Abstract
The traditional antiviral assays for the determination of interferon potency are reported to have considerable variability between and within assays. Although several reporter gene assays based on interferon-inducible promoter activities have been reported, data from comprehensive validation studies are lacking and few studies have been conducted to analyze the variant forms of interferons, which could have undesirable clinical implications. Here, a reporter gene assay employing a HEK293 cell line stably transfected with luciferase gene under the control of interferon-stimulated response element promoter was developed and validated. The assay was found to be more sensitive, with a larger detection range than the antiviral assay. Several cytokines tested did not interfere with the test, suggesting the assay possesses a certain degree of selectivity. Moreover, the robustness of the assay was demonstrated by minimal variations in the results generated by different analysts and cell passage number (up to 52 passages). Finally, the method was employed to analyze several interferon variants (interferon-α 2a) and we found that the aggregated form has completely lost its potency; while a modest loss of bioactivity in oxidized interferon was observed (approx. 23%), the deamidated form essentially retained its activity.
1. Introduction
Interferons (IFNs) are produced in response to bacterial and viral infections, and play essential roles in host defence [1]. In addition, they have also been found to possess anticancer and immunomodulatory effects [1–4]. They are classified based on cellular origin and on the type of receptors to which they bind. Those classified to date include more than 10 IFN-α and a single β species (termed collectively IFN-α/β), IFN-γ and 3 IFN-λ species. In humans and mice, the α and β species are encoded by genes clustered in regions of chromosomes 9 and 4 [5, 6]. IFN-α/β are classified as type I since they bind to IFN cell surface receptors type 1. Although they have different binding affinities, they have similar biological effects. IFN-γ belongs to type II since it binds only to type 2 receptors. Various types of cells including dendritic cells, leukocytes, fibroblasts, epithelial cells, and macrophages, are known to produce IFN-α and IFN-β while activated T-cells and natural killer cells have been reported to produce IFN-γ [5, 6]. Only type I and type II IFNs have been approved for use in humans; the 3 IFN-λ species are newly classified [7], and have not been approved for human therapy.
Recombinant IFNs for human therapies are produced in mammalian cells or bacteria and have been used over the last decades to treat infectious and autoimmune diseases and in the treatment of selected types of cancer [8–11]. Like many other approved therapeutic products on the market, IFNs have undisputable therapeutic benefits but are also linked to the occurrence of adverse drug reactions (ADRs) or lack of efficacy. Therefore, it is important to determine the bioactivity or potency of IFN prior to its use in humans. The potency of IFN has been determined traditionally by the antiviral assay (AVA) [12, 13], in which the activity of IFN is measured based on its inhibitory effects on viral replication. However, AVA is inherently disadvantageous because of higher assay variations, the requirement for working with virus in Biosafety level-2 (BSL-2) laboratories, and the need to titrate the virus [14–17]. In recent years, potency assays based on IFN-inducible gene expression have been extensively studied as a potential replacement for the traditional AVA [13, 18]. The principle of these reporter gene assays (RGA) is very straightforward, that is, quantitative determination of responses to IFN treatment in indicator cell lines stably transfected with IFN-inducible promoter driven reporter gene products. A variety of IFN-inducible promoters have been investigated including Mx, IFN-stimulated response element (ISRE) or glial fibrillary acidic protein (GFAP), so have the various quantifiable gene products such as Beta Galactosidase (β-Gal), chloramphenicol acetyltransferase (CAT), Secreted Alkaline Phosphatase (SEAP) or luciferase [14–26]. These previous studies suggest that the RGA is generally simpler and faster than AVA. Yet, data are limited in terms of reproducibility, precision and accuracy in addition to the absence of data from analyses of chemically modified IFNs. Currently, the European Pharmacopoeia monograph for IFN bulk solution requires potency determination on the basis of tests for protection of cells against a viral cytopathic effect. Here we report the development of a RGA based on ISRE-driven luciferase activity measurements and subsequent validation of the assay with respect to sensitivity, specificity, reproducibility, precision and robustness in addition to the application of this assay to the potency determination of physically or chemically modified IFNs.
2. Materials and Methods
2.1. IFNs and Other Cytokines
Reference reagent is used in the validation studies, Gb23-902-531 (IFN-β) which was obtained from the National Institute of Health through BEI Resources branch of ATCC. Glo lysis buffer (E266A) and Bright Glo luciferase assay reagent (E2620) were purchased from Promega (Ottawa, ON, Canada), IL-1β (407615), IL-2 (407623), IL-6 (407652) and TNF-α (654205) were obtained from Calbiochem (San Jose, CA). IFN-α 2a and 2b reference standards were obtained from EDQM (European Directorate for the Quality of Medicines, Strasbourg, France). Various IFN-α and IFN-β samples were kindly provided by various manufacturers (see below). The activities of the in-house reference standards or samples used in these designed studies were predetermined by traditional antiviral assays using WHO standards unless otherwise specified.
2.2. Development of Stable Cell Lines Expressing Luciferase under the Control of ISRE (HEK293-ISRE-Luc)
ISRE-Luc reporter plasmid and pIRESp2 encoding puromycin-resistant gene (Clonetech, Mountain View, CA) (ratio 1 : 10) were introduced into HEK293 cells (ATCC, Manassas, VA) by the calcium phosphate method. Cells were selected beginning at 48 hours after transfection for approximately 2 weeks in MEM containing 10% fetal calf serum, antibiotics, and 2 μg/mL puromycin (Sigma). Puromycin-resistant cells were then cloned twice by limiting dilutions to select single cell clones. After that the selected clones were routinely maintained in MEM in the presence of 2 μg/mL of puromycin and 10% fetal calf serum (Gibco, Burlington, ON). Cell passaging is achieved by detaching the cells in 0.25% Trypsin-EDTA and splitting the cells every 3 days.
