Abstract
To assay the efficiency of the FLP/FRT site-specific recombination system in Danio rerio, a construct consisting of a muscle-specific promoter driving EGFP flanked by FRT sites was developed. FLPe capped RNA was microinjected into transgenic single cell stage zebrafish embryos obtained by crossing hemizygous transgenic males with wild-type females. By 48 h post fertilization (hpf), the proportion of embryos displaying green fluorescence following FLPe RNA microinjection was significantly lower (7.7%; P < 0.001) than would be expected from a cross in the absence of the recombinase (50%). Embryos that retained fluorescence displayed marked mosaicism. Inheritance of the excised transgene in non-fluorescent, transgenic embryos was verified by PCR analysis and FLPe-mediated recombination was confirmed by DNA sequencing. Sperm derived from confirmed transgenic males in these experiments was used to fertilize wild-type eggs to determine whether germline excision of the transgene had occurred. Clutches sired by FLPe-microinjected males contained 0–4% fluorescent embryos. Transgenic males that were phenotypically wild-type produced no fluorescent progeny, demonstrating complete excision of the transgene from their germline. FLPe microinjected males that retained some fluorescent muscle expression produced a small proportion of fluorescent offspring, suggesting that in mosaic males not all germline cells had undergone FLPe-mediated transgene excision. Our results show that FLPe, which is derived from Saccharomyces cerevisiae, is an efficient recombinase in zebrafish maintained at 28.5°C.
Keywords: FLP, FLPe, Recombinase, Zebrafish
Introduction
Already established as an important developmental model, zebrafish (Danio rerio) are emerging as a powerful genetic model as well. Zebrafish possess certain characteristics that make them ideally suited for developmental and genetic studies, namely short generation time, large number of offspring, and optical clarity of externally fertilized embryos. Embryogenesis is finished in about 24 h, organogenesis is completed in about 5 days, and sexual maturity is reached at 3 months. A single female can provide hundreds of embryos at a time, either by natural crosses or by in vitro fertilization. Eggs are externally fertilized, develop synchronously outside of the mother and remain optically transparent up to 72 hpf, allowing for easy visualization of internal processes as well as the expression of fluorescently tagged transgenes and other reporter constructs.
Twelve years after the first successful in vivo demonstration of Cre/loxP in transgenic mice (Lakso et al. 1992; Orban et al. 1992), the Cre/loxP site-specific recombination system was introduced into zebrafish (Dong and Stuart 2004). The extensive use of Cre recombinase in mice has been proven to be a powerful tool for the study of mammalian genetics. Cre (cyclization recombination) is a 38-kDa protein from bacteriophage P1 that can recombine target DNA recognition sequences called loxP (locus of crossing over [x] in P1) sites. The loxP site is a 34-bp DNA sequence that consists of two 13-bp inverted repeats, which act as binding sites for Cre, flanking an asymmetrical 8-bp core region. Depending on the orientation and location of the recombinase recognition sites, Cre can catalyze excision, integration, inversion, or translocation of DNA segments. The biological role of Cre/loxP in the circular plasmid form of bacteriophage P1 is to resolve DNA dimers into plasmid monomers for equal partitioning to daughter cells during cell division (Austin et al. 1981; Sternberg and Hamilton 1981). Gene targeting in mouse embryonic stem cells can add recombinase recognition sites to specific locations in the genome, enabling a variety of sophisticated genetic manipulations, including chromosomal inversions, site-specific transgene insertion, conditional transgenesis and conditional gene inactivation (reviewed in Nagy 2000). A database has been designed recently to keep track of the over 500 different Cre-expressing transgenic mice (Nagy et al. 2009).
