Abstract
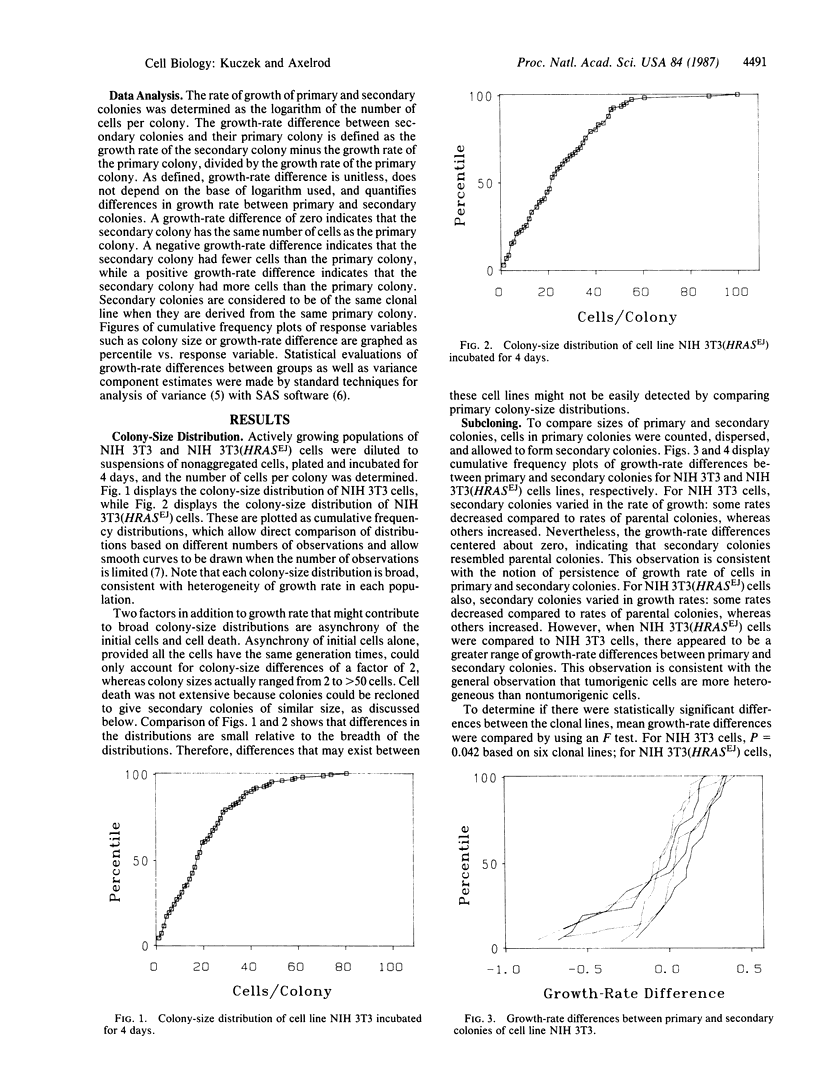
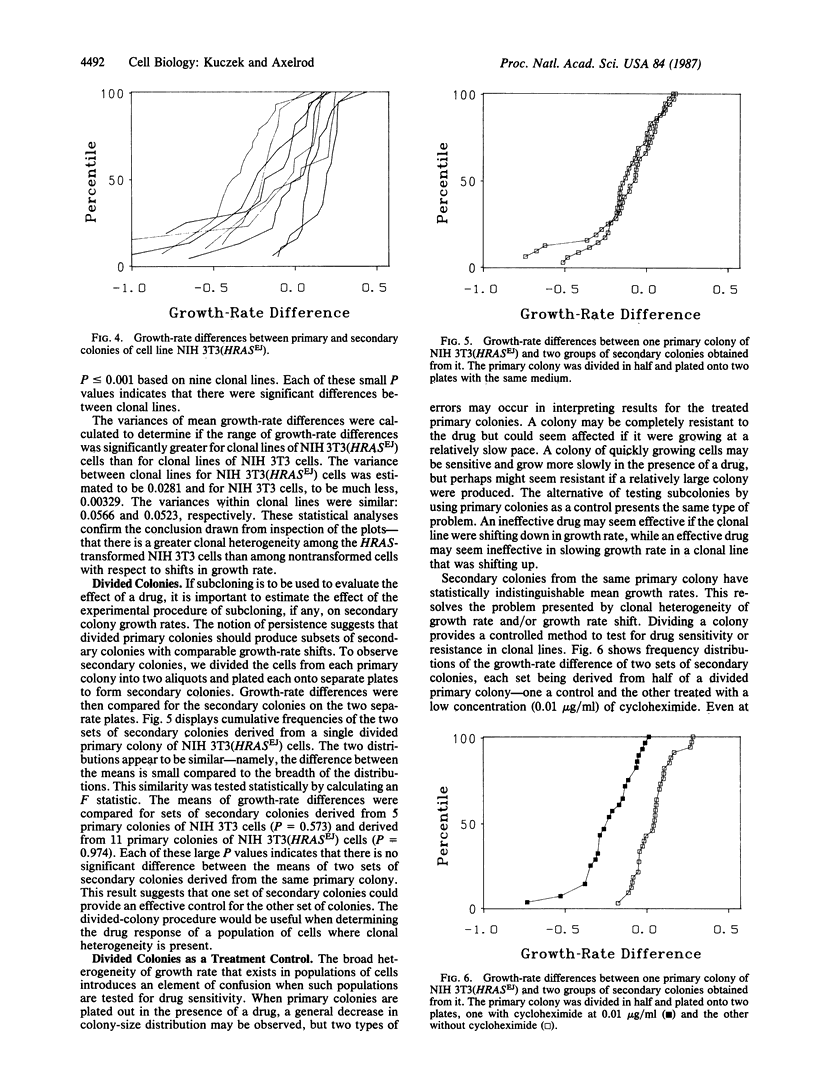
In vitro tests for predicting the response of tumors to chemotherapeutic agents might be improved if they were modified to take into account tumor-cell heterogeneity. We have studied the heterogeneity of cellular growth rate and drug response in mouse fibroblast NIH 3T3 cells and in NIH 3T3 cells transformed with the human HRAS gene (homologue of the Harvey sarcoma virus oncogene v-Ha-ras) from the EJ human bladder carcinoma cell line. Growth-rate heterogeneity was detected as a broad distribution of numbers of cells per colony. In spite of this heterogeneity, secondary colonies have numbers of cells per colony that resemble that of the primary colony from which they were derived. The variance between unrelated secondary colonies is increased by HRASEJ. Colony-size measurements are reliable because primary colonies divided in half formed two groups of secondary colonies (on two separate plates) that had indistinguishable mean colony sizes. Based on these observations, a divided-colony procedure was devised to detect the drug response of heterogeneous cell populations. Primary colonies are divided into two groups of cells, one of which is treated with a drug and the other is left untreated as a control. The size distribution of treated secondary colonies is then compared to that of the untreated control and to that of the primary colony from which it was derived. The divided-colony procedure is proposed as a modification of the human-tumor-cloning system to increase the sensitivity and reliability of in vitro procedures used to determine the drug response of heterogeneous tumor-cell populations.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alder S., Ciampi A., McCulloch E. A. A kinetic and clonal analysis of heterogeneity in K562 cells. J Cell Physiol. 1984 Feb;118(2):186–192. doi: 10.1002/jcp.1041180211. [DOI] [PubMed] [Google Scholar]
- Bernstein S. C., Weinberg R. A. Expression of the metastatic phenotype in cells transfected with human metastatic tumor DNA. Proc Natl Acad Sci U S A. 1985 Mar;82(6):1726–1730. doi: 10.1073/pnas.82.6.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondy G. P., Wilson S., Chambers A. F. Experimental metastatic ability of H-ras-transformed NIH3T3 cells. Cancer Res. 1985 Dec;45(12 Pt 1):6005–6009. [PubMed] [Google Scholar]
- Bradley M. O., Kraynak A. R., Storer R. D., Gibbs J. B. Experimental metastasis in nude mice of NIH 3T3 cells containing various ras genes. Proc Natl Acad Sci U S A. 1986 Jul;83(14):5277–5281. doi: 10.1073/pnas.83.14.5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castor L. N. A G1 rate model accounts for cell-cycle kinetics attributed to 'transition probability'. Nature. 1980 Oct 30;287(5785):857–859. doi: 10.1038/287857a0. [DOI] [PubMed] [Google Scholar]
- Dairkee S. H., Glaser D. A. A mutagen-testing assay based on heterogeneity in diameter and integrated optical density of mammalian cell colonies. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2112–2116. doi: 10.1073/pnas.81.7.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Der C. J., Krontiris T. G., Cooper G. M. Transforming genes of human bladder and lung carcinoma cell lines are homologous to the ras genes of Harvey and Kirsten sarcoma viruses. Proc Natl Acad Sci U S A. 1982 Jun;79(11):3637–3640. doi: 10.1073/pnas.79.11.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eva A., Tronick S. R., Gol R. A., Pierce J. H., Aaronson S. A. Transforming genes of human hematopoietic tumors: frequent detection of ras-related oncogenes whose activation appears to be independent of tumor phenotype. Proc Natl Acad Sci U S A. 1983 Aug;80(16):4926–4930. doi: 10.1073/pnas.80.16.4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita J., Srivastava S. K., Kraus M. H., Rhim J. S., Tronick S. R., Aaronson S. A. Frequency of molecular alterations affecting ras protooncogenes in human urinary tract tumors. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3849–3853. doi: 10.1073/pnas.82.11.3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallick G. E., Kurzrock R., Kloetzer W. S., Arlinghaus R. B., Gutterman J. U. Expression of p21ras in fresh primary and metastatic human colorectal tumors. Proc Natl Acad Sci U S A. 1985 Mar;82(6):1795–1799. doi: 10.1073/pnas.82.6.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldie J. H., Coldman A. J. A mathematic model for relating the drug sensitivity of tumors to their spontaneous mutation rate. Cancer Treat Rep. 1979 Nov-Dec;63(11-12):1727–1733. [PubMed] [Google Scholar]
- Goldie J. H., Coldman A. J., Gudauskas G. A. Rationale for the use of alternating non-cross-resistant chemotherapy. Cancer Treat Rep. 1982 Mar;66(3):439–449. [PubMed] [Google Scholar]
- Greig R. G., Koestler T. P., Trainer D. L., Corwin S. P., Miles L., Kline T., Sweet R., Yokoyama S., Poste G. Tumorigenic and metastatic properties of "normal" and ras-transfected NIH/3T3 cells. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3698–3701. doi: 10.1073/pnas.82.11.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUGHES W. H. The inheritance of differences in growth rate in Escherichia coli. J Gen Microbiol. 1955 Apr;12(2):265–268. doi: 10.1099/00221287-12-2-265. [DOI] [PubMed] [Google Scholar]
- Heppner G. H., Dexter D. L., DeNucci T., Miller F. R., Calabresi P. Heterogeneity in drug sensitivity among tumor cell subpopulations of a single mammary tumor. Cancer Res. 1978 Nov;38(11 Pt 1):3758–3763. [PubMed] [Google Scholar]
- Heppner G. H. Tumor heterogeneity. Cancer Res. 1984 Jun;44(6):2259–2265. [PubMed] [Google Scholar]
- Hersh R. T., Kitos P. A. Is G1 normally distributed? J Theor Biol. 1980 Sep 7;86(1):117–122. doi: 10.1016/0022-5193(80)90069-7. [DOI] [PubMed] [Google Scholar]
- Humphries R. K., Eaves A. C., Eaves C. J. Self-renewal of hemopoietic stem cells during mixed colony formation in vitro. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3629–3633. doi: 10.1073/pnas.78.6.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel M., Darzynkiewicz Z., Arino O., Traganos F. Analysis of a cell cycle model based on unequal division of metabolic constituents to daughter cells during cytokinesis. J Theor Biol. 1984 Oct 21;110(4):637–664. doi: 10.1016/s0022-5193(84)80149-6. [DOI] [PubMed] [Google Scholar]
- Krontiris T. G., DiMartino N. A., Colb M., Parkinson D. R. Unique allelic restriction fragments of the human Ha-ras locus in leukocyte and tumour DNAs of cancer patients. 1985 Jan 31-Feb 6Nature. 313(6001):369–374. doi: 10.1038/313369a0. [DOI] [PubMed] [Google Scholar]
- Kurzrock R., Gallick G. E., Gutterman J. U. Differential expression of p21ras gene products among histological subtypes of fresh primary human lung tumors. Cancer Res. 1986 Mar;46(3):1530–1534. [PubMed] [Google Scholar]
- Nedelman J., Rubinow S. I. Investigation into the experimental kinetic support of the two-state model of the cell cycle. Cell Biophys. 1980 Sep;2(3):207–231. doi: 10.1007/BF02790450. [DOI] [PubMed] [Google Scholar]
- POWELL E. O. An outline of the pattern of bacterial generation times. J Gen Microbiol. 1958 Apr;18(2):382–417. doi: 10.1099/00221287-18-2-382. [DOI] [PubMed] [Google Scholar]
- POWELL E. O., ERRINGTON F. P. Generation times of individual bacteria: some corroborative measurements. J Gen Microbiol. 1963 May;31:315–327. doi: 10.1099/00221287-31-2-315. [DOI] [PubMed] [Google Scholar]
- Peterson J. A. Analysis of variability in albumin content of sister hepatoma cells and a model for geometric phenotypic variability (quantitative shift model). Somat Cell Mol Genet. 1984 Jul;10(4):345–357. doi: 10.1007/BF01535630. [DOI] [PubMed] [Google Scholar]
- Pharr P. N., Nedelman J., Downs H. P., Ogawa M., Gross A. J. A stochastic model for mast cell proliferation in culture. J Cell Physiol. 1985 Dec;125(3):379–386. doi: 10.1002/jcp.1041250304. [DOI] [PubMed] [Google Scholar]
- Pozzatti R., Muschel R., Williams J., Padmanabhan R., Howard B., Liotta L., Khoury G. Primary rat embryo cells transformed by one or two oncogenes show different metastatic potentials. Science. 1986 Apr 11;232(4747):223–227. doi: 10.1126/science.3456644. [DOI] [PubMed] [Google Scholar]