2.3. RGA Procedure
The procedure is comprised of three simple steps, that is, seeding the cells in the presence of IFN at specified concentrations into 96-well plates, followed by an overnight incubation of the cultures and direct measurement of luciferase activity in the cell lysates. In brief, 96-well Costar plates were seeded with 0.04 × 106 cells in the presence of IFN or controls in a total volume of 100 μL per well. The plate was incubated at 37°C with 5% CO2 for 21 hours. The culture supernatants were then removed by aspiration and the cells washed once with PBS, followed by addition of 70 μL Promega Glo Lysis buffer. The plates were subsequently shaken for 5 minutes on a titre-plate shaker (Fisher Scientific, Ottawa, ON), followed by the addition of 70 μL of Promega Bright Glo Luciferase Assay reagent. Luciferase activity was then determined by reading on a Luminoscan Ascent plate reader and data analysed using Combistat (see below “data analyses”).
2.4. Matrix Interference (Selectivity)
The specificity of the RGA was determined by performing matrix interference experiments. To this end, we used IL-2, IL-1β, IL-6, and TNF-α to treat the cells under the same condition as described above for the analyses of IFN and determined whether the presence of these cytokines could interfere with luciferase output.
2.5. Preparation and Fractionation of Modified IFN Variants
The preparation of modified forms of human IFN (aggregated, deamidated, or oxidized) was carried out as described elsewhere [27] (Diress et al. unpublished data). In brief, the aggregated forms were prepared by incubating the samples at 37–80°C. If samples contained precipitates and/or a milky suspension formed during the incubation, the precipitate was removed by centrifugation. Denaturation and reduction of samples were carried out by incubating IFN-α 2a sample at 25°C in a solution containing 0.01 M sodium phosphate (pH 7.0), 6.0 M guanidine hydrochloride (GdnHCl) or 1, 4-dithiothreitol (DTT), respectively. Oxidation of IFN-α 2a products was carried out according to the procedure described in the European Pharmacopoeia monograph for interferon alpha-2a (2007 #26). Accordingly, IFN-α 2a sample (0.1 mg/mL) was incubated with 0.05% (v/v) hydrogen peroxide (H2O2) at 37°C from 3 to 18 hours. The oxidation was stopped by adding L-methionine (12 mg/mL) and incubating the sample at room temperature for 1 hour. Deamidation was performed by incubating IFN-α 2a (0.1 mg/mL) samples in 20 mM sodium phosphate (pH 8.0) or 20 mM Tris-HCl (pH 8.0) buffers. The samples were incubated at 37°C for a period of one day to seven weeks. Pure fractions containing native, aggregated, denatured and reduced forms were collected from stock IFN-α samples using size exclusion high performance liquid chromatography (SE-HPLC) at room temperature as described by us recently [27]. Oxidized and deamidated variants were collected according to methods described elsewhere (Diress et al. unpublished data). Fractions eluting from the HPLC columns were collected manually based on the elution time on the detector signal and the fractions were >85% pure. Preparation, modification and fraction collection of the samples were performed under sterile conditions to avoid contamination and growth of bacteria.
2.6. AVA for IFN Potency Determination
The antiviral activity of IFN is used in our routine procedure for the determination of IFN potency. In brief, WISH cells (ATCC, Manassas, VA) were seeded in 96-well plates at a concentration of approximately 3 × 105 cells/mL (100 μL per well). The plates were incubated at 37°C in the presence of 5% CO2 for 4–6 hours. In the meantime the standards and test samples were prepared (6 replicates for 3 replicate plates of each dilution). The supernatants from the 96-wells were then removed and the diluted standards and test samples were added. The plates were incubated again for 18–24 hours. The supernatants were then removed and vesicular stomatitis virus (VSV) was added at 100 TCID50 for an additional incubation of 24 hours. The virus-containing supernatants were then removed, followed by staining of the cell layers with crystal violet staining solution (0.5 mg in 20% ethanol) for 30 minutes at room temperature. The staining solution was then carefully aspirated. Destaining solution (50% ethanol/0.1% acetic acid) was subsequently pipetted into the wells and incubated for 4 minutes at room temperature. The plates were read at 630 nm/570 nm in a plate reader (Fisher Scientific, Ottawa, On). The potency of the test sample (IU/mL) was calculated using 4-parameter logistic (4-PL) model.
2.7. Statistical Analyses
Analyses of the data consisted of statistical models used to calculate the relative potencies as well as statistical techniques for method validation. In order to calculate the relative potencies, dose response and linear range, we used the 4-PL model (Section 5.3 of European Pharmacopoeia Statistical Analyses of results of the biological assays and tests). Statistical techniques for method validation employed summary statistics such as mean, standard deviation, coefficient of variation (CV) as well as statistical techniques such as ANOVA and confidence intervals. All statistical tests were carried out at a 5% significance level. Analyses were carried out using Combistats for relative potency calculations and SAS for method validation.
3. Results
3.1. Response of HEK293-ISRE-Luciferase Cells to the Treatments of Various Types of IFN
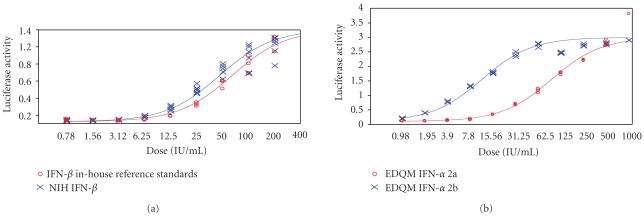
As the assay is to be used for potency determination of human interferon, HEK293 cells were chosen since they are of human origin. Nineteen clones of cells expressing luciferase under the control of ISRE were isolated and subcloned twice by limiting dilution in the presence of puromycin (see Section 2). Eight clones were found to be expressing high levels of luciferase activity in response to IFN treatment in the initial screening process. As no significant difference was found with respect to the responses to IFN treatment among these eight clones, one of them was selected for further studies as the cell line for the subsequent RGA. We first determined how the luciferase activity in the HEK293-ISRE-Luciferase cell clones (denoted as HEK293-ISRE-Luc) responded to various forms of human interferons. Type 1 IFN (α and β) was found to induce luciferase activity in a dose-response fashion (Figure 1), with panels (a) and (b) for IFN-β and IFN-α, respectively. As expected, IFN-γ failed to induce detectable luciferase activity. For potency analysis of IFN-γ, a GAS element-driven reporter gene system should be considered [1–4, 28]. These results are consistent with previous observations that type I and type II IFNs exert their functional activities through different pathways and type I IFN is known to activate the ISRE-driven transcription [2, 3]. We also conducted a time course and found that the highest luciferase activity was obtained approximately 21 hours after the IFN treatment; the luciferase activity begins to decrease after 21–24 hours, with significant decreased levels of luciferase activity detected after 48 hours (not shown). Noticeably, linear ranges for IFN-α and β were found to be 1.5–50 IU/mL and 6.25 to 200 IU/mL, respectively. Given that linear range is generally less heteroscedastic, we decided to analyze the potencies of the IFN in this range for all subsequent experiments unless otherwise specified (See below).
Figure 1.
HEK293-ISRE-Luc cell line responses to treatment with various types of interferons. Panel (a) represents dose response curve obtained with IFN-β. Panel (b) represents dose response curve obtained with IFN-α. The assay was performed as described in “Section 2”. The cells were treated with type I IFN, followed by the analyses of luciferase activity. As expected, although IFN-α and IFN-β are both type I IFNs that drive the ISRE, the signalling pathway used is slightly different and thus we obtain a different dose response curve.
3.2. Precision
3.2.1. Intra-Assay Variation
The experiment consisted of three replicates tested in five different plates. Such design made it possible to better understand the plate-to-plate variability as well as the variation between the three samples. The activity of the samples used in these studies were pre-determined by traditional antiviral assays using international standards (see below). Respective potencies were calculated for each plate. We then calculated summary statistics using the obtained potencies. Our analyses resulted in a CV less than 8% for all three samples (CV% being 6.00, 6.76, and 7.35%, resp.). These results suggest, as shown in Figure 2, that the three lots of samples behave nearly indistinguishably as compared with the standards.
Figure 2.
Intra-assay variation. Three samples were used. Sample 1 was the In-House reference standard (a), with sample #2 and #3 being final products reconstituted from final drug products in syringes (b, c). Each dose was run in triplicate and tested on 5 different plates. The graphs depict the linear range of the tested doses as determined by comparison to the NIH IFN-β standard.
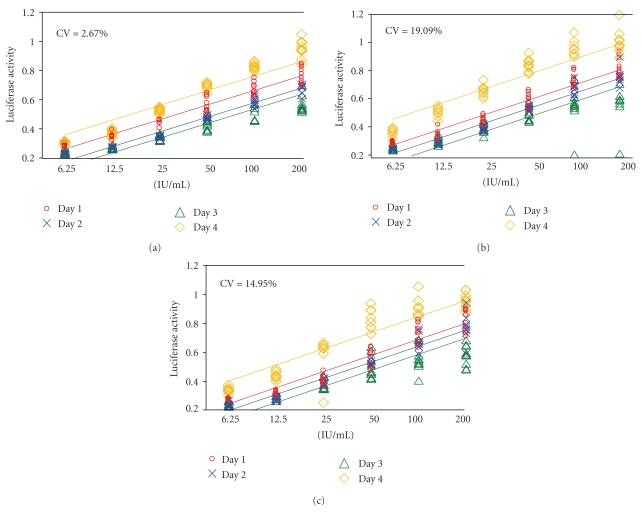
3.2.2. Inter-Assay Variation
In order to determine the inter-assay variation, we ran the assay on four different days. On each day, we performed three replicates in the same manner as explained in the previous section. It is important to note that the only condition changed in all assays was the day on which the assays were performed. Respective potencies were calculated for each day. Our analyses resulted in CV values less than 20% (2.67%, 19.09%, and 14.95%, resp.) for all three samples, suggesting that the inter-assay variation is negligible. In addition, ANOVA suggested no statistically significant day effect (P-values: .273, .087, and .0685 for in-house standard, sample 1 and sample 2, resp., against the NIH reference standards). These findings are confirmed as presented in Figure 3.
Figure 3.
Inter-assay variations (different runs). Three samples were used. Sample 1 was the in-house reference standard (a), with sample #2 and #3 being final products (b,c). Each dose was run in triplicate and tested on 3 separate plates. The graphs represent the linear range of the tested doses as determined by comparison to the international reference standard. The experiments were conducted on 4 different days.
3.2.3. The Effects of Different Analysts on the Assay
Next we wanted to know whether there was any significant difference in results if the experiments were run by different laboratory staff with no prior training for this assay. We asked four technologists to perform the assay based on the standard operating procedure. It is of note that the only condition changed in all assays was the analyst (Figure 4). Respective potencies were calculated on data generated by the four analysts. For sample 2, we obtained CV values less than 20% (14.6%). However, sample 1 gave us a CV value of 22.25% suggesting that prior training should be considered if the laboratory staff is inexperienced in performing RGAs (for more details see “Section 4”). Nevertheless, we observed no statistically significant difference amongst the four analysts (P = .266 and .1816 for sample 1 and sample 2, resp.).
Figure 4.
Determination of variability in experiments conducted by 4 different analysts. Two samples were used (both final products of IFN-β 1a). The analysts who had no prior experiences of conducting the assay were instructed to conduct the experiments by following the protocol. Each sample was run in triplicate and tested on 3 different plates. The graphs represent the linear range of the tested doses as determined by comparison to the international reference standard.
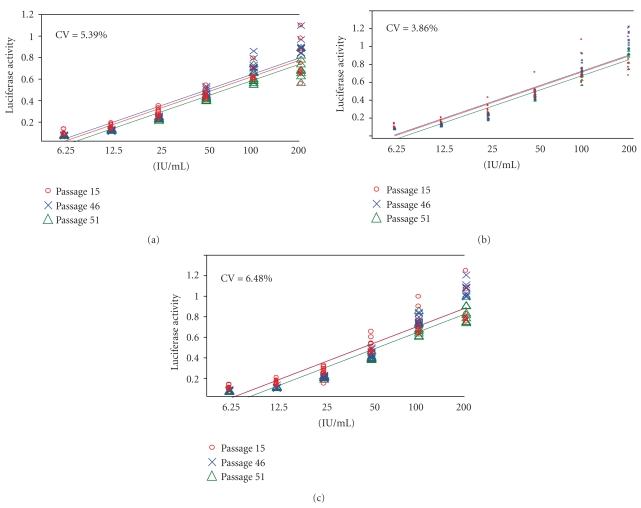
3.3. The Stability of the Cell Lines
In order to evaluate the stability of cell lines, we subsequently tested how cells respond to the IFN treatment at various passage numbers (15, 46 and 51, resp.). Each cell passage was tested in triplicate on three separate plates (Figure 5). CV values based on the calculated potencies for each passage number are very small (5.39%, 3.86%, and 6.48% resp.) which suggest that there is no effect of passage number on responsiveness to IFN in this assay. Furthermore, results from all three samples tested in the stability assays also support the notion that the cells are stable (P > .05).
Figure 5.
Test of the stability of the cell lines. Cells at three different stages (passage #15, 46, 51) were tested in this validation, which involved 3 replicates for each dose and in 3 replicate plates. Sample 1 was the in-house reference standard (a), with sample #2 and #3 being final drug products from syringes (b,c). Each sample was run in triplicate and tested on 3 different plates. The graphs represent the linear range of the tested doses as determined by comparison to the international reference standard.
3.4. Selectivity of HEK293-ISRE-Luc in Response to Different Cytokines (Matrix Interferences)
Experiments were next conducted to assess whether the presence of other cytokines (i.e., matrix interferences) could affect the expression of luciferase activity in the HEK293-ISRE-Luc cells. Four cytokines available in the laboratory were chosen including IL-1β, IL-2, IL-6 and TNF-α. These cytokines were spiked into the medium along with IFN-β and then used to treat the cells. The potency values were compared to those obtained with IFN-β treatment only. As shown in Table 1, no interference was found with the treatments of IL-1β (950000 WHO U/mL), IL-2 (0.5 μg), IL-6 (35000 WHO U/mL) or TNF-α (5000 ng/mL) as the relative potencies are close to 100% with corresponding confidence intervals narrower than 80 to 120%. We also tested wider concentrations of these cytokines (IL-1β: 40–400 U/mL; IL-2: 200–2000 IU/mL; IL-6: 200–2000 IU/mL and TNF-α: 100–1000 ng/mL) and found no significant interference (data not shown). No study has been conducted to measure the interference of other cytokines.
Table 1.
Matrix interference.
| LL of estimated potency | Estimated potency | UL estimated Potency | LL of relative potency (%) | Relative potency | UL of relative potency | |
|---|---|---|---|---|---|---|
| IL-2 | 0.962461 | 1.01024 | 1.06045 | 96.2 | 101.00 | 106.00 |
| IL-6 | 0.929288 | 0.975531 | 1.02393 | 92.9 | 97.60 | 102.40 |
| IL-1β | 0.944126 | 0.991046 | 1.04024 | 94.4 | 99.10 | 104.00 |
| TNFα | 1.0784 | 1.13208 | 1.18928 | 107.8 | 113.20 | 118.90 |
The table shows data from one concentration of IL-2, IL-6, IL-1β, or TNF-α. Wider concentrations of IL-2, IL-6, IL-1β and TNF-α found no interference with our assay (not shown in the table). The tested ranges are the following: IL-2: 200–2000 IU/mL; IL-6: 200–2000 IU/mL; IL-1β: 40–400 U/mL; TNF-α: 100–1000 ng/mL. The values in bold face represent relative potency compared with the reference standards as determined by statistical analyses. LL stands for lower limits of the 95% interval while UL denotes upper limits of the 95% interval.
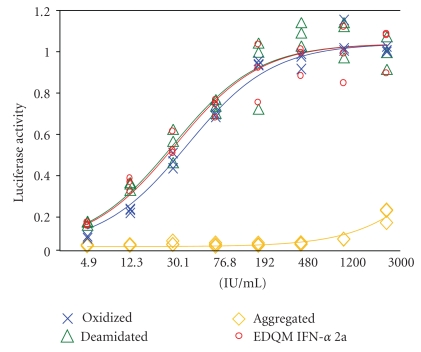
3.5. Analyses of IFN Variants Using the RGA
Unlike drugs derived from small molecules, therapeutic proteins are prone to a number of changes during preparation, formulation or storage. These changes could involve modifications such as oxidation, deamidation, glycosylation, isomerization, aggregation, misfolding, adsorption, or precipitation [29–31]. These modifications could lead to potential loss of therapeutic efficacy or unwanted immune reactions (i.e., ADRs). For instance, aggregation reduces the efficacy of human calcitonin [32] whereas aggregates of IFN-α were found to enhance immunogenicity [33, 34]. So we investigated next whether the RGA could be used to test the potency of modified interferon variants including oxidized, deamidated and aggregated forms. To this end, the variants were prepared and purified as described elsewhere [27] (Diress et al. unpublished data). The IFNs were then compared with the EDQM reference standard (IFN-α 2a). As shown in Figure 6, the deamidated and the oxidized samples behave similarly to the reference standard (in fact it is very hard to distinguish between the standard and the deamidated sample while we can notice a small difference between the standard and the oxidized form). On the other hand, the aggregated form shows no activity. Thus, these findings suggest no significant loss of potency with deamidated IFN while approximately 23% loss of potency was observed with the oxidized form. In addition, the aggregated form completely lost its potency (refer to Table 2). These results suggest that the RGA could be further explored to analyze the potency of the variants.
Figure 6.
Analyses of IFN variants. The variants (IFN-α 2a) were prepared as described in [27] and (Diress et al. unpublished data). The variants included aggregated, deamidated or oxidized forms. The data showed that no significant loss of potency was observed with deamidated IFN while approximately 23% loss of potency was observed with the oxidized form; moreover, the aggregated form completely lost its bioactivity. Each sample was tested in triplicate and potency was determined by comparison to the EDQM IFN-α 2a reference standard.
Table 2.
Relative potency determination of IFN variants compared with the reference standards.
| LL of estimated potency* | Estimated potency* | UL of estimated potency* | LL of relative potency (%) | Relative potency (%)# | UL of relative potency (%) | |
|---|---|---|---|---|---|---|
| Oxidized | 2655720 | 20936900 | 26468300 | 61.30 | 77.50 | 98.00 |
| Deaminated | 22397100 | 28327600 | 35830700 | 83.00 | 104.90 | 132.70 |
| Aggregated | 33793 | 58875 | 99115 | 0.10 | 0.220 | 0.4 |
*:Actual reading of luciferase activity;
#:Relative potency compared with the reference standards as determined by statistical analyses (bold face). The values in bold face refer to the relative potency of the samples with respect to the reference standards (EDQM IFN-α 2a). LL stands for lower limits of the 95% interval while UL denotes upper limits of the 95% interval.
3.6. Validation of RGA and Comparison with the AVA
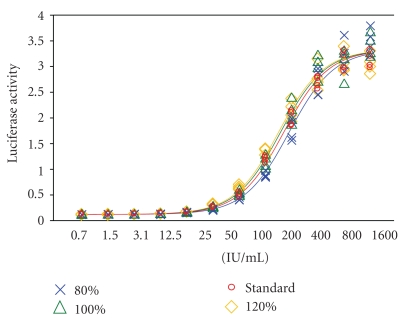
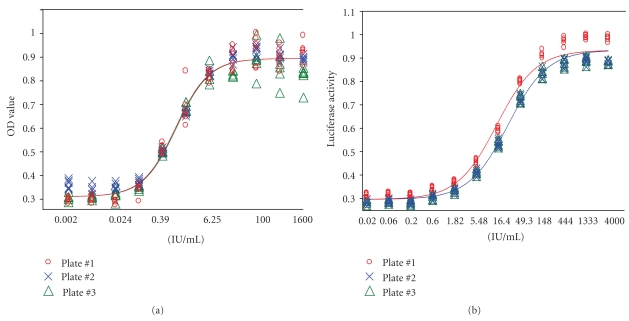
To validate the RGA, known amounts of IFN-β predetermined by the routine AVA were spiked into the culture media and subjected to the RGA for the determination of potency in comparison with the reference standards. Three expected potency ratios, (i.e., 80%, 100% and 120%) of the internal standard concentrations were employed. As shown in Figure 7, the dose response curves for the three “spiked” IFN-β samples satisfy 4-PL model assumptions (i.e., regression, linearity, and parallelism). Most importantly, the concentrations of IFN-β yield the expected potency ratios of 0.8, 1.0, and 1.2, with very narrow 95% confidence intervals (Table 3). Experiments were then conducted to do a head-to-head comparison between RGA and AVA with respect to the detection range and variability. In this study, the IFN-β was tested for 12 different serial dilutions, with each dose containing 6 replicates in three separate plates. We found that both, AVA and RGA, satisfied the 4-PL model assumptions (Figure 8). RGA displayed even better curve fit than AVA did. Moreover, within the replicates for each concentration of the sample, RGA is much less variable than AVA, as demonstrated by smaller CV% (1.89 versus 4.35). Furthermore, it is worth mentioning that we also investigated the plate-to-plate variations, and the variations amongst the three plates are not statistically different between plates (P = .32) although visual inspection of the results might suggest that AVA (Figure 8(a) performs better than RGA (Figure 8(b)). But the truth is that RGA has a much wider detection range (0.24 IU–1600 IU) compared with the AVA (0.098 IU–100 IU) and also shows less heteroscedasticity, that is, the OD values are less variable in response to lower and higher concentrations of IFN (Figure 8). Taken together, these data obtained from the above studies suggest that the new method (RGA) has in general a better performance than the traditional method (AVA).
Figure 7.
Validation of the RGA. Known amounts of IFN-β pre-determined by routine AVA were spiked into the culture media and subjected to RGA. Three expected potency ratios of the internal standard concentrations were employed, that is, 80%, 100%, and 120%. Samples were run as four replicates and potency was determined by comparing to the in-house reference standard.
Table 3.
Relative potency of IFN-β as determined by RGA.
| IFN-β samples (%) | Mean estimated potency | Relative to assessment (%) | Lower and upper limits (95% CI) (%) |
|---|---|---|---|
| 80 | 25875.9 | 86.8% | 79.4–94.9 |
| 100 | 32462.7 | 108.9% | 99.6–119.1 |
| 120 | 34193.8 | 114.7% | 104.9–125.5 |
The IFN-β samples were pre-determined by AVA, followed by spiking into media at 80, 100, and 120% of the expected values. The “spiked” samples were then subjected to RGA analyses in comparison with the reference standards.
Figure 8.
Comparison between AVA and RGA. Panel (a) represents dose response curve from the AVA for the potency determination of IFN-β 1b. Panel (b) represents dose response curve from the RGA for the potency determination of IFN-β 1b. For both methods, each dose of the sample was analyzed in six replicates on three different plates. Data shown here represents one of several repeated experiments.
4. Discussion
IFNs have been prescribed in the last few decades for the treatment of infectious and autoimmune diseases and selected types of cancer [8–11]. Accurate determination of the potency of therapeutic IFNs is crucial for the safety and efficacy of the drug. It is of note that adverse reactions caused by therapeutic interferon have been reported [35, 36]. Its potency is traditionally determined by AVAs, in which the inhibitory activity of IFNs on viral replication in cell cultures is measured. AVA is known to be a laborious assay associated with poor reproducibility. It also requires maintenance of a permissive cell line, as well as manipulation of the viruses in a biocontainment laboratory [14–17, 19, 20, 37–40]. Various bioassays used for interferon potency determinations have been discussed [12, 13, 18]. Specifically, several types of alternative potency assays for IFN have been reported including antiproliferative assays, quantitative reverse transcription polymerase chain reaction (qPCR) and RGAs. The anti-proliferative assays appear to be less sensitive than AVA, display marked variations between the different cell lines and require radio-isotope use in some cases [13]. Recently, some investigators reported the use of qPCR to determine the potency of IFN through quantification of IFN-responsive genes [39, 40]. Moore et al. developed two assays based on qPCR and cDNA for the measurements of IFN bioactivity and adapted for the analyses of neutralizing antibodies (NAbs) to type I IFN [38]. Alternative assays based on quantification of IFN-inducible gene products, that is, IFN-responsive promoter-driven reporter gene, have recently received great attention because of their simplicity, rapidity and reproducibility. These RGAs employed a variety of IFN responsive promoters including Mx, ISRE, GFAP [16–26]. Using Mx2/Luc reporter cell line, Seo et al. [23] reported the RGA is even more sensitive than the ISRE/Luc reporter cell line. While previous studies have provided strong evidences that RGA is advantageous compared to the traditional AVA with respect to simplicity, rapidity and reproducibility, optimization and/or modification of the RGAs also appear to be necessary since some systems are associated with narrow detection range or require extra steps after the addition of IFN. In addition, no RGAs have been used to study the chemically modified IFNs, which could potentially result in a loss of therapeutic efficacy or increased immunogenicity of the therapeutic proteins [24–26, 29–34]. In this communication, we studied the suitability of HEK293 cells stably transfected with ISRE-driven luciferase construct for the analyses of native and variant forms of IFN and the validation of the assay in a wider biological and statistical context. The choice of HEK293 is largely due to the fact that HEK293 cells are of human origin [41], easy to be cultured and reported to be suitable for human IFN potency testing [13–17]. Indeed, the RGA described here is in general agreement with previous RGAs, particularly with the degree of variability (CV) [16–26]. Here we summarize our observation in comparison with the data reported by others in the literatures: (1) unlike most others, our validation of the stability of the cell line exceeded more than 51 passages, with no significant difference found in cell line stability even after 51 passages in relation to IFN responsiveness. It is unclear to us how stable the other cell lines are as most reports did not reveal data from studies on cell stability beyond 51 passages except that of Smilović et al. [22] who reported that the reporter gene/CHO cell line was stable for over 1 year. (2) Assays based on luciferase measurements like this one reported here are simpler than reporter assays using CAT since luciferase-based RGAs do not need ELISA to quantify CAT expression; noticeably, SEAP-based RGAs have also been reported to be less sensitive than RGAs based on luciferase; however, the SEAP substrate (p-nitrophenylphosphate, NPP) may be directly added to the wells [13]. (3) Our studies using RGA yielded some interesting observations, specifically, the aggregated form of IFN losses it's activity, consistent with data from Caserman et al. [19]; we also observed a modest loss of activity in the oxidized form of IFN and a nearly “full” activity of deamidated IFN, suggesting that bioactivity measurement may not be enough to identify the deamidated or oxidized variants. For this purpose, we have developed reverse phase HPLC and ion exchange HPLC to identify those variants (Diress et al, manuscript in preparation). (4) Lastly, the inclusion of four analysts and head-to-head comparison in the validation studies revealed that although RGA is much simpler, prior training for staff who have never performed luciferase assays is still recommended, given the relatively larger CV% values found in those analysts (Figure 4). In our experience however, RGA procedure enabled laboratory staff to become proficient in conducting RGAs much quicker than they do for the AVA. Our cell lines reported here would be made available to any investigator who is interested in using these stable cells for their own validation studies.
Acknowledgments
The authors thank Dr. Richard Isbrucker, Matthew Alteen, Ben Leung, Jaquie Fildes, Lori Schauland, Jessica Querry-Goski, and Diane Grummisch (Health Canada) for technical assistance and consultations, Drs. Evangelos Bakopanos, Chantal Cazeault, Habiba Chakir, Dexi Dai, Deqi Huang, and William L Casley (Health Canada) for helpful discussions. Jennifer Whitteker, Bozena Jaentschke, Monika Tocchi, and Matt LeBrun (Health Canada) are acknowledged for their participation in the study involving multiple analysts. They also appreciate critical reviews of the manuscript by Dr. Michael Rosu-Myles and Dr. Shiv Prasad and editorial review by Mrs. Monika Tocchi (Health Canada). They are indebted to several manufacturers for providing samples for their validation studies.
References
- 1.Pestka S, Langer JA, Zoon KC, Samuel CE. Interferons and their actions. Annual Review of Biochemistry. 1987;56:727–777. doi: 10.1146/annurev.bi.56.070187.003455. [DOI] [PubMed] [Google Scholar]
- 2.Stark GR, Kerr IM, Williams BRG, Silverman RH, Schreiber RD. How cells respond to interferons. Annual Review of Biochemistry. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 3.Biron CA. Interferons α and β as immune regulators—a new look. Immunity. 2001;14(6):661–664. doi: 10.1016/s1074-7613(01)00154-6. [DOI] [PubMed] [Google Scholar]
- 4.Chawla-Sarkar M, Leaman DW, Borden EC. Preferential induction of apoptosis by interferon (IFN)-β compared with IFN-α2: correlation with TRAIL/Apo2L induction in melanoma cell lines. Clinical Cancer Research. 2001;7(6):1821–1831. [PubMed] [Google Scholar]
- 5.De Veer MJ, Holko M, Frevel M, et al. Functional classification of interferon-stimulated genes identified using microarrays. Journal of Leukocyte Biology. 2001;69(6):912–920. [PubMed] [Google Scholar]
- 6.Taki S. Type I interferons and autoimmunity: lessons from the clinic and from IRF-2-deficient mice. Cytokine and Growth Factor Reviews. 2002;13(4-5):379–391. doi: 10.1016/s1359-6101(02)00023-0. [DOI] [PubMed] [Google Scholar]
- 7.Ank N, Paludan SR. Type III IFNs: new layers of complexity in innate antiviral immunity. BioFactors. 2009;35(1):82–87. doi: 10.1002/biof.19. [DOI] [PubMed] [Google Scholar]
- 8.Weinstock-Guttman B, Jacobs LD. What is new in the treatment of multiple sclerosis? Drugs. 2000;59(3):401–410. doi: 10.2165/00003495-200059030-00002. [DOI] [PubMed] [Google Scholar]
- 9.Goodkin DE. Interferon β therapy for multiple sclerosis. The Lancet. 1998;352(9139):1486–1487. doi: 10.1016/S0140-6736(98)00057-9. [DOI] [PubMed] [Google Scholar]
- 10.Quesada JR, Gutterman JU, Hersh EM. Treatment of hairy cell leukemia with alpha interferons. Cancer. 1986;57(8, supplement):1678–1680. doi: 10.1002/1097-0142(19860415)57:8+<1678::aid-cncr2820571308>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 11.Baron S, Tyring SK, Fleischmann WR, et al. The interferons: mechanisms of action and clinical applications. Journal of the American Medical Association. 1991;266(10):1375–1383. doi: 10.1001/jama.266.10.1375. [DOI] [PubMed] [Google Scholar]
- 12.Meager A, Gaines Das R, Zoon K, Mire-Sluis A. Establishment of new and replacement World Health Organization International Biological Standards for human interferon alpha and omega. Journal of Immunological Methods. 2001;257(1-2):17–33. doi: 10.1016/s0022-1759(01)00460-4. [DOI] [PubMed] [Google Scholar]
- 13.Meager A. Biological assays for interferons. Journal of Immunological Methods. 2002;261(1-2):21–36. doi: 10.1016/s0022-1759(01)00570-1. [DOI] [PubMed] [Google Scholar]
- 14.Giard DJ, Fleischaker RJ. A study showing a high degree of interlaboratory variation in the assay of human interferon. Journal of Biological Standardization. 1984;12(3):265–269. doi: 10.1016/s0092-1157(84)80005-0. [DOI] [PubMed] [Google Scholar]
- 15.Fray MD, Mann GE, Charleston B. Validation of an Mx/CAT reporter gene assay for the quantification of bovine type-I interferon. Journal of Immunological Methods. 2001;249(1-2):235–244. doi: 10.1016/s0022-1759(00)00359-8. [DOI] [PubMed] [Google Scholar]
- 16.Lleonart R, Näf D, Browning H, Weissmann C. A novel, quantitative bioassay for type I interferon using a recombinant indicator cell line. Nature Biotechnology. 1990;8(12):1263–1267. doi: 10.1038/nbt1290-1263. [DOI] [PubMed] [Google Scholar]
- 17.Canosi U, Mascia M, Gazza L, et al. A highly precise reporter gene bioassay for type I interferon. Journal of Immunological Methods. 1996;199(1):69–76. doi: 10.1016/s0022-1759(96)00168-8. [DOI] [PubMed] [Google Scholar]
- 18.Meager A, Das RG. Biological standardization of human interferon beta: establishment of a replacement world health organization international biological standard for human glycosylated interferon beta. Journal of Immunological Methods. 2005;306(1-2):1–15. doi: 10.1016/j.jim.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 19.Caserman S, Menart V, Gaines Das R, Williams S, Meager A. Thermal stability of the WHO international standard of interferon alpha 2b (IFN-α2b): application of new reporter gene assay for IFN-α2b potency determinations. Journal of Immunological Methods. 2007;319(1-2):6–12. doi: 10.1016/j.jim.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 20.Hammerling U, Bongcam-Rudloff E, Setterblad N, et al. The β-gal interferon assay: a new, precise, and sensitive method. Journal of Interferon and Cytokine Research. 1998;18(7):451–460. doi: 10.1089/jir.1998.18.451. [DOI] [PubMed] [Google Scholar]
- 21.Däubener W, Wanagat N, Pilz K, Seghrouchni S, Fischer HG, Hadding U. A new, simple, bioassay for human IFN-γ. Journal of Immunological Methods. 1994;168(1):39–47. doi: 10.1016/0022-1759(94)90207-0. [DOI] [PubMed] [Google Scholar]
- 22.Smilović V, Caserman S, Fonda I, Gaberc-Porekar V, Menart V. A novel reporter gene assay for interferons based on CHO-K1 cells. Journal of Immunological Methods. 2008;333(1-2):192–196. doi: 10.1016/j.jim.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 23.Seo YJ, Kim GIH, Kwak HOJ, et al. Validation of a HeLa Mx2/Luc reporter cell line for the quantification of human type i interferons. Pharmacology. 2009;84(3):135–144. doi: 10.1159/000235158. [DOI] [PubMed] [Google Scholar]
- 24.Lallemand C, Meritet JF, Erickson R, et al. Quantification of neutralizing antibodies to human type I interferons using division-arrested frozen cells carrying an interferon-regulated reporter-gene. Journal of Interferon and Cytokine Research. 2008;28(6):393–404. doi: 10.1089/jir.2007.0142. [DOI] [PubMed] [Google Scholar]
- 25.Lam R, Farrell R, Aziz T, et al. Validating parameters of a luciferase reporter gene assay to measure neutralizing antibodies to IFNβ in multiple sclerosis patients. Journal of Immunological Methods. 2008;336(2):113–118. doi: 10.1016/j.jim.2008.03.014. [DOI] [PubMed] [Google Scholar]
- 26.Lallemand C, Meritet JF, Blanchard B, Lebon P, Tovey MG. One-step assay for quantification of neutralizing antibodies to biopharmaceuticals. Journal of Immunological Methods. 2010;356(1-2):18–28. doi: 10.1016/j.jim.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 27.Diress A, Lorbetskie B, Larocque L, et al. Study of aggregation, denaturation and reduction of interferon alpha-2 products by size-exclusion high-performance liquid chromatography with fluorescence detection and biological assays. Journal of Chromatography A. 2010;1217(19):3297–3306. doi: 10.1016/j.chroma.2009.10.081. [DOI] [PubMed] [Google Scholar]
- 28.Ramana CV, Chatterjee-Kishore M, Nguyen H, Stark GR. Complex roles of Stat1 in regulating gene expression. Oncogene. 2000;19(21):2619–2627. doi: 10.1038/sj.onc.1203525. [DOI] [PubMed] [Google Scholar]
- 29.Bucciantini M, Giannoni E, Chiti F, et al. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 2002;416(6880):507–511. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- 30.Wang W. Protein aggregation and its inhibition in biopharmaceutics. International Journal of Pharmaceutics. 2005;289(1-2):1–30. doi: 10.1016/j.ijpharm.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 31.Booth DR, Sunde M, Bellotti V, et al. Instability, unfolding and aggregation of human lysozyme variants underlying amyloid fibrillogenesis. Nature. 1997;385(6619):787–793. doi: 10.1038/385787a0. [DOI] [PubMed] [Google Scholar]
- 32.Cudd A, Arvinte T, Das GRE, Chinni C, MacIntyre I. Enhanced potency of human calcitonin when fibrillation is avoided. Journal of Pharmaceutical Sciences. 1995;84(6):717–719. doi: 10.1002/jps.2600840610. [DOI] [PubMed] [Google Scholar]
- 33.Braun A, Kwee L, Labow MA, Alsenz J. Protein aggregates seem to play a key role among the parameters influencing the antigenicity of interferon alpha (IFN-α) in normal and transgenic mice. Pharmaceutical Research. 1997;14(10):1472–1478. doi: 10.1023/a:1012193326789. [DOI] [PubMed] [Google Scholar]
- 34.Hermeling S, Aranha L, Damen JMA, et al. Structural characterization and immunogenicity in wild-type and immune tolerant mice of degraded recombinant human interferon alpha2b. Pharmaceutical Research. 2005;22(12):1997–2006. doi: 10.1007/s11095-005-8177-9. [DOI] [PubMed] [Google Scholar]
- 35.Meca-Lallana JE, de Mingo-Casado P, Amorin-Díaz M, Martínez-Navarro ML, Barreiro AF. Effects of glatiramer acetate on spasticity in previously interferon-β-treated and treatment-naive patients with relapsing-remitting multiple sclerosis: a prospective, nonrandomized, open-label, uncontrolled, observational pilot study. Clinical Therapeutics. 2010;32(6):1061–1066. doi: 10.1016/j.clinthera.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 36.Kato H, Fujishiro T, Narita M. A case of severe thrombocytopenia in a patient with chronic hepatitis C caused by a single administration of pegylated interferon alpha 2a subsequent to 48 weeks of pegylated interferon alpha 2b plus ribavirin therapy. Internal Medicine. 2010;49(16):1741–1744. doi: 10.2169/internalmedicine.49.3586. [DOI] [PubMed] [Google Scholar]
- 37.François C, Bernard I, Castelain S, et al. Quantification of different human alpha interferon subtypes and pegylated interferon activities by measuring MxA promoter activation. Antimicrobial Agents and Chemotherapy. 2005;49(9):3770–3775. doi: 10.1128/AAC.49.9.3770-3775.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moore M, Meager A, Wadhwa M, Burns C. Measurement of neutralising antibodies to type I interferons by gene expression assays specific for type 1 interferon-inducible 6-16 mRNA. Journal of Pharmaceutical and Biomedical Analysis. 2009;49(2):534–539. doi: 10.1016/j.jpba.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 39.Bertolotto A, Sala A, Caldano M, et al. Development and validation of a real time PCR-based bioassay for quantification of neutralizing antibodies against human interferon-beta. Journal of Immunological Methods. 2007;321(1-2):19–31. doi: 10.1016/j.jim.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 40.Gilli F, van Beers M, Marnetto F, Jiskoot W, Bertolotto A, Schellekens H. Development of a bioassay for quantification of neutralising antibodies against human interferon-beta in mouse sera. Journal of Immunological Methods. 2008;336(2):119–126. doi: 10.1016/j.jim.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 41.Merla G, Micale L, Muscarella LA, et al. VHL frameshift mutation as target of nonsense-mediated mRNA decay in drosophila melanogaster and human HEK293 cell line. Journal of Biomedicine and Biotechnology. 2009;2009 doi: 10.1155/2009/860761. Article ID 860761. [DOI] [PMC free article] [PubMed] [Google Scholar]