging results from recent studies, this mini review will discuss the current role and future potential of somatic mutation testing from circulating or cell free DNA (cfDNA) derived from the blood of patients with solid tumours.
Circulating free DNA (cfDNA) and its potential
Circulating nucleic acids in humans were first described in 1948 by Mandel and Metais (1948). Characterization of these nucleic acids in later studies revealed that they are predominantly double stranded DNAs, circulating in complexes with histones as mono or oligonuleosomes (Stroun et al. 1987; Rumore and Steinman 1990). Low levels of cfDNA can be detected in healthy individuals but increased levels can be found in patients with a number of diseases including patients with cancers (Leon et al. 1977). Four decades after Mandel and Metais’s seminal discovery, it was shown that tumour specific molecular characteristics could be tested using cfDNA isolated from the plasma of cancer patients (Stroun et al. 1987, 1989) providing a potential source of tumour specific information that could be utilized for cancer diagnosis, personalized medicine and cancer prognosis. However, and perhaps surprisingly, the mechanism and source of cfDNA release remains largely uncertain. It has been suggested that cfDNA originates from either malignant or haemopoietic apoptotic and necrotic cells, from the lysis of circulating tumour cells or from active secretion of nucleic acids by tumour cells (Stroun et al. 2000, 2001). The detection of tumour specific DNA alterations such as mutations and methylation in cfDNA confirm that, at least in part, cfDNA is tumour derived and provides a less invasive, more easily accessible source of DNA for genetic analysis than tumour biopsies. This is of increasing clinical importance in cancer medicine with development of targeted agents, the benefit of which is often determined by the presence or absence of genetic mutations within the tumour cells (Van Cutsem et al. 2009; Mok et al. 2009).
Recent advances in PCR based technology have now allowed the analysis of point mutations in EGFR, KRAS, BRAF and PIK3CA genes from cfDNA isolated from patients’ plasma or serum (Kimura et al. 2007; Hodgson et al. 2010; Board et al. 2009, 2010). These genes are of particular importance in determining response to a variety of novel agents in clinical use and in development for the treatment of cancer. The optimal methodology for cfDNA isolation and mutation detection is still unclear with a variety of different techniques and technologies used in the literature. However, combined with recent progress made in this research field and further advances in technology, somatic mutation testing from cfDNA has huge future potential from a cancer therapeutic perspective.
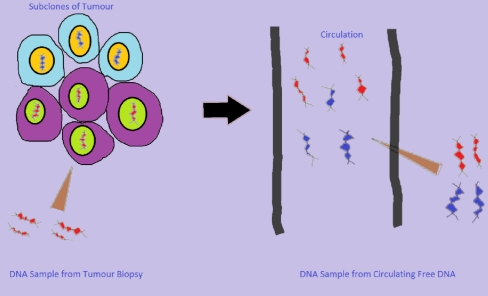
As a biopsy is usually taken from one small part of the tumour, it is debatable whether it represents the whole tumour (Fleischhacker and Schmidt 2008). It has been argued that analysis of cfDNA, on the other hand, might yield information about all subclones within the tumour (Fig. 1; Fleischhacker and Schmidt 2008). Moreover, real time monitoring of the evolution of a tumour is desirable for understanding of genotypic changes that are responsible for cancer recurrence, progression and development of drug resistance. In routine clinical practice, performing serial tumour biopsies is seldom practical or justified and to achieve real time monitoring of tumour dynamics, mutation testing should be minimally invasive and easily repeatable. Blood based somatic mutation testing could potentially meet these requirements. It could also be envisaged that, to prove the mechanism of a drug targeted towards a particular clone of mutant tumour cells, it would be needed to demonstrate that these mutant cells are eradicated after treatment. If a particular tumour derived mutation(s) can be tested from circulating free DNA, disappearance of these mutations from the circulation may confirm that the drug is hitting its intended target. Furthermore, reappearance of these mutations in the blood stream may herald disease recurrence or drug resistance. Lastly, whole genome profiling of cfDNA by next generation sequencing might lead to discovery of novel driver mutations in cancer candidate genes and could significantly advance our understanding of tumour biology.
Fig. 1.
Harvesting tumour derived DNA. Circulating free DNA could represent genetic profile of the tumour better than DNA from biopsy. A primary tumour could contain many subclones of tumour cells, which have different genetic profiles, and as biopsy material is usually obtained from only one small part of the tumour, it might not contain materials from all the subclones (adapted from Fleishhacker et al. 2008)
Current status of somatic mutation testing from cfDNA in cancer
Currently there is no routinely used blood based test for somatic mutation detection in patients with solid tumours. Wide variations in methodologies in analyzing cancer specific mutations in cfDNA in the published studies indicate that reproducibility remains a major issue and technology platforms still need improvement. Standardization of techniques will be necessary before this alternative approach of somatic mutation testing could be incorporated effectively into clinical trials and routine clinical practice.
Lung cancer
Mutation in EGFR occurs in ~35% of NSCLC patients of East Asian origin and ~16% in Western populations (Mok et al. 2009; Rosell et al. 2009). Multiple in-frame deletions in exon 19 and the p.L858R missense mutation in exon 21 comprise 90% of the mutations detected (Kosaka et al. 2004). Studies have confirmed EGFR mutations as a predictive biomarker of treatment response to tyrosine kinase inhibitors, gefitinib and erlotinib (Yang et al. 2008; Sequist et al. 2008; Inoue et al. 2006; Tsao et al. 2005). As such screening for EFGR mutations in NSCLC patients is deemed necessary before offering these drugs to patients. However, NSCLC is frequently inaccessible to tumour biopsy and diagnosis is often based on cytology or fine needle aspirations, from which sufficient DNA for mutation analysis can rarely be obtained. Previous studies investigating mutation status of NSCLC demonstrated that significant proportion of patients will have unknown mutation status because tumour biopsy material is not available or insufficient for genotyping (Riely et al. 2006; Jackman et al. 2009). For this reason cfDNA from patients with NSCLC offers a useful alternative for mutation detection where tumour data is unavailable. Kimura et al. (2006) first reported the feasibility of detection of EGFR mutations in cfDNA extracted from serum of patients with NSCLC. Two years later, the same group published results from EGFR mutation analysis of 42 paired tumour and serum samples (Kimura et al. 2007) using allele-specific amplification refractory mutation testing system combined with scorpion probes (Scorpion-ARMS). It was demonstrated that EGFR mutation status was consistent in 39 (93%) of the 42 paired samples tested. However, this encouraging result was not confirmed in another study conducted by Maheswaran et al., which reported EGFR mutations in only 33% of plasma-derived cfDNA samples from 12 patients whose tumour was positive for EGFR mutation by using Scorpion-ARMS (Maheswaran et al. 2008). More recently, the Spanish Lung Cancer Group reported that of 164 patients with EGFR mutations in tumours, 97 (59%) had an EGFR mutation in serum tested by protein nucleic acids mediated PCR analysis (Rosell et al. 2009). Almost all of the patients who initially responded to gefitinib and erlotinib will eventually develop resistance to the drugs. Development of a secondary EGFR mutation, p.T790M, which inhibits binding of these drugs to the ATP binding pocket of EGFR, accounts for acquired resistance to EGFR targeted treatment in 50% of patients (Engelman and Jänne 2008). Kuang et al. demonstrated that p.T790M mutation can be tested from plasma derived cfDNA highlighting the potential role of mutation testing from cfDNA in monitoring secondary resistance to anticancer treatment (Kuang et al. 2009).