Although the use of Cre recombinase in zebrafish is still in its infancy when compared to the mouse, sophisticated implementation of the Cre/loxP system, such as site-directed gene integration at mutant lox sites (Liu et al. 2007) and heat shock-inducible gene activation (Langenau et al. 2005; Thummel et al. 2005; Feng et al. 2007; Hans et al. 2009), have been recently reported. To further enhance the zebrafish genetic toolkit, the addition of a second site-specific recombination system, such as FLP/FRT, would be beneficial and allow for more elegant experimentation. FLP (flipase) is a 43-kDa protein from the 2-μm circle of Saccharomyces cerevisiae. Like Cre, FLP can recombine 34-bp DNA recognition sequences called FRT (FLP recognition target) sites, consisting of two imperfect 13-bp inverted repeats flanking an asymmetrical 8-bp core region (Fig. 1b). The biological role of FLP/FRT in S. cerevisiae is to maintain a high copy number of the 2-μm circle by inverting half of the plasmid during replication such that a large multimeric plasmid is created which is later resolved into individual monomers (Futcher 1986). An in vitro study by Buchholz et al. (1996) determined that the most favorable temperature for FLP activity was near 30°C, consistent with the yeast origin of the FLP/FRT site-specific recombination system. An enhanced version of FLP recombinase, FLPe, was developed by cycling mutagenesis to improve its thermolability so as to prevent its inactivation at higher temperatures (Buchholz et al. 1998). Although predominantly used in Drosophila for mitotic recombination and gene activation (reviewed in Bischof and Basler 2008), the FLP/FRT recombination system has also been used in mice, both separately (Ludwig et al. 1996; Dymecki 1996; Rodriguez et al. 2000; Kanki et al. 2006) and in conjunction with Cre/loxP (Meyers et al. 1998; Turakainen et al. 2009; Yamamoto et al. 2009). To assay the efficiency of the FLP/FRT site-specific recombination system in Danio rerio, FLPe capped RNA was microinjected into transgenic zebrafish embryos carrying an FRT-flanked EGFP expression cassette. Injected fish were observed for muscle-specific GFP expression, and outcrossed to wild-type zebrafish to assess germline excision efficiency.
Fig. 1.
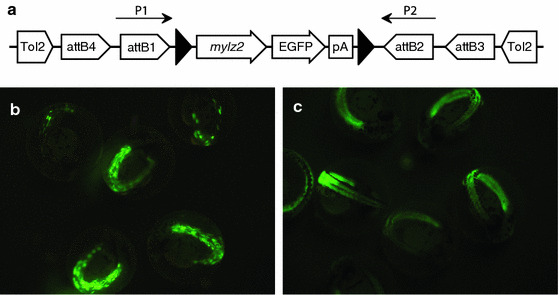
a Schematic drawing of FRT mylz2-EGFP construct for assaying FLPe-mediated site-specific recombination. Black triangles represent FRT sites. The Tol2Kit (Kwan et al. 2007), which utilizes Gateway cloning (Invitrogen), was used to create this construct. Tol2 sites at either end of the construct facilitated zebrafish transgenesis. b Founder zebrafish embryos microinjected with the construct, FRT mylz2-EGFP, displayed mosaicism by 48 hpf. c Hemizygous F1 embryos derived from mosaic founder fish that genotyped positive for the transgene in their sperm displayed even distribution of EGFP expression throughout
Materials and methods
Zebrafish care
The *AB line of wild-type zebrafish were raised and spawned as previously described (Westerfield 2000). Zebrafish were maintained at a water temperature of 28.5°C in an air temperature controlled room with a 14 h light/10 h dark photoperiod. Zebrafish were fed a combination of paramecia, Artemia nauplii (Argent Laboratories, Redmond, WA) and commercial fish food twice daily.
DNA transgene construction
mylz2-EGFP (Xu et al. 1999), was amplified from pMLC2f-EGFP1 with primers containing FRT sites (underlined): 5′- ggggacaagtttgtacaaaaaagcaggctgaagttcctattctctagaaagtataggaacttcattcgccacagaggaatgagccacc-3′ and 5′- ggggaccactttgtacaagaaagctgggtgaagttcctatactttctagagaataggaacttctttatttgtgaaatttgtgatgcta-3′. The primers also contained attB sites (italicized) such that the PCR product would be compatible with the middle entry clone of the Tol2Kit (Kwan et al. 2007). A 5′ entry clone and a 3′ entry clone were combined in an LR reaction with the FRT-flanked mylz2-EGFP middle entry clone and a pDestTol2pA2 destination vector (Kwan et al. 2007) to produce FRT mylz2-EGFP (Fig. 1a).
In vitro synthesis of Tol2 RNA
Tol2 transposase RNA was generated from pCS2FA-transposase (Kwan et al. 2007). The plasmid was linearized with NotI and capped RNA was synthesized using the mMESSAGE mMACHINE SP6 Kit (Ambion, Austin, TX). RNA was purified with an RNeasy Mini Kit (Qiagen, Valencia, CA) and LiCl precipitated.
Microinjection and production of a stable transgenic zebrafish line
Tol2 transposon-mediated transgenesis methods (Kwan et al. 2007) were used to produce stable lines of transgenic zebrafish. Immediately prior to microinjection, FRT mylz2-EGFP plasmid DNA and Tol2 transposase RNA were combined to a final concentration of 5 ng/μl each. The DNA/RNA mixture was microinjected into the blastodisc of single cell-stage wild-type zebrafish embryo. Injected embryos were then moved to a 28.5°C incubator and screened for EGFP by 48 hpf. Germline positive transgenic fish were identified as those which produced EGFP positive T1 embryos.
Fluorescent microscopy
Microinjected zebrafish were anesthetized in tricaine/MS-222 and observed under a MZ16F stereomicroscope (Leica, Bannockburn, IL) with a GFP2 filter set. Photographs were obtained with a 12-megapixel DFC 500 digital camera (Leica).
In vitro synthesis of FLPe RNA and microinjection
FLPe RNA was generated from pCSFLPe (Werdien et al. 2001). The plasmid was linearized with NotI and capped RNA was synthesized using the mMESSAGE mMACHINE SP6 Kit (Ambion). RNA was purified with an RNeasy Mini Kit (Qiagen) and LiCl precipitated. FLPe RNA was quantified and diluted down to 75 ng/μl before being microinjected into single cell transgenic zebrafish embryos. T2 single-cell embryos from multiple clutches derived from outcrossing hemizygous transgenic T1 male zebrafish were microinjected with approximately 2 nl of FLPe capped RNA over the course of several days. Fish that had inherited the transgene, as evidenced by a fluorescent phenotype or by the presence of recombinase-excision footprint, were raised to sexual maturity and sperm from males was genotyped and used to fertilize eggs from wild-type females to determine whether excision of the transgene had occurred in the germline.
PCR analysis and DNA sequencing
FLPe excision primers, 5′-CTCGGAATGTCAATACACCTTGCAGAG-3′ and 5′-GTTGATGCCGGTGAACGTGCAAA-3′, were used to verify site-specific excision from FLPe RNA-injected transgenic zebrafish. Genomic DNA from finclips of FLPe RNA-injected zebrafish was used as the template for PCR. PCR products were gel purified with a QIAquick Gel Extraction Kit (Qiagen) and cloned into a pCR2.1-TOPO vector (Invitrogen). Colonies were grown in liquid LB with the appropriate antibiotic and plasmid DNA was obtained with a QIAprep Spin Miniprep Kit (Qiagen). DNA sequence was determined on a 3730 Capillary Electrophoresis Genetic Analyzer (Applied Biosystems, Foster City, CA).
Results and discussion
Production of a stable transgenic zebrafish line containing FRT mylz2-EGFP
To create a stable transgenic line of zebrafish containing FRT mylz2-EGFP (Fig. 1a), 716 wild-type zebrafish eggs were microinjected with the DNA construct/transposase RNA mix. Of the 240 embryos that survived past day five, 39 were mosaic for EGFP expression (Fig. 1b). At sexual maturity, sperm from the 27 EGFP mosaic zebrafish males was obtained and a PCR screen for the transgene was performed to identify male transgenic germline positive founders. One male genotyped positive for the transgene (data not shown) and was used for in vitro fertilization of wild-type zebrafish eggs. Of the 387 wild-type zebrafish eggs that were in vitro fertilized with the PCR-positive sperm, 78 T1 embryos were hemizygous for EGFP (Fig. 1c).
An advantage of using the Tol2 transposon-mediated transgenesis method is that transgene insertions are always single copy (Kawakami 2005; Kwan et al. 2007) unlike the concatemers that are common with traditional microinjection methods. A 1:1 ratio of GFP+ to GFP− fish (30:32, 12:9, 10:11, 89:95, 29:24, 20:20, 28:27, 8:8) was observed when transgenic T1 offspring were outcrossed to wild-type zebrafish and suggested that the transgene was segregating as a single locus.
Site-specific excision from FLPe RNA microinjection
Hemizygous transgenic T1 male zebrafish were outcrossed to wild-type female zebrafish to produce T2 embryos for FLPe RNA microinjection. A clutch of embryos obtained by outcrossing a hemizygous transgenic fish would be expected to yield 50% transgenic fish (EGFP+) and 50% wild-type fish (EGFP-). Survival and proportion of fluorescent embryos did not differ between clutches in the same treatment group (P > 0.05; data not shown) and so data for all clutches in each treatment group were pooled. At 48 h after FLPe RNA injection, only 7.7% (17/220) embryos showed any level of muscle-specific EGFP expression (Fig. 2a, b). The remaining 203 embryos appeared wild-type (Table 1). A control hemizygous transgenic outcross that was not microinjected with FLPe RNA produced a clutch with an expected 1:1 ratio of EGFP + to EGFP- fish (Fig. 2c, d; Table 1).
Fig. 2.
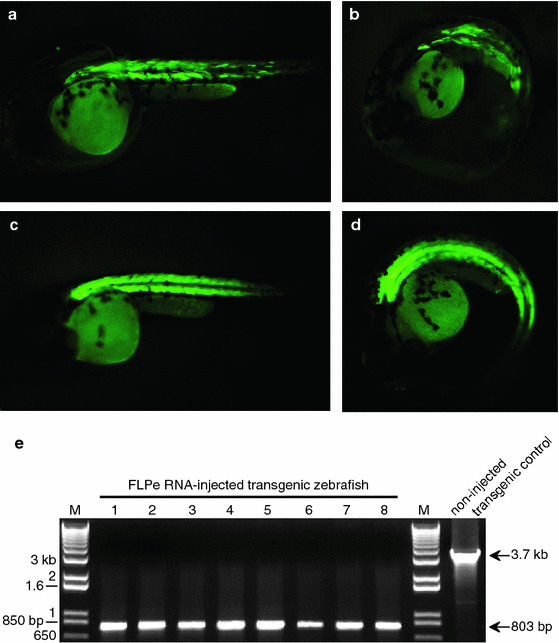
Mosaic EGFP expression after FLPe recombinase RNA microinjection. Forty-eight hours post fertilization, mosaic expression of fluorescence can be observed in FLPe RNA-injected transgenic zebrafish embryos (a, b). Individual muscle cells that have not had the FRT-flanked transgene excised can be seen fluorescing green. In contrast, non-injected transgenic controls (c, d), show evenly distributed EGFP expression throughout the muscles of the zebrafish. e PCR confirmation of recombinase-mediated site-specific recombination. Genomic DNA from finclips of FLPe RNA-injected transgenic zebrafish was used as a DNA template for PCR to confirm excision of the FRT-flanked EGFP transgene. In FLPe-injected zebrafish (1–8), site-specific recombination produced an 803-bp fragment, indicative of excision. In the non-injected transgenic control, an unexcised 3.7-kb band is expected. Excision products were cloned and sequenced to verify precise FLPe-mediated site-specific recombination in zebrafish
Table 1.
Proportion of fish displaying muscle-specific fluorescence following microinjection of FLPe-capped RNA into single cell embryos derived from outcrossing hemizygous FRT mylz2-EGFP transgenic fish with wild-type fish
| Recombinase microinjected | EGFP+a | EGFP− | Total | χ2 | P |
|---|---|---|---|---|---|
| FLPe | 17 (7.7%) | 203 (92.3%) | 220 | 157.25 | <0.001 |
| Control | 28 (49.1%) | 29 (50.9%) | 57 | 0.017 | >0.995 |
a Includes fish that displayed any evidence of EGFP expression
To confirm FLPe-mediated excision of the EGFP transgene from FLPe RNA-injected embryos, PCR was performed on genomic DNA obtained from finclips of injected zebrafish with primers P1 and P2 (Fig. 1a). Site-specific recombination between the FRT sites was expected to produce an 803-bp fragment. As shown in Fig. 2e, finclips collected from eight FLPe RNA-injected transgenic zebrafish with a mosaic GFP phenotype produced an 800-bp fragment indicative of successful FLPe-mediated excision. A 3.7-kb intact fragment was not observed in these samples possibly due to a reduction in the amount of non-recombined template available for amplification as well as preferential amplification of the shorter PCR product. In non-injected transgenic zebrafish controls, a 3.7-kb intact fragment was produced with primers P1 and P2 (Fig. 1a). To verify that the excised PCR fragments were indeed due to FLPe-mediated site-specific recombination, PCR products were sequenced and were found to contain a single 34-bp FRT footprint (data not shown).
To ascertain whether excision had occurred in the germline, FLPe RNA-injected T2 transgenic fish were raised to sexual maturity and outcrossed to wild-type zebrafish. If excision had occurred in the germline, a deviation from the 1:1 ratio of transgenic to wild-type offspring would be expected. Only 4.0% (34/842) of embryos derived from outcrosses of FLPe RNA-injected fish that exhibited a mosaic phenotype appeared EGFP + by 48 hpf (Table 2). When multiple FLPe RNA-injected transgenic fish that lacked a fluorescent phenotype (EGFP-) were outcrossed to wild-type zebrafish, 0.0% (0/339) of the embryos were positive for EGFP expression (Table 2) suggesting that these fish had successfully excised the transgene from the germ cell lineage.
Table 2.
Segregation ratios observed in the offspring derived from outcrossing confirmed FRT mylz2-EGFP transgenic fish that had been microinjected with FLPe-capped RNA at the single cell stage
| Recombinase microinjected | Parental phenotype | Offspring phenotype | Total | χ2 | P | |
|---|---|---|---|---|---|---|
| EGFP+ | EGFP− | |||||
| FLPe | Mosaic muscle-specific fluorescence | 34 (4.04%) | 806 (95.96%) | 842 | 711.5 | <0.001 |
| FLPe | No fluorescence | 0 (0%) | 339 (100%) | 339 | 339 | <0.001 |
During zebrafish development, primordial germ cells can be specified as those containing vasa, a molecular marker for germline cells. Vasa transcript can be found as early as the two-cell stage and the number of vasa-containing cells (4) remains constant up to the 1,000-cell stage (3 hpf) (Raz 2002). However, somites, which eventually differentiate into sclerotome (vertebrae), dermatome (dermis), and myotome (skeletal muscle), do not begin to appear until the segmentation stage (10–24 hpf) (Kimmel et al. 1995). The fact that none of the FLPe RNA-injected transgenic fish that lacked a fluorescent phenotype were germ cell-mosaic suggests that FLPe excision frequently occurred at a very early stage, thereby excising the transgene from all cell lineages.
One other study has examined FLPe site-specific recombination in zebrafish (Boniface et al. 2009). In that study transgenic zebrafish expressing Cre or FLPo (a mouse codon-optimized version of FLPe) under the control of a heat shock promoter were crossed to a recombinase reporter line and inversion efficiencies were recorded. The authors reported that following an hour long heat shock (38°C) induction, Cre expression inverted 82% of the germline reporter construct whereas FLPo expression inverted only 31% of the germline reporter construct. Given FLP activity is optimal near 30°C (Buchholz et al. 1996), it would be interesting to compare the efficiency of these two recombinases, and that of wild-type FLP in zebrafish maintained in ambient water temperatures of 28.5°C.
In conclusion, we found that the FLP/FRT site-specific recombination system from S. cerevisiae was very active in zebrafish maintained at a water temperature of 28.5°C. FLPe-mediated excision occurred rapidly following RNA microinjection into single-cell embryos, and frequently removed the FRT-flanked transgene from the germ cell lineage of transgenic fish. Used in conjunction with Cre/loxP, FLP/FRT will allow zebrafish researchers to design more elegant experiments similar to those already seen in mice (Kondo et al. 2006) and plants (Djukanovic et al. 2008).
Acknowledgments
We would like to thank Dr. Ryffel for his kind gift of pCSFLPe and Dr. Gong for pMLC2f-EGFP1. This research was supported by a grant from the United States Department of Agriculture Biotechnology Risk Assessment Grant (USDA BRAG) #2005-33120-16451 to A.L.V.
Abbreviations
- EGFP
Enhanced green fluorescent protein
- FLPe
Enhanced FLP recombinase
- FRT
FLP recognition target
- hpf
Hours post fertilization
References
- Austin S, Ziese M, Sternberg N. A novel role for site-specific recombination in maintenance of bacterial replicons. Cell. 1981;25:729–736. doi: 10.1016/0092-8674(81)90180-X. [DOI] [PubMed] [Google Scholar]
- Bischof J, Basler K. Recombinases and their use in gene activation, gene inactivation, and transgenesis. Methods Mol Biol. 2008;420:175–195. doi: 10.1007/978-1-59745-583-1_10. [DOI] [PubMed] [Google Scholar]
- Boniface EJ, Lu J, Victoroff T, Zhu M, Chen W. FlEx-based transgenic reporter lines for visualization of Cre and Flp activity in live zebrafish. Genesis. 2009;47:484–491. doi: 10.1002/dvg.20526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz F, Ringrose L, Angrand PO, Rossi F, Stewart AF. Different thermostabilities of FLP and Cre recombinases: implications for applied site-specific recombination. Nucleic Acids Res. 1996;24:4256–4262. doi: 10.1093/nar/24.21.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz F, Angrand PO, Stewart AF. Improved properties of FLP recombinase evolved by cycling mutagenesis. Nat Biotechnol. 1998;16:657–662. doi: 10.1038/nbt0798-657. [DOI] [PubMed] [Google Scholar]
- Djukanovic V, Lenderts B, Bidney D, Lyznik LA. A Cre::FLP fusion protein recombines FRT or loxP sites in transgenic maize plants. Plant Biotechnol J. 2008;6:770–781. doi: 10.1111/j.1467-7652.2008.00357.x. [DOI] [PubMed] [Google Scholar]
- Dong J, Stuart GW. Transgene manipulation in zebrafish by using recombinases. Methods Cell Biol. 2004;77:363–379. doi: 10.1016/S0091-679X(04)77020-X. [DOI] [PubMed] [Google Scholar]
- Dymecki SM. Flp recombinase promotes site-specific DNA recombination in embryonic stem cells and transgenic mice. Proc Natl Acad Sci U S A. 1996;93:6191–6196. doi: 10.1073/pnas.93.12.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Langenau DM, Madge JA, Quinkertz A, Gutierrez A, Neuberg DS, Kanki JP, Look AT. Heat-shock induction of T-cell lymphoma/leukaemia in conditional Cre/lox-regulated transgenic zebrafish. Br J Haematol. 2007;138:169–175. doi: 10.1111/j.1365-2141.2007.06625.x. [DOI] [PubMed] [Google Scholar]
- Futcher AB. Copy number amplification of the 2 micron circle plasmid of Saccharomyces cerevisiae. J Theor Biol. 1986;119:197–204. doi: 10.1016/S0022-5193(86)80074-1. [DOI] [PubMed] [Google Scholar]
- Hans S, Kaslin J, Freudenreich D, Brand M. Temporally-controlled site-specific recombination in zebrafish. PLoS ONE. 2009;4:e4640. doi: 10.1371/journal.pone.0004640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki H, Suzuki H, Itohara S. High-efficiency CAG-FLPe deleter mice in C57BL/6 J background. Exp Anim. 2006;55:137–141. doi: 10.1538/expanim.55.137. [DOI] [PubMed] [Google Scholar]
- Kawakami K. Transposon tools and methods in zebrafish. Dev Dyn. 2005;234:244–254. doi: 10.1002/dvdy.20516. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullman B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kondo S, Takahashi Y, Shiozawa S, Ichise H, Yoshida N, Kanegae Y, Saito I. Efficient sequential gene regulation via flp-and cre-recombinase using adenovirus vector in mammalian cells including mouse es cells. Microbiol Immunol. 2006;50:831–843. doi: 10.1111/j.1348-0421.2006.tb03850.x. [DOI] [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien CB. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- Lakso M, Sauer B, Mosinger B, Jr, Lee EJ, Manning RW, Yu SH, Mulder KL, Westphal H. Targeted oncogene activation by site-specific recombination in transgenic mice. Proc Natl Acad Sci U S A. 1992;89:6232–6236. doi: 10.1073/pnas.89.14.6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenau DM, Feng H, Berghmans S, Kanki JP, Kutok JL, Look AT. Cre/lox-regulated transgenic zebrafish model with conditional myc-induced T cell acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2005;102:6068–6073. doi: 10.1073/pnas.0408708102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WY, Wang Y, Qin Y, Wang YP, Zhu ZY. Site-directed gene integration in transgenic zebrafish mediated by Cre recombinase using a combination of mutant lox sites. Mar Biotechnol. 2007;9:420–428. doi: 10.1007/s10126-007-9000-x. [DOI] [PubMed] [Google Scholar]
- Ludwig DL, Stringer JR, Wight DC, Doetschman HC, Duffy JJ. FLP-mediated site-specific recombination in microinjected murine zygotes. Transgenic Res. 1996;5:385–395. doi: 10.1007/BF01980203. [DOI] [PubMed] [Google Scholar]
- Meyers EN, Lewandoski M, Martin GR. An Fgf8 mutant allelic series generated by Cre- and Flp-mediated recombination. Nat Genet. 1998;18:136–141. doi: 10.1038/ng0298-136. [DOI] [PubMed] [Google Scholar]
- Nagy A. Cre recombinase: the universal reagent for genome tailoring. Genesis. 2000;26:99–109. doi: 10.1002/(SICI)1526-968X(200002)26:2<99::AID-GENE1>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Nagy A, Mar L, Watts G. Creation and use of a cre recombinase transgenic database. Methods Mol Biol. 2009;530:365–378. doi: 10.1007/978-1-59745-471-1_19. [DOI] [PubMed] [Google Scholar]
- Orban PC, Chui D, Marth JD. Tissue- and site-specific DNA recombination in transgenic mice. Proc Natl Acad Sci U S A. 1992;89:6861–6865. doi: 10.1073/pnas.89.15.6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz E. Primordial germ cell development in zebrafish. Semin Cell Dev Biol. 2002;13:489–495. doi: 10.1016/S1084952102001027. [DOI] [PubMed] [Google Scholar]
- Rodriguez CI, Buchholz F, Galloway J, Sequerra R, Kasper J, Ayala R, Stewart AF, Dymecki SM. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat Genet. 2000;25:139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- Sternberg N, Hamilton D. Bacteriophage P1 site-specific recombination. I. Recombination between loxP sites. J Mol Biol. 1981;150:467–486. doi: 10.1016/0022-2836(81)90375-2. [DOI] [PubMed] [Google Scholar]
- Thummel R, Burket CT, Brewer JL, Sarras MP, Jr, Li L, Perry M, McDermott JP, Sauer B, Hyde DR, Godwin AR. Cre-mediated site-specific recombination in zebrafish embryos. Dev Dyn. 2005;233:1366–1377. doi: 10.1002/dvdy.20475. [DOI] [PubMed] [Google Scholar]
- Turakainen H, Saarimaki-Vire J, Sinjushina N, Partanen J, Savilahti H. Transposition-based method for the rapid generation of gene-targeting vectors to produce Cre/Flp-modifiable conditional knock-out mice. PLoS ONE. 2009;4:e4341. doi: 10.1371/journal.pone.0004341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werdien D, Peiler G, Ryffel GU. FLP and Cre recombinase function in Xenopus embryos. Nucl Acids Res. 2001;29:e53. doi: 10.1093/nar/29.11.e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book—a guide for the labratory use of zebrafish (Danio rerio) Eugene, Oregon: University of Oregon Press; 2000. [Google Scholar]
- Xu Y, He J, Tian HL, Chan CH, Liao J, Yan T, Lam TJ, Gong Z. Fast skeletal muscle-specific expression of a zebrafish myosin light chain 2 gene and characterization of its promoter by direct injection into skeletal muscle. DNA Cell Biol. 1999;18:85–95. doi: 10.1089/104454999315655. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Shook NA, Kanisicak O, Yamamoto S, Wosczyna MN, Camp JR, Goldhamer DJ. A multifunctional reporter mouse line for Cre- and FLP-dependent lineage analysis. Genesis. 2009;47:107–114. doi: 10.1002/dvg.20474. [DOI] [PMC free article] [PubMed] [Google Scholar]