- Schlag P., Schreml W. Heterogeneity in growth pattern and drug sensitivity of primary tumor and metastases in the human tumor colony-forming assay. Cancer Res. 1982 Oct;42(10):4086–4089. [PubMed] [Google Scholar]
- Sekiya T., Fushimi M., Hori H., Hirohashi S., Nishimura S., Sugimura T. Molecular cloning and the total nucleotide sequence of the human c-Ha-ras-1 gene activated in a melanoma from a Japanese patient. Proc Natl Acad Sci U S A. 1984 Aug;81(15):4771–4775. doi: 10.1073/pnas.81.15.4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skipper H. E. The forty-year-old mutation theory of Luria and Delbrück and its pertinence to cancer chemotherapy. Adv Cancer Res. 1983;40:331–363. doi: 10.1016/s0065-230x(08)60683-1. [DOI] [PubMed] [Google Scholar]
- Stamatoyannopoulos G., Kurnit D. M., Papayannopoulou T. Stochastic expression of fetal hemoglobin in adult erythroid cells. Proc Natl Acad Sci U S A. 1981 Nov;78(11):7005–7009. doi: 10.1073/pnas.78.11.7005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TILL J. E., MCCULLOCH E. A., SIMINOVITCH L. A STOCHASTIC MODEL OF STEM CELL PROLIFERATION, BASED ON THE GROWTH OF SPLEEN COLONY-FORMING CELLS. Proc Natl Acad Sci U S A. 1964 Jan;51:29–36. doi: 10.1073/pnas.51.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanigawa N., Mizuno Y., Hashimura T., Honda K., Satomura K., Hikasa Y., Niwa O., Sugahara T., Yoshida O., Kern D. H. Comparison of drug sensitivity among tumor cells within a tumor, between primary tumor and metastases, and between different metastases in the human tumor colony-forming assay. Cancer Res. 1984 Jun;44(6):2309–2312. [PubMed] [Google Scholar]
- Theillet C., Lidereau R., Escot C., Hutzell P., Brunet M., Gest J., Schlom J., Callahan R. Loss of a c-H-ras-1 allele and aggressive human primary breast carcinomas. Cancer Res. 1986 Sep;46(9):4776–4781. [PubMed] [Google Scholar]
- Tsuruo T., Fidler I. J. Differences in drug sensitivity among tumor cells from parental tumors, selected variants, and spontaneous metastases. Cancer Res. 1981 Aug;41(8):3058–3064. [PubMed] [Google Scholar]
- Wu A. M. Regulation of self-renewal of human T lymphocyte colony-forming units (TL-CFUs). J Cell Physiol. 1983 Oct;117(1):101–108. doi: 10.1002/jcp.1041170114. [DOI] [PubMed] [Google Scholar]
- Yokota J., Tsunetsugu-Yokota Y., Battifora H., Le Fevre C., Cline M. J. Alterations of myc, myb, and rasHa proto-oncogenes in cancers are frequent and show clinical correlation. Science. 1986 Jan 17;231(4735):261–265. doi: 10.1126/science.3941898. [DOI] [PubMed] [Google Scholar]
- Yung W. K., Shapiro J. R., Shapiro W. R. Heterogeneous chemosensitivities of subpopulations of human glioma cells in culture. Cancer Res. 1982 Mar;42(3):992–998. [PubMed] [Google Scholar]