As KRAS mutations are also negative predictors of response to anti-EGFR treatment in lung cancer, recent studies also explored KRAS mutation testing from cfDNA (Ramirez et al. 2003; Wang et al. 2010). Wang et al. (2010) reported their findings from KRAS mutation analysis of DNA extracted from 273 plasma samples and matched tumour tissues from advanced NSCLC patients of East Asian origin. PCR-restriction fragment length polymorphism (PCR–RFLP) combined with denaturing high performance liquid chromatography was used for mutational analysis. KRAS mutation was found in 35 (13%) plasma samples and 30 (11%) tumours. Concordance of mutations between plasma and tumour was 77%. The fact that more mutations were found in plasma compared to tumour raises the possibility of false positive results in plasma samples by the technique employed. However, the heterogeneity of the primary tumours could also potentially explain this discordant result as it is possible that a biopsy did not contain all mutant subclones (Fig. 1). Moreover, cfDNA could also be shed from metastatic disease sites and heterogeneity between primary and metastatic tumours should also be taken into consideration in interpreting the results of the study.
Colorectal cancer
The genetic basic of colorectal cancer is well characterized and importance of genetic biomarkers has been recognized since late 1990s. Mutations in tumour suppressor genes, APC and p53, and proto-oncogene, KRAS, are all implicated in colorectal carcinogenesis. KRAS mutations are detected in up to 40% of colorectal cancers (CRC) (Bos et al. 1987). In CRC, KRAS mutations confer resistance to treatment with EGFR antibodies and only patients with wild type KRAS tumours obtain benefit from these agents (Van Cutsem et al. 2009; Bokemeyer et al. 2009). It is therefore vital that the KRAS mutation status of a patient’s colorectal tumour can be detected to allow patients access to treatment to which there is increased likelihood of benefit.
Several studies have tried to establish the presence of somatic mutations in cfDNA from patients with CRC. Although there is a wide variation in techniques employed, earlier studies have demonstrated; (1) cfDNA can be detected in plasma of CRC patients and its level increases with stage of disease (Diehl et al. 2005), (2) APC, p53 and KRAS mutations can all be detected in cfDNA and could serve as circulating biomarkers in CRC (Wang et al. 2004) and (3) there is a potentially prognostic value of the peresence of cfDNA in resected CRC during postoperative follow up (Ryan et al. 2003). More recently, Diehl et al. (2008) demonstrated that circulating mutant DNA can be used to assess tumour dynamics in patients undergoing multimodality therapies for CRC. Although increasing clinical relevance of BRAF and PIK3CA mutations in CRC is currently being recognized (Di Nicolantonio et al. 2008; Sartore-Bianchi et al. 2009; Perrone et al. 2009), there is no study reporting significance of detecting BRAF and PIK3CA mutations in plasma or serum in CRC patients.
Pancreatic cancer
Although mutation in codon 12 of KRAS is a very common event, occurring in up to 90% of pancreatic cancers, its biological relevance in anticancer drug treatment resistance is still not completely clear. However, considering the scarcity of tumour materials for research in metastatic pancreatic cancer, circulating biomarkers might be able to help elucidate its biology. Preliminary studies demonstrated the feasibility of detecting KRAS mutations in plasma or serum of pancreatic cancer patients (Fleischhacker and Schmidt 2007). Only three important studies, which reflect the current status of cfDNA testing in pancreatic cancer, will be discussed in this review. Castells et al. (1999) studied KRAS mutations in 44 consecutive patients with histologically confirmed primary pancreatic ductal carcinoma using PCR–RFLP in both primary tumours and plasma samples. A control group of 37 patients with chronic pancreatitis was also included in the study. Out of 39 patients in whom both plasma and tissue samples were available, 28 patients (72%) had KRAS mutations in their primary tumours and 9 (23%) had mutations detectable in plasma. Intriguingly, presence of KRAS mutations in plasma was identified as the only independent predictive factor of survival. However, plasma KRAS mutation was also detected in two patients with chronic pancreatitis. Dianxu et al. (2002) tried to establish the diagnostic value of codon 12 KRAS mutations in plasma combined with serum CA19-9 in 58 consecutive patients with a suspected pancreatic mass. Forty-one patients were subsequently diagnosed with pancreatic adenocarcinoma. Mutations in KRAS codon 12 were found in 29 (71%) patients in plasma using PCR–RFLP whereas elevated CA19-9 was found in 30 patients with pancreatic adenocarcinoma demonstrating that plasma KRAS mutation does not have an advantage over CA 19-9 as a diagnostic marker in this setting. Perhaps more encouragingly, circulating KRAS codon 12 mutations could serve as predictive biomarkers in locally advanced pancreatic cancer patients undergoing combined modalities treatment with chemo-radiotherapy and gefitinib in a phase I study (Olsen et al. 2009). KRAS mutations were detected in 5 of 11 patients enrolled in the pre-gefitinib plasma; of those five patients, KRAS mutations became undetectable post therapy in three patients and had overall survival of 8, 11, and 21 months. In contrast, two patients whose post-treatment plasma still had mutant KRAS survived only 2 and 4 months. This study demonstrated that incorporating KRAS status into early phase trials using currently available techniques could produce potentially useful information.
Breast cancer
Somatic PIK3CA mutations occur in ~25% of breast cancer (Bachman et al. 2004; Lee et al. 2005; Levine et al. 2005). As such, mutated PI3K has become an attractive therapeutic target in breast cancer therapy and a number of agents targeting the PIK pathway are currently in clinical development. Parallel with this development, our group demonstrated the feasibility of PIK3CA testing in cfDNA. CfDNA was extracted from plasma and serum samples of 46 patients and four hot spot mutations in PIK3CA gene, p.H1047R, p.H1047L, p.E545K and p.E542K, were analyzed with Scorpion-ARMS. Matched tumour and plasma data was available for 41 cases. Ten (24%) mutations were detected in tumour and of those ten patients, 8 (80%) had mutations in cfDNA isolated from plasma and 6 (60%) had mutations in cfDNA isolated from serum. Concordance between matched tumour and cell free DNA data was 95% (95%CI: 83–99%) and 88% (95% CI: 73–95%) for plasma derived cfDNA and serum derived cfDNA respectively.
