Abstract
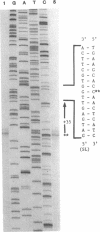
Three critical enzymes catechol oxygenase II (chlorocatechol dioxygenase), muconate cycloisomerase II, and dienelactone hydrolase, are involved in the degradation of chlorocatechols, which are obligatory intermediates in the catabolism of chlorinated aromatic compounds. The organization and complete nucleotide sequence of the genes for these enzymes have been determined on a 4.2-kilobase-pair (kbp) Bgl II fragment cloned from the plasmid pAC27, based on the agreement of open reading frame lengths with apparent mobilities of polypeptides expressed in Escherichia coli maxicells, predicted N-terminal amino acid sequences with those of the purified proteins, and predicted total amino acid compositions with those of the purified proteins. The 4.2-kbp fragment contains the three genes and ribosome binding sites for those genes but no promoter. When placed downstream of the tac promoter in the broad-host-range plasmid pMMB22, this fragment directs the synthesis of all three enzymes in both E. coli and Pseudomonas putida only on induction with isopropyl beta-D-thiogalactopyranoside, suggesting that the gene cluster is regulated as a single unit under the control of a single promoter. Endogenous transcription initiation of the gene cluster on pAC27, however, occurs from a site present within a 386-bp Bgl II fragment upstream of the 4.2-kbp fragment, and sequences 5' to that site are similar to the sequences of other positively controlled Pseudomonas promoters occurring on the TOL and NAH plasmids.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aldrich T. L., Frantz B., Gill J. F., Kilbane J. J., Chakrabarty A. M. Cloning and complete nucleotide sequence determination of the catB gene encoding cis,cis-muconate lactonizing enzyme. Gene. 1987;52(2-3):185–195. doi: 10.1016/0378-1119(87)90045-x. [DOI] [PubMed] [Google Scholar]
- Burke J. F. High-sensitivity S1 mapping with single-stranded [32P]DNA probes synthesized from bacteriophage M13mp templates. Gene. 1984 Oct;30(1-3):63–68. doi: 10.1016/0378-1119(84)90105-7. [DOI] [PubMed] [Google Scholar]
- Chatterjee D. K., Chakrabarty A. M. Genetic homology between independently isolated chlorobenzoate-degradative plasmids. J Bacteriol. 1983 Jan;153(1):532–534. doi: 10.1128/jb.153.1.532-534.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee D. K., Chakrabarty A. M. Genetic rearrangements in plasmids specifying total degradation of chlorinated benzoic acids. Mol Gen Genet. 1982;188(2):279–285. doi: 10.1007/BF00332688. [DOI] [PubMed] [Google Scholar]
- Dale R. M., McClure B. A., Houchins J. P. A rapid single-stranded cloning strategy for producing a sequential series of overlapping clones for use in DNA sequencing: application to sequencing the corn mitochondrial 18 S rDNA. Plasmid. 1985 Jan;13(1):31–40. doi: 10.1016/0147-619x(85)90053-8. [DOI] [PubMed] [Google Scholar]
- Deretic V., Gill J. F., Chakrabarty A. M. Gene algD coding for GDPmannose dehydrogenase is transcriptionally activated in mucoid Pseudomonas aeruginosa. J Bacteriol. 1987 Jan;169(1):351–358. doi: 10.1128/jb.169.1.351-358.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Don R. H., Pemberton J. M. Properties of six pesticide degradation plasmids isolated from Alcaligenes paradoxus and Alcaligenes eutrophus. J Bacteriol. 1981 Feb;145(2):681–686. doi: 10.1128/jb.145.2.681-686.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz B., Ngai K. L., Chatterjee D. K., Ornston L. N., Chakrabarty A. M. Nucleotide sequence and expression of clcD, a plasmid-borne dienelactone hydrolase gene from Pseudomonas sp. strain B13. J Bacteriol. 1987 Feb;169(2):704–709. doi: 10.1128/jb.169.2.704-709.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosal D., You I. S., Chatterjee D. K., Chakrabarty A. M. Genes specifying degradation of 3-chlorobenzoic acid in plasmids pAC27 and pJP4. Proc Natl Acad Sci U S A. 1985 Mar;82(6):1638–1642. doi: 10.1073/pnas.82.6.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Nakazawa A., Nakazawa T. Nucleotide sequence of the promoter region of the xylDEGF operon on TOL plasmid of Pseudomonas putida. Gene. 1984 Sep;29(3):323–330. doi: 10.1016/0378-1119(84)90061-1. [DOI] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Raibaud O., Schwartz M. Positive control of transcription initiation in bacteria. Annu Rev Genet. 1984;18:173–206. doi: 10.1146/annurev.ge.18.120184.001133. [DOI] [PubMed] [Google Scholar]
- Reineke W., Knackmuss H. J. Hybrid pathway for chlorobenzoate metabolism in Pseudomonas sp. B13 derivatives. J Bacteriol. 1980 May;142(2):467–473. doi: 10.1128/jb.142.2.467-473.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A., Hack A. M., Rupp W. D. Simple method for identification of plasmid-coded proteins. J Bacteriol. 1979 Jan;137(1):692–693. doi: 10.1128/jb.137.1.692-693.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell M. A. Homology between nucleotide sequences of promoter regions of nah and sal operons of NAH7 plasmid of Pseudomonas putida. Proc Natl Acad Sci U S A. 1986 Jan;83(2):369–373. doi: 10.1073/pnas.83.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanier R. Y., Palleroni N. J., Doudoroff M. The aerobic pseudomonads: a taxonomic study. J Gen Microbiol. 1966 May;43(2):159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- Wheelis M. L., Ornston L. N. Genetic control of enzyme induction in the -ketoadipate pathway of Pseudomonas putida: deletion mapping of cat mutations. J Bacteriol. 1972 Feb;109(2):790–795. doi: 10.1128/jb.109.2.790-795.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]