Abstract
In preparation for mitosis, the centrosome doubles once and only once to provide the two poles of the mitotic spindle. The presence of more than two centrosomes increases the chances that mitosis will be multipolar, and chromosomes will be distributed unequally. Since the number of mother–daughter centriole pairs determines the number of centrosomes, it is important that only one daughter centriole is assembled at, but slightly separated from, the proximal end of each mother centriole. This numerical and spatial specificity has led to the belief that a ‘template’ on the mother centriole provides a unique site for procentriole assembly. We review observations that are leading to the demise of this intuitively attractive idea. In its place, we are left with the notion that pericentriolar material at the wall of the mother centriole provides a local environment that promotes the assembly of a macromolecular complex that seeds the daughter centriole. Even though the system normally behaves in a digital fashion to go from zero to just one daughter centriole per mother, this behaviour appears to be based in the precise analogue control of multiple proteins, their activities, and the structure provided by the mother centriole.
Keywords: cell cycle, centriole, centrosome, duplication, singularity, template
The centrosome and its duplication
The centrosome in mammals serves as the primary MTOC (microtubule-organizing centre) of the cell, and in that capacity, it has a profound influence on all microtubule-dependent processes. The interphase centrosome of mammalian somatic cells seen at the ultrastructural level consists of a pair of centrioles associated with a seemingly amorphous cloud of electron-dense material known as the PCM (pericentriolar material) (see Brinkley et al., 1967; Vorobjev et al., 1982; Wheatley, 1982; Bornens et al., 1987; Preble et al., 2000; for reviews of centrosome ultrastructure, see Figure 1). One centriole (the mother) is a cell generation older than the other (the daughter), and in many cell types, it organizes the axoneme of the primary cilium (Wheatley, 1982; Beisson and Wright, 2003; Sluder, 2004). The PCM consists of a fibrous meshwork that binds a variety of proteins, including the gamma tubulin ring complexes that nucleate microtubules during interphase and mitosis (Figures 2a, 2b) (Gould and Borisy, 1977; Zimmerman et al., 1999; Palazzo et al., 2000; Andersen et al., 2003). In many cell types, the centrioles are in close proximity to each other during early interphase (for example, see Kuriyama and Borisy, 1981; Rieder and Borisy, 1982), whereas in others, they can be widely separated (Piel et al., 2000). Whether the centrioles are close together or separated, both are associated with PCM, though typically, there is more around the older centriole (Rieder and Borisy, 1982; Piel et al., 2000).
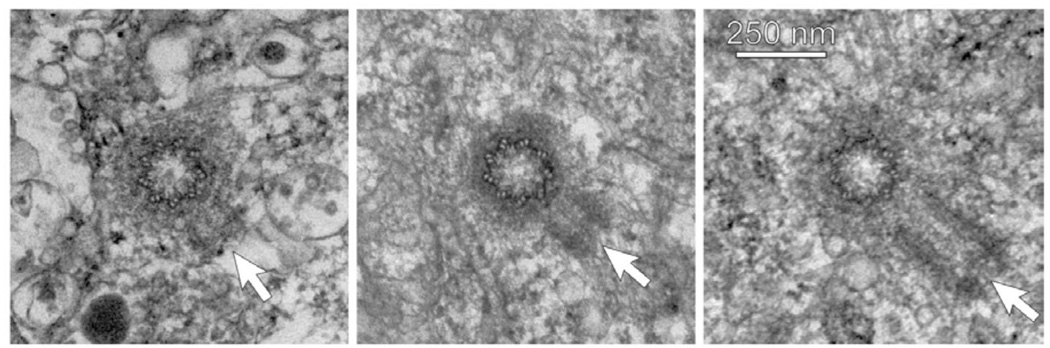
Figure 1. Centrosome ultrastructure in HeLa cells at times during S-phase arrest.
First frame: the mother centriole is cut in cross-section revealing the nine triplet microtubule barrel of the centriole surrounded by a diffuse cloud of pericentriolar material. The procentriole (cut in longitudinal section – arrow) typically assembles at right angles to and at a slight distance from the wall of the mother centriole. In early S-phase, the procentriole is short. Second and third frames: Later times in S-phase-arrested cells showing elongation of the daughter centrioles (arrows). Images taken from Figure S1 of Loncarek J, Hergert P, Magidson V, Khodjakov A. Control of daughter centriole formation by the pericentriolar material. Nat Cell Biol 2008;10:322–8 with permissions of the authors and Nature Press.
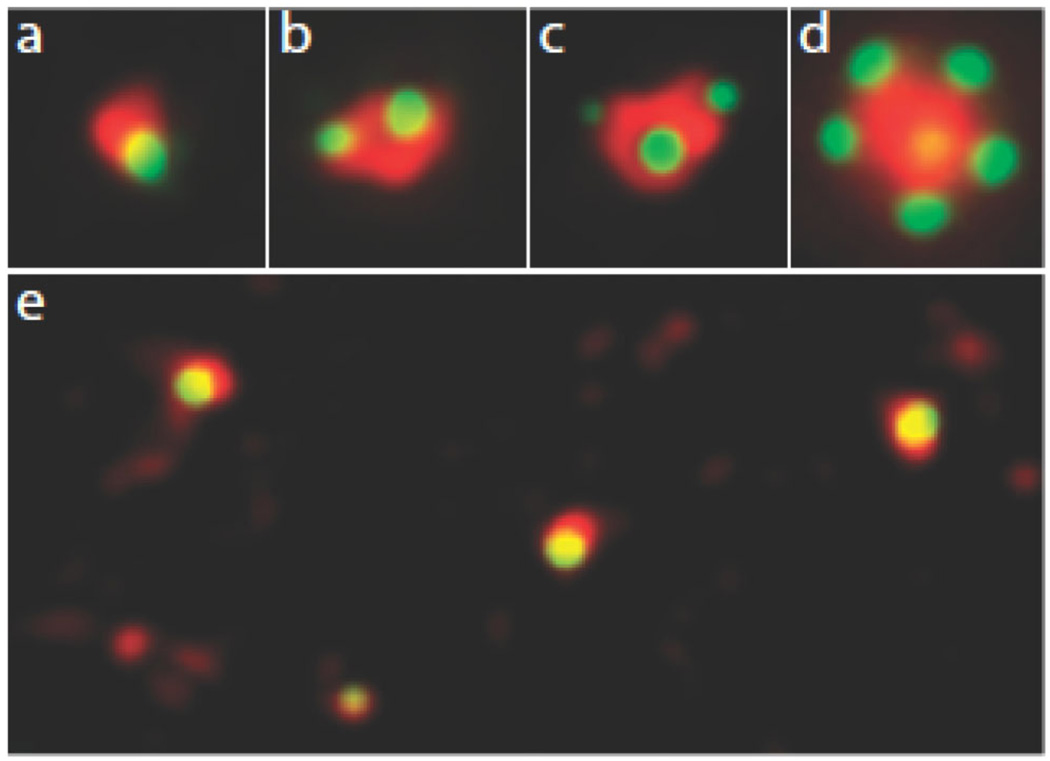
Figure 2. Fluorescence microscopic images of centrosomes in mammalian somatic cells.
The green contrast shows the localization of GFP-centrin 1, which is concentrated in the distal lumen of centrioles and thus serves as a centriole marker. The red contrast is gamma tubulin located in the pericentriolar material. (a) A single centriole associated with pericentriolar material during G1. (b) A centrosome in G2 containing a mother centriole (larger centrin dot) and a daughter centriole. The pair of centrioles is ofter referred to as a diplosome. (c) Spontaneous occurrence of two daughter centrioles with one mother centriole (a triplosome). This is seen at low frequency in some cell types such as CHO cells. (d) Overduplication of a mother centriole when Plk 4 is overexpressed. The mother centriole (seen weakly as the yellow dot in the middle of the gamma tubulin cloud) is driven to make more than one procentriole at one time giving a figure that resembles a rosette. (e) Multiple centrioles assembled de novo in a cell whose resident centrioles were ablated with a laser microbeam. Note the smaller size of the pericentriolar material and foci of gamma tubulin without centrioles.
The centrosome must precisely double in preparation for mitosis to provide the two poles of the mitotic spindle (Figure 3). The events of centrosome duplication described below begin at about the time of S-phase onset. In G2, the centrosome, as a whole, splits, and the two sister centrosomes, each with a pair of mother–daughter centrioles, start to separate around the nucleus. After nuclear envelope breakdown, these sister centrosomes nucleate the astral arrays that contribute most of the microtubules to the formation of the spindle. Centrosomes, through these astral microtubules, act in a dominant fashion to determine spindle polarity, spindle position/orientation in the cell and the plane of cleavage (Khodjakov and Rieder, 2001). Since centrioles attract and localize the PCM that acts as a MTOC, the cycle of centriole duplication and separation determines the reproduction of the centrosome as a whole (Sluder and Rieder, 1985; Bobinnec et al., 1998; Sluder, 2004).
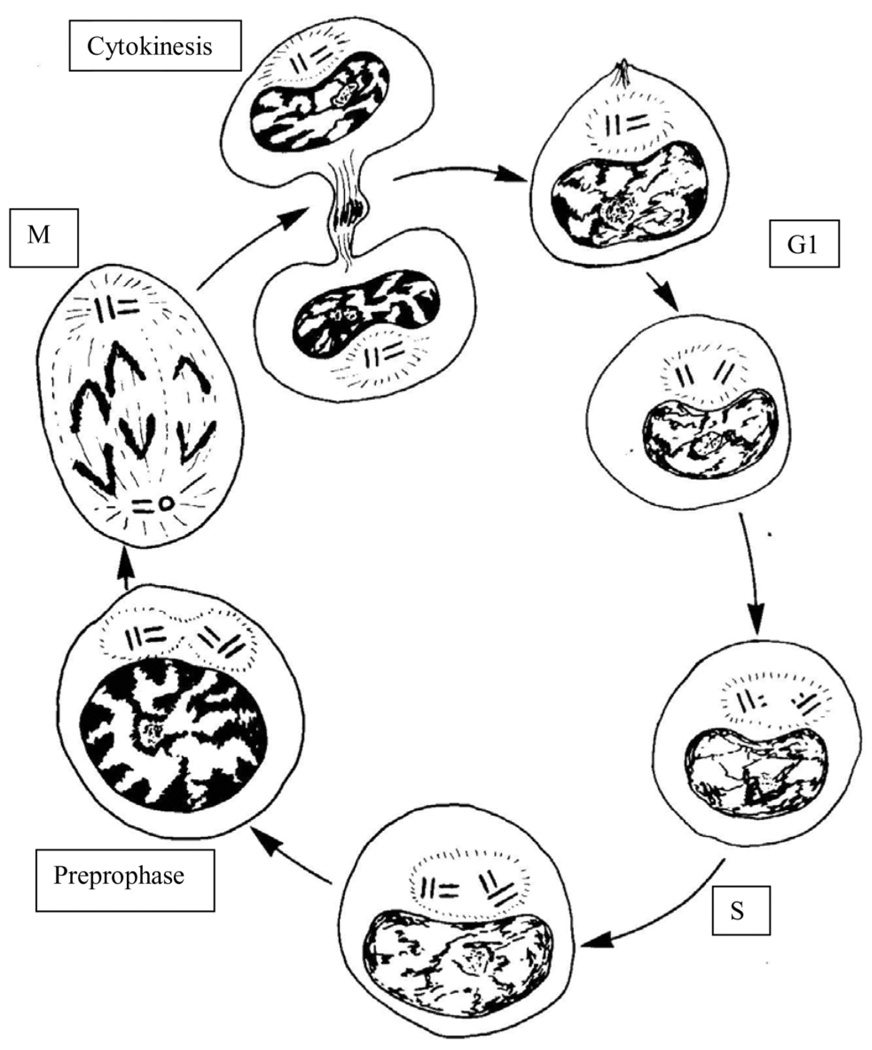
Figure 3.
Schematic representation of the centriole cycle during the cell cycle in mammalian cells. Centrosomes are shown as a hollow circle of fine lines enclosing the centrioles, which are represented by paired parallel lines as if in longitudinal section. Cell at 1 o’clock is a G1 daughter cell that has inherited a single centrosome