Chen et al. (2009) demonstrated in a proof of principle study that tumour specific p53 mutations in plasma could be used to monitor patients’ response to chemotherapy in six patients with stage II and III breast cancer, who underwent neo-adjuvant chemotherapy. Based on these studies, a mutation signature in plasma would be a useful predictive circulating biomarker in breast cancer and confirmation of these early results in lager studies are needed.
Cutaneous melanoma
In cutaneous melanoma, the BRAF gene is mutated in ~60% of cases and p.V600E (c.1799T > A) accounts for more than 90% of BRAF mutations (Brose et al. 2002). In 2007, two studies reported the feasibility of BRAF mutation testing from cfDNA (Daniotti et al. 2007; Yancovitz et al. 2007). Shinozaki et al. (2007) were the first to assess the value of BRAF mutations in serum derived cfDNA as a pharmacodynamic marker to biochemotherapy. Forty-eight patients with stage IV disease were included in the study. Out of 24 responders, 10 (41%) had BRAF mutations in serum before biochemotherapy detected by using a peptide nucleic acid (PNA) clamp and locked nucleic acid (LNA) mediated quantitative real time PCR. In 9 of those 10 patients, BRAF mutations was no longer detectable in serum within 4 weeks of the last cycle of treatment demonstrating potential value of circulating BRAF mutations in treatment response monitoring in metastatic cutaneous melanoma; presence of BRAF mutations in serum was associated with significantly worse overall survival in patients receiving biochemotherapy.
Our group investigated the clinical utility of cfDNA from serum as an alternative source of BRAF mutation testing in 126 metastatic melanoma patients who participated in a phase II study testing the efficacy of AZD6244, a specific MEK1/2 inhibitor (Board et al. 2009). Responses to AZD6244 were observed only in patients whose tumours harboured BRAF mutations. ARMS was used for both mutation testing in tumours and serum. Matched tumour and serum samples were available in 96 cases and 45 (47%) patients have BRAF mutation in tumour and 25 (27%) patients have BRAF mutation in serum derived cfDNA. Based on those data, concordance in BRAF mutation detection was 76% (95% CI 66–84%) and pick up rate in cfDNA was 56% (95% CI 40–70%). Further studies include collection of cfDNA to optimize the use of BRAF mutation analysis in cfDNA to preselect patients for treatment with this targeted therapy. In a separate study, absolute concentration of BRAF p.V600E mutant copies in plasma distinguished patients with invasive melanomas (n = 55) from healthy controls (n = 18) with 97% sensitivity and 83% specificity and concordance between mutations detected in plasma and those of tumour tissue was 80% (Pinzani et al. 2010).
Thyroid cancer
The BRAF p.V600E mutation occurs in ~40% of patients with papillary thyroid carcinoma and is associated with a more aggressive disease (Cohen et al. 2004; Xing et al. 2005). BRAF mutation can be detected in serum derived DNA from patients with papillary thyroid carcinomas (Chuang et al. 2010) and further studies will be needed to determine the clinical utility of mutation testing from cfDNA in this patient group.
Hepatocellular carcinoma
Mutation in TP53 is implicated in pathogenesis of HCC (Hussain et al. 2007). It occurs in more than 50% of patients with HCC from high incidence areas such as sub-Saharan Africa and East Asia (Hainaut and Hollstein 2000) and Ser-249 TP53 mutation, a missense mutation resulting in substitution of arginine with serine at codon 249, reflects high dietary exposure to aflatoxins (Hsu et al. 1991). A few studies reported the feasibility of detection of Ser-249 mutation in cfDNA derived from plasma or serum of HCC patients (Szymańska et al. 2004; Kirk et al. 2005; Hosny et al. 2008) highlighting the potential role of using circulating mutant DNA in diagnosis of HCC.
Recent studies that investigated the concordance between somatic mutations detected in cfDNA and those detected from tumours and early exploratory studies that tested the pharmacodynamic value of circulating mutant DNA in patients with cancer are summarized in Tables 1 and 2 respectively.
Table 1.
Summary of cfDNA studies that investigated the concordance between mutations detected in cfDNA and matched tumour
| Tumour | Mutation | Study | Number of matched samples | Technique used | Medium | Mutation rate in plasma/serum | Mutation rate in tumour | Concordance between plasma/serum and tumour (%) |
|---|---|---|---|---|---|---|---|---|
| Non-small cell lung cancer | EGFR (Exon 19 deletions and p.L858R) | Kimura et al. (2007) | 42 | Scorpion-ARMS | Serum | 19% | 21.4% | 92.9 |
| Maheswaran et al. (2008) | 12a | Scorpion-ARMS | Plasma | N/A | N/A | 33 | ||
| Rosell et al. (2009) | 164a | PNA mediated PCR | Serum | N/A | N/A | 59 | ||
| KRAS | Wang et al. (2010) | 273 | PCR–RFLP | Plasma | 12.8% | 11% | 76.7 | |
| Pancreatic cancer | KRAS | Castells et al. (1999) | 39 | PCR–RFLP | Plasma | 23% | 72% | 32 |
| Breast cancer | PIK3CA | Board et al. (2010) | 41 | Scorpion-ARMS | Plasma | 19.5% | 24% | 95 |
| Angenendt et al. (2010) | 50 | BEAMing | Plasma | 30% | 30% | 100 | ||
| Cutaneous melanoma | BRAF | Board et al. (2009) | 96 | ARMS | Serum | 26.6% | 47% | 76 |
| Pinzani et al. (2010) | 56 | Allele specific real time PCR | Plasma | 42% | 51% | 80 |
ARMS amplification refractory mutation testing system, EGFR epidermal growth factor receptor, PNA protein nucleic acid, RFLP restriction fragment length polymorphism, BEAMing beads, emulsions, amplification and magnetics, NA not applicable
aAll patients had EGFR mutations in tumour
Table 2.
Summary of exploratory studies that tested pharmacodynamic value of ctDNA
| Tumour | Study | Patients | Treatment | Mutation | Summary of the findings |
|---|---|---|---|---|---|
| Colorectal | Diehl et al. (2008) | 18 (16 stage IV, 1 stage III, 1 stage II) | Surgery and/or chemotherapy | APC, KRAS, p53 | ctDNA reflects the tumour burden |
| ctDNA is more reliable and sensitive indicator of disease than CEA | |||||
| Pancreatic | Olsen et al. (2009) | 11 (locally advanced) | Chemo-radiotherapy and gefitinib | KRAS | Five patients had ctDNA in pretreatment plasma |
| Of those five patients, ctDNA became undetectable in 3 patients post-treatment and they had overall survival of 8, 11, and 21 months | |||||
| Breast | Chen et al. (2009) | 6 (non-metastatic) | Neo-adjuvant chemotherapy | p53 | ctDNA were undetectable after treatment in four women consistent with clinical remission |
| Cutaneous melanoma | Shinozaki et al. (2007) | 48 (stage IV) | Bio-chemotherapy | BRAF | Twenty-four patients responded to treatment |
| Ten responders had ctDNA in pretreatment serum and nine of those had undetectable ctDNA in serum within 4 weeks of the last cycle of treatment | |||||
| BRAF mutation in serum is associated with significantly worse overall survival |
ctDNA circulating mutant DNA, CEA carcino-embryonic antigen
Novel techniques for mutation testing from cf DNA
Currently, Sanger sequencing is the most widely used technique for screening of somatic mutations in solid tumours. However, this can detect a mutation of interest only when a sample contains 15–20% of mutant alleles (Tsiatis et al. 2010). In contrast, pyrosequencing, which is a more sensitive sequencing platform, detects down to 5% of mutant alleles (Tsiatis et al. 2010). ARMS-Scorpion PCR assay offers better sensitivity with an ability to detect the presence of a mutant at 1% of the total DNA (Board et al. 2008), albeit with a substantial extra financial cost. However, as well as tumour derived mutant DNA, wild type DNA also circulates in the blood and the tumour derived mutant DNA fraction in cfDNA could be significantly less than 1% (Diehl et al. 2005; Angenendt et al. 2010). This imposes a difficult technical challenge in developing a sensitive cfDNA assay for mutation detection. As summarised in Table 1, for most tumour types, sensitivity remains the major issue for currently available cfDNA assays and implementation of novel technology platforms will be necessary to improve the clinical utility of somatic mutation detection from cfDNA. This review will outline newer technology platforms that may offer advantages over currently available techniques and could improve mutation detection in cfDNA.
Microfluidic digital PCR (Biomark System)
This technique was first used by Yung et al. (2009) in detection of EGFR mutations in plasma derived cfDNA of patients with NSCLC. A major advantage of this technique over other PCR based techniques is that mutant copy numbers can also be counted. The Biomark Digital Array Chip consists of twelve panels with each panel further partitioned into 765 reaction chambers. One chip can perform 9180 reactions in a single run. PCR reactions from each chamber yield a fluorescent signal and the colour of the signal depends on whether there is any mutant copy in the chamber. By counting different color coded signals from reaction wells, it is possible to count mutant and wild type copies contained in a sample. This technique could detect one mutant copy in the background of 1,000 wild type copies. The results from Yung et al., in a small study, are superior to those seen previously with ARMS and other technologies. Furthermore, the quantification of mutant sequences possible with this technique allowed demonstration of reduced mutant sequences in those patients with partial or complete responses to EGFR inhibitors and raises the possibility of developing cfDNA as a pharmacodynamic biomarker.
BEAMing (beads, emulsion, amplifications and magnetics)
BEAMing is a novel, highly sensitive PCR based technique for DNA mutation detection (Diehl et al. 2006). PCR products are amplified and tagged before being attached to beads during a water-in-oil emulsion step. A single base extension at the position of the mutation is performed on the beads to label mutant and wild type beads with different fluorescence. The beads are analysed by flow cytometry and sequencing can be performed from single beads to confirm the presence of wild type or mutant sequences. This technique is reported to be able to detect a single mutant sequence in 10,000 wild type sequences (Li et al. 2006). Using this technology mutant APC sequences can be detected in cfDNA from patients with CRC, with an average of 11% (1.9–27.0%) mutant sequences in patients with advanced (Dukes D) CRC compared to 0.9% (0.03–1.75%) mutant sequences in Dukes B CRC (Diehl et al. 2005). This technique has been applied subsequently to cfDNA from patients undergoing chemotherapy and surgery for CRC and has demonstrated that sequential measurement of mutant DNA fragments in plasma can be used as an indicator of disease response or relapse (Diehl et al. 2008). The major disadvantage of this technology for using in the disease monitoring is that a specific marker assay has to be developed for each patient and tumour biopsy is required to screen for markers in order to do this. This restricts the use of BEAMing to cases where tumour biopsies are available or where BEAMing is used to monitor disease post surgery. It is not currently suitable for screening asymptomatic individuals for cancer where the specific DNA alteration is unknown. However, a major advantage of this technique is its sensitivity and one recent study reported that in a cohort of 50 metastatic breast cancer patients, PIK3CA mutations could be tested reliably from plasma derived cfDNA by using BEAMing with concordance between mutations detected from plasma and tumour of 100% (Angenendt et al. 2010).
Next generation sequencing
The advent of massively parallel DNA sequencing platforms has changed the landscape of cancer genomic research. Currently, there are three next generation platforms, Roche 454 Genome Sequencer FLX Titanium, Illumina Genome Analyzer II and ABI SOLiD. These technologies have effectively been used for whole cancer genome sequencing for analysis of single nucleotide variants, whole genome analysis of somatic rearrangements and whole transcriptome sequencing (Shah et al. 2009; Pleasance et al. 2009; Maher et al. 2009). Leary et al. (2010) have recently demonstrated that a specific somatic rearrangement signature could be found in cancer patients using massively parallel sequencing and these signatures could be detected in patients’ plasma by using digital PCR approach. Van der Vaart et al. (2009) also reported characterisation of cfDNA from 12 prostate cancer patients and 10 healthy controls by parallel tagged sequencing on the 454 platform.
With the exception of the few small studies outlined above, these technologies have yet to be applied in large-scale trials of cfDNA. The main drawback to these platforms is that they work optimally with non-degraded, micromolar input of DNA which is not a characteristic of cfDNA. The sensitivity of these technologies is yet to be established but will ultimately be dependant upon the DNA input (for example, a sensitivity of less than 0.1% is theoretically impossible if less than 1,000 template copies are available). A pre-amplification step can be used prior to sequencing but whole genome amplification of cfDNA in cancer may not be representative of the initial DNA template. Despite these caveats, with further technological advances, next generation sequencing platforms are likely to be increasingly applied to cfDNA and could revolutionise cancer molecular genetics.
Conclusion
Since the discovery of circulating nucleic acids in human by Mandel and Metais in 1948, circulating or cell free DNA research has progressed to the point where introduction into routine clinical practice is a real possibility. The main challenge remains to make cancer genomic data applicable to the management of cancer patients and testing genetic biomarkers more accessible in a minimally invasive way. This tremendous challenge will require concerted efforts of cancer genomic researchers, translational research community and cancer physicians. Well designed translational research studies are urgently needed for qualification of genetic biomarkers which are already available from recent studies of whole genome sequencing of cancer. From this perspective, looking into cancer genomic landscapes through the windows of cfDNA analysis is an exciting prospect.
Acknowledgments
K. A. is supported by Cancer Research UK and AstraZeneca via their Clinical Pharmacology Fellowship Scheme. W. N. is supported by National Institutes of Health Research at the Manchester Biomedical Research Centre.
References
- Angenendt P, David K, Juhl H, Diehl F (2010) Detection of phosphoinositide-3-kinase, catalytic, and alpha polypeptide (PIK3CA) mutations in matched tissue and plasma samples from patients with metastatic breast cancer. J Clin Oncol (Meeting Abstracts) 28:15s (suppl 10502)
- Bachman KE, Argani P, Samuels Y, Silliman N, Ptak J, Szabo S, Konishi H, Karakas B, Blair BG, Lin C, Peters BA, Velculescu VE, Park BH. The PIK3CA gene is mutated with high frequency in human breast cancers. Cancer Biol Ther. 2004;3(8):772–775. doi: 10.4161/cbt.3.8.994. [DOI] [PubMed] [Google Scholar]
- Board RE, Thelwell NJ, Ravetto PF, Little S, Ranson M, Dive C, Hughes A, Whitcombe D. Multiplexed assays for detection of mutations in PIK3CA. Clin Chem. 2008;54(4):757–760. doi: 10.1373/clinchem.2007.098376. [DOI] [PubMed] [Google Scholar]
- Board RE, Ellison G, Orr MC, Kemsley KR, McWalter G, Blockley LY, Dearden SP, Morris C, Ranson M, Cantarini MV, Dive C, Hughes A. Detection of BRAF mutations in the tumour and serum of patients enrolled in the AZD6244 (ARRY-142886) advanced melanoma phase II study. Br J Cancer. 2009;101(10):1724–1730. doi: 10.1038/sj.bjc.6605371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Board RE, Wardley AM, Dixon JM, Armstrong AC, Howell S, Renshaw L, Donald E, Greystoke A, Ranson M, Hughes A, Dive C. Deteection of PIK3CA mutations in circulating free DNA in patients with breast cancer. Breast Cancer Res Treat. 2010;120(2):461–467. doi: 10.1007/s10549-010-0747-9. [DOI] [PubMed] [Google Scholar]
- Bokemeyer C, Bondarenko I, Makhson A, Hartmann JT, Aparicio J, Braud F, Donea S, Ludwig H, Schuch G, Stroh C, Loos AH, Zubel A, Koralewski P. Fluorouracil, leucovorin, and oxaliplatin with and without cetuximab in the first-line treatment of metastatic colorectal cancer. J Clin Oncol. 2009;27(5):663–671. doi: 10.1200/JCO.2008.20.8397. [DOI] [PubMed] [Google Scholar]
- Bos JL, Fearon ER, Hamilton SR, Verlaan-de Vries M, Boom JH, Eb AJ, Vogelstein B. Prevalence of ras gene mutations in human colorectal cancers. Nature. 1987;327(6120):293–297. doi: 10.1038/327293a0. [DOI] [PubMed] [Google Scholar]
- Brose MS, Volpe P, Feldman M, Kumar M, Rishi I, Gerrero R, Einhorn E, Herlyn M, Minna J, Nicholson A, Roth JA, Albelda SM, Davies H, Cox C, Brignell G, Stephens P, Futreal PA, Wooster R, Stratton MR, Weber BL. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res. 2002;62(23):6997–7000. [PubMed] [Google Scholar]
- Castells A, Puig P, Móra J, Boadas J, Boix L, Urgell E, Solé M, Capellà G, Lluís F, Fernández-Cruz L, Navarro S, Farré A. K-ras mutations in DNA extracted from the plasma of patients with pancreatic carcinoma: diagnostic utility and prognostic significance. J Clin Oncol. 1999;17(2):578–584. doi: 10.1200/JCO.1999.17.2.578. [DOI] [PubMed] [Google Scholar]
- Chen Z, Feng J, Buzin CH, Liu Q, Weiss L, Kernstine K, Somlo G, Sommer SS. Analysis of cancer mutation signatures in blood by a novel ultra-sensitive assay: monitoring of therapy or recurrence in non-metastatic breast cancer. PLoS One. 2009;4(9):e7220. doi: 10.1371/journal.pone.0007220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang TC, Chuang AY, Poeta L, Koch WM, Califano JA, Tufano RP. Detectable BRAF mutation in serum DNA samples from patients with papillary thyroid carcinomas. Head Neck. 2010;32(2):229–234. doi: 10.1002/hed.21178. [DOI] [PubMed] [Google Scholar]
- Cohen Y, Rosenbaum E, Clark DP, Zeiger MA, Umbricht CB, Tufano RP, Sidransky D, Westra WH. Mutational analysis of BRAF in fine needle aspiration biopsies of the thyroid: a potential application for the preoperative assessment of thyroid nodules. Clin Cancer Res. 2004;10(8):2761–2765. doi: 10.1158/1078-0432.CCR-03-0273. [DOI] [PubMed] [Google Scholar]
- Daniotti M, Vallacchi V, Rivoltini L, Patuzzo R, Santinami M, Arienti F, Cutolo G, Pierotti MA, Parmiani G, Rodolfo M. Detection of mutated BRAFV600E variant in circulating DNA of stage III–IV melanoma patients. Int J Cancer. 2007;120(11):2439–2444. doi: 10.1002/ijc.22598. [DOI] [PubMed] [Google Scholar]
- Di Nicolantonio F, Martini M, Molinari F, Sartore-Bianchi A, Arena S, Saletti P, Dosso S, Mazzucchelli L, Frattini M, Siena S, Bardelli A. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008;26(35):5705–5712. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- Dianxu F, Shengdao Z, Tianquan H, Yu J, Lei Ruoqing, Zurong Y, Xuezhi W. A prospective study of detection of pancreatic carcinoma by combined plasma K-ras mutations and serum CA19-9 analysis. Pancreas. 2002;25(4):336–341. doi: 10.1097/00006676-200211000-00003. [DOI] [PubMed] [Google Scholar]
- Diehl F, Li M, Dressman D, He Y, Shen D, Szabo S, Diaz LA, Jr, Goodman SN, David KA, Juhl H, Kinzler KW, Vogelstein B. Detection and quantification of mutations in the plasma of patients with colorectal tumours. Proc Natl Acad Sci U S A. 2005;102(45):16368–16373. doi: 10.1073/pnas.0507904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl F, Li M, He Y, Kinzler KW, Vogelstein B, Dressman D. BEAMing: single-molecule PCR on microparticles in water-in-oil emulsions. Nat Methods. 2006;3(7):551–559. doi: 10.1038/nmeth898. [DOI] [PubMed] [Google Scholar]
- Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA, Kinzler KW, Vogelstein B, Diaz LA., Jr Circulating mutant DNA to assess tumour dynamics. Nat Med. 2008;14(9):985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA, Jänne PA. Mechanisms of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Clin Cancer Res. 2008;14(10):2895–2899. doi: 10.1158/1078-0432.CCR-07-2248. [DOI] [PubMed] [Google Scholar]
- Fleischhacker M, Schmidt B. Circulating nucleic acids (CNAs) and cancer—a survey. Biochim Biophys Acta. 2007;1775(1):181–232. doi: 10.1016/j.bbcan.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Fleischhacker M, Schmidt B. Cell-free DNA resuscitated for tumour testing. Nat Med. 2008;14(9):914–915. doi: 10.1038/nm0908-914. [DOI] [PubMed] [Google Scholar]
- Gatalica Z, Torlakovic E. Pathology of the hereditary colorectal carcinoma. Fam Cancer. 2008;7(1):15–26. doi: 10.1007/s10689-007-9146-8. [DOI] [PubMed] [Google Scholar]
- Hainaut P, Hollstein M. p53 and human cancer: the first ten thousand mutations. Adv Cancer Res. 2000;77:81–137. doi: 10.1016/S0065-230X(08)60785-X. [DOI] [PubMed] [Google Scholar]
- Hodgson DR, Wellings R, Orr MC, McCormack R, Malone M, Board RE, Cantarini MV. Circulating tumour-derived predictive biomarkers in oncology. Drug Discov Today. 2010;15(3–4):98–101. doi: 10.1016/j.drudis.2009.12.006. [DOI] [PubMed] [Google Scholar]
- Hosny G, Farahat N, Tayel H, Hainaut P. Ser-249 TP53 and CTNNB1 mutations in circulating free DNA of Egyptian patients with hepatocellular carcinoma versus chronic liver diseases. Cancer Lett. 2008;264(2):201–208. doi: 10.1016/j.canlet.2008.01.031. [DOI] [PubMed] [Google Scholar]
- Hsu IC, Metcalf RA, Sun T, Welsh JA, Wang NJ, Harris CC. Mutational hotspot in the p53 gene in human hepatocellular carcinomas. Nature. 1991;350(6317):427–428. doi: 10.1038/350427a0. [DOI] [PubMed] [Google Scholar]
- Hussain SP, Schwank J, Staib F, Wang XW, Harris CC. TP53 mutations and hepatocellular carcinoma: insights into the etiology and pathogenesis of liver cancer. Oncogene. 2007;26(15):2166–2176. doi: 10.1038/sj.onc.1210279. [DOI] [PubMed] [Google Scholar]
- Inoue A, Suzuki T, Fukuhara T, Maemondo M, Kimura Y, Morikawa N, Watanabe H, Saijo Y, Nukiwa T. Prospective phase II study of gefitinib for chemotherapy-naive patients with advanced non-small-cell lung cancer with epidermal growth factor receptor gene mutations. J Clin Oncol. 2006;24(21):3340–3346. doi: 10.1200/JCO.2005.05.4692. [DOI] [PubMed] [Google Scholar]
- Jackman DM, Miller VA, Cioffredi LA, Yeap BY, Jänne PA, Riely GJ, Ruiz MG, Giaccone G, Sequist LV, Johnson BE. Impact of epidermal growth factor receptor and KRAS mutations on clinical outcomes in previously untreated non-small cell lung cancer patients: results of an online tumor registry of clinical trials. Clin Cancer Res. 2009;15(16):5267–5273. doi: 10.1158/1078-0432.CCR-09-0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H, Kasahara K, Kawaishi M, Kunitoh H, Tamura T, Holloway B, Nishio K. Detection of epidermal growth factor receptor mutations in serum as a predictor of the response to gefitinib in patients with non-small-cell lung cancer. Clin Cancer Res. 2006;12(13):3915–3921. doi: 10.1158/1078-0432.CCR-05-2324. [DOI] [PubMed] [Google Scholar]
- Kimura H, Suminoe M, Kasahara K, Sone T, Araya T, Tamori S, Koizumi F, Nishio K, Miyamoto K, Fujimura M, Nakao S. Evaluation of epidermal growth factor receptor mutation status in serum DNA as a predictor of response to gefitinib (IRESSA) Br J Cancer. 2007;97(6):778–784. doi: 10.1038/sj.bjc.6603949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk GD, Lesi OA, Mendy M, Szymañska K, Whittle H, Goedert JJ, Hainaut P, Montesano R. 249(ser) TP53 mutation in plasma DNA, hepatitis B viral infection, and risk of hepatocellular carcinoma. Oncogene. 2005;24(38):5858–5867. doi: 10.1038/sj.onc.1208732. [DOI] [PubMed] [Google Scholar]
- Kopnin BP. Targets of oncogenes and tumor suppressors: key for understanding basic mechanisms of carcinogenesis. Biochemistry (Mosc) 2000;65(1):2–27. [PubMed] [Google Scholar]
- Kosaka T, Yatabe Y, Endoh H, Kuwano H, Takahashi T, Mitsudomi T. Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res. 2004;64(24):8919–8923. doi: 10.1158/0008-5472.CAN-04-2818. [DOI] [PubMed] [Google Scholar]
- Kuang Y, Rogers A, Yeap BY, Wang L, Makrigiorgos M, Vetrand K, Thiede S, Distel RJ, Jänne PA. Noninvasive detection of EGFR T790M in gefitinib or erlotinib resistant non-small cell lung cancer. Clin Cancer Res. 2009;15(8):2630–2636. doi: 10.1158/1078-0432.CCR-08-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leary RJ, Kinde I, Diehl F, Schmidt K, Clouser C, Duncan C, Antipova A, Lee C, McKernan K, De La Vega FM, Kinzler KW, Vogelstein B, Diaz LA, Jr, Velculescu VE. Development of personalized tumour biomarkers using massively parallel sequencing. Sci Transl Med. 2010;2(20):20ra14. doi: 10.1126/scitranslmed.3000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Soung YH, Kim SY, Lee HW, Park WS, Nam SW, Kim SH, Lee JY, Yoo NJ, Lee SH. PIK3CA gene is frequently mutated in breast carcinomas and hepatocellular carcinomas. Oncogene. 2005;24(8):1477–1480. doi: 10.1038/sj.onc.1208304. [DOI] [PubMed] [Google Scholar]
- Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37(3):646–650. [PubMed] [Google Scholar]
- Levine DA, Bogomolniy F, Yee CJ, Lash A, Barakat RR, Borgen PI, Boyd J. Frequent mutation of the PIK3CA gene in ovarian and breast cancers. Clin Cancer Res. 2005;11(8):2875–2878. doi: 10.1158/1078-0432.CCR-04-2142. [DOI] [PubMed] [Google Scholar]
- Li M, Diehl F, Dressman D, Vogelstein B, Kinzler KW. BEAMing up for detection and quantification of rare sequence variants. Nat Methods. 2006;3(2):95–97. doi: 10.1038/nmeth850. [DOI] [PubMed] [Google Scholar]
- Maher CA, Kumar-Sinha C, Cao X, Kalyana-Sundaram S, Han B, Jing X, Sam L, Barrette T, Palanisamy N, Chinnaiyan AM. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009;458(7234):97–101. doi: 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, Inserra E, Diederichs S, Iafrate AJ, Bell DW, Digumarthy S, Muzikansky A, Irimia D, Settleman J, Tompkins RG, Lynch TJ, Toner M, Haber DA. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359(4):366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel P, Metais P. Les Acides Nucleiques Du Plasma Sanguin Chez L’Homme. P. C R Acad Sci Paris. 1948;142:241–243. [PubMed] [Google Scholar]
- Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, Nishiwaki Y, Ohe Y, Yang JJ, Chewaskulyong B, Jiang H, Duffield EL, Watkins CL, Armour AA, Fukuoka M. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361(10):947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- Olsen CC, Schefter TE, Chen H, Kane M, Leong S, McCarter MD, Chen Y, Mack P, Eckhardt SG, Stiegmann G, Raben D. Results of a phase I trial of 12 patients with locally advanced pancreatic carcinoma combining gefitinib, paclitaxel, and 3-dimensional conformal radiation: report of toxicity and evaluation of circulating K-ras as a potential biomarker of response to therapy. Am J Clin Oncol. 2009;32(2):115–121. doi: 10.1097/COC.0b013e318180baa3. [DOI] [PubMed] [Google Scholar]
- Perrone F, Lampis A, Orsenigo M, Bartolomeo M, Gevorgyan A, Losa M, Frattini M, Riva C, Andreola S, Bajetta E, Bertario L, Leo E, Pierotti MA, Pilotti S. PI3KCA/PTEN deregulation contributes to impaired responses to cetuximab in metastatic colorectal cancer patients. Ann Oncol. 2009;20(1):84–90. doi: 10.1093/annonc/mdn541. [DOI] [PubMed] [Google Scholar]
- Pinzani P, Salvianti F, Cascella R, Massi D, Giorgi V, Pazzagli M, Orlando C. Allele specific Taqman-based real-time PCR assay to quantify circulating BRAFV600E mutated DNA in plasma of melanoma patients. Clin Chim Acta. 2010;411(17–18):1319–1324. doi: 10.1016/j.cca.2010.05.024. [DOI] [PubMed] [Google Scholar]
- Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin ML, Ordóñez GR, Bignell GR, Ye K, Alipaz J, Bauer MJ, Beare D, Butler A, Carter RJ, Chen L, Cox AJ, Edkins S, Kokko-Gonzales PI, Gormley NA, Grocock RJ, Haudenschild CD, Hims MM, James T, Jia M, Kingsbury Z, Leroy C, Marshall J, Menzies A, Mudie LJ, Ning Z, Royce T, Schulz-Trieglaff OB, Spiridou A, Stebbings LA, Szajkowski L, Teague J, Williamson D, Chin L, Ross MT, Campbell PJ, Bentley DR, Futreal PA, Stratton MR. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462(7276):1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plesec TP, Hunt JL. KRAS mutation testing in colorectal cancer. Adv Anat Pathol. 2009;16(4):196–203. doi: 10.1097/PAP.0b013e3181a9d4ed. [DOI] [PubMed] [Google Scholar]
- Ramirez JL, Sarries C, Castro PL, Roig B, Queralt C, Escuin D, Aguirre I, Sanchez JM, Manzano JL, Margelí M, Sanchez JJ, Astudillo J, Taron M, Rosell R. Methylation patterns and K-ras mutations in tumour and paired serum of resected non-small-cell lung cancer patients. Cancer Lett. 2003;193(2):207–216. doi: 10.1016/S0304-3835(02)00740-1. [DOI] [PubMed] [Google Scholar]
- Riely GJ, Politi KA, Miller VA, Pao W. Update on epidermal growth factor receptor mutations in non-small cell lung cancer. Clin Cancer Res. 2006;12(24):7232–7241. doi: 10.1158/1078-0432.CCR-06-0658. [DOI] [PubMed] [Google Scholar]
- Rosell R, Moran T, Queralt C, Porta R, Cardenal F, Camps C, Majem M, Lopez-Vivanco G, Isla D, Provencio M, Insa A, Massuti B, Gonzalez-Larriba JL, Paz-Ares L, Bover I, Garcia-Campelo R, Moreno MA, Catot S, Rolfo C, Reguart N, Palmero R, Sánchez JM, Bastus R, Mayo C, Bertran-Alamillo J, Molina MA, Sanchez JJ, Taron M, Spanish Lung Cancer Group Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009;361(10):958–967. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- Rumore PM, Steinman CR. Endogenous circulating DNA in systemic lupus erythematosus. Occurrence as multimeric complexes bound to histone. J Clin Invest. 1990;86(1):69–74. doi: 10.1172/JCI114716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan BM, Lefort F, McManus R, Daly J, Keeling PW, Weir DG, Kelleher D. A prospective study of circulating mutant KRAS2 in the serum of patients with colorectal neoplasia: strong prognostic indicator in postoperative follow up. Gut. 2003;52(1):101–108. doi: 10.1136/gut.52.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartore-Bianchi A, Martini M, Molinari F, Veronese S, Nichelatti M, Artale S, Di Nicolantonio F, Saletti P, Dosso S, Mazzucchelli L, Frattini M, Siena S, Bardelli A. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res. 2009;69(5):1851–1857. doi: 10.1158/0008-5472.CAN-08-2466. [DOI] [PubMed] [Google Scholar]
- Sequist LV, Martins RG, Spigel D, Grunberg SM, Spira A, Jänne PA, Joshi VA, McCollum D, Evans TL, Muzikansky A, Kuhlmann GL, Han M, Goldberg JS, Settleman J, Iafrate AJ, Engelman JA, Haber DA, Johnson BE, Lynch TJ. First-line gefitinib in patients with advanced non-small-cell lung cancer harboring somatic EGFR mutations. J Clin Oncol. 2008;26(15):2442–2449. doi: 10.1200/JCO.2007.14.8494. [DOI] [PubMed] [Google Scholar]
- Sequist LV, Engelman JA, Lynch TJ. Toward noninvasive genomic screening of lung cancer patients. J Clin Oncol. 2009;27(16):2589–2591. doi: 10.1200/JCO.2008.20.4875. [DOI] [PubMed] [Google Scholar]
- Shah SP, Morin RD, Khattra J, Prentice L, Pugh T, Burleigh A, Delaney A, Gelmon K, Guliany R, Senz J, Steidl C, Holt RA, Jones S, Sun M, Leung G, Moore R, Severson T, Taylor GA, Teschendorff AE, Tse K, Turashvili G, Varhol R, Warren RL, Watson P, Zhao Y, Caldas C, Huntsman D, Hirst M, Marra MA, Aparicio S. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461(7265):809–813. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- Shinozaki M, O’Day SJ, Kitago M, Amersi F, Kuo C, Kim J, Wang HJ, Hoon DS. Utility of circulating B-RAF DNA mutation in serum for monitoring melanoma patients receiving biochemotherapy. Clin Cancer Res. 2007;13(7):2068–2074. doi: 10.1158/1078-0432.CCR-06-2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluiter MD, van Rensburg EJ. Large genomic rearrangements of the BRCA1 and BRCA2 genes: review of the literature and report of a novel BRCA1 mutation. Breast Cancer Res Treat. 2011;125(2):325–349. doi: 10.1007/s10549-010-0817-z. [DOI] [PubMed] [Google Scholar]
- Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458(7239):719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroun M, Anker P, Lyautey J, Lederrey C, Maurice PA. Isolation and characterization of DNA from the plasma of cancer patients. Eur J Cancer Clin Oncol. 1987;23(6):707–712. doi: 10.1016/0277-5379(87)90266-5. [DOI] [PubMed] [Google Scholar]
- Stroun M, Anker P, Maurice P, Lyautey J, Lederrey C, Beljanski M. Neoplastic characteristics of the DNA found in the plasma of cancer patients. Oncology. 1989;46(5):318–322. doi: 10.1159/000226740. [DOI] [PubMed] [Google Scholar]
- Stroun M, Maurice P, Vasioukhin V, Lyautey J, Lederrey C, Lefort F, Rossier A, Chen XQ, Anker P. The origin and mechanism of circulating DNA. Ann NY Acad Sci. 2000;906:161–168. doi: 10.1111/j.1749-6632.2000.tb06608.x. [DOI] [PubMed] [Google Scholar]
- Stroun M, Lyautey J, Lederrey C, Olson-Sand A, Anker P. About the possible origin and mechanism of circulating DNA apoptosis and active DNA release. Clin Chim Acta. 2001;313(1–2):139–142. doi: 10.1016/S0009-8981(01)00665-9. [DOI] [PubMed] [Google Scholar]
- Szymańska K, Lesi OA, Kirk GD, Sam O, Taniere P, Scoazec JY, Mendy M, Friesen MD, Whittle H, Montesano R, Hainaut P. Ser-249TP53 mutation in tumour and plasma DNA of hepatocellular carcinoma patients from a high incidence area in the Gambia, West Africa. Int J Cancer. 2004;110(3):374–379. doi: 10.1002/ijc.20103. [DOI] [PubMed] [Google Scholar]
- Tsao MS, Sakurada A, Cutz JC, Zhu CQ, Kamel-Reid S, Squire J, Lorimer I, Zhang T, Liu N, Daneshmand M, Marrano P, Cunha Santos G, Lagarde A, Richardson F, Seymour L, Whitehead M, Ding K, Pater J, Shepherd FA. Erlotinib in lung cancer—molecular and clinical predictors of outcome. N Engl J Med. 2005;353(2):133–144. doi: 10.1056/NEJMoa050736. [DOI] [PubMed] [Google Scholar]
- Tsiatis AC, Norris-Kirby A, Rich RG, Hafez MJ, Gocke CD, Eshleman JR, Murphy KM. Comparison of Sanger sequencing, pyrosequencing, and melting curve analysis for the detection of KRAS mutations: diagnostic and clinical implications. J Mol Diagn. 2010;12(4):425–432. doi: 10.2353/jmoldx.2010.090188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutsem E, Köhne CH, Hitre E, Zaluski J, Chang Chien CR, Makhson A, D’Haens G, Pintér T, Lim R, Bodoky G, Roh JK, Folprecht G, Ruff P, Stroh C, Tejpar S, Schlichting M, Nippgen J, Rougier P. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009;360(14):1408–1417. doi: 10.1056/NEJMoa0805019. [DOI] [PubMed] [Google Scholar]
- Vaart M, Semenov DV, Kuligina EV, Richter VA, Pretorius PJ. Characterisation of circulating DNA by parallel tagged sequencing on the 454 platform. Clin Chim Acta. 2009;409(1–2):21–27. doi: 10.1016/j.cca.2009.08.011. [DOI] [PubMed] [Google Scholar]
- Wang JY, Hsieh JS, Chang MY, Huang TJ, Chen FM, Cheng TL, Alexandersen K, Huang YS, Tzou WS, Lin SR. Molecular detection of APC, K-ras, and p53 mutations in the serum of colorectal cancer patients as circulating biomarkers. World J Surg. 2004;28(7):721–726. doi: 10.1007/s00268-004-7366-8. [DOI] [PubMed] [Google Scholar]
- Wang S, An T, Wang J, Zhao J, Wang Z, Zhuo M, Bai H, Yang L, Zhang Y, Wang X, Duan J, Wang Y, Guo Q, Wu M. Potential clinical significance of a plasma-based KRAS mutation analysis in patients with advanced non-small cell lung cancer. Clin Cancer Res. 2010;16(4):1324–1330. doi: 10.1158/1078-0432.CCR-09-2672. [DOI] [PubMed] [Google Scholar]
- Xing M, Westra WH, Tufano RP, Cohen Y, Rosenbaum E, Rhoden KJ, Carson KA, Vasko V, Larin A, Tallini G, Tolaney S, Holt EH, Hui P, Umbricht CB, Basaria S, Ewertz M, Tufaro AP, Califano JA, Ringel MD, Zeiger MA, Sidransky D, Ladenson PW. BRAF mutation predicts a poorer clinical prognosis for papillary thyroid cancer. J Clin Endocrinol Metab. 2005;90(12):6373–6379. doi: 10.1210/jc.2005-0987. [DOI] [PubMed] [Google Scholar]
- Yancovitz M, Yoon J, Mikhail M, Gai W, Shapiro RL, Berman RS, Pavlick AC, Chapman PB, Osman I, Polsky D. Detection of mutant BRAF alleles in the plasma of patients with metastatic melanoma. J Mol Diagn. 2007;9(2):178–183. doi: 10.2353/jmoldx.2007.060135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CH, Yu CJ, Shih JY, Chang YC, Hu FC, Tsai MC, Chen KY, Lin ZZ, Huang CJ, Shun CT, Huang CL, Bean J, Cheng AL, Pao W, Yang PC. Specific EGFR mutations predict treatment outcome of stage IIIB/IV patients with chemotherapy-naive non-small-cell lung cancer receiving first-line gefitinib monotherapy. J Clin Oncol. 2008;26(16):2745–2753. doi: 10.1200/JCO.2007.15.6695. [DOI] [PubMed] [Google Scholar]
- Yung TK, Chan KC, Mok TS, Tong J, To KF, Lo YM. Single-molecule detection of epidermal growth factor receptor mutations in plasma by microfluidics digital PCR in non-small cell lung cancer patients. Clin Cancer Res. 2009;15(6):2076–2084. doi: 10.1158/1078-0432.CCR-08-2622. [DOI] [PubMed] [Google Scholar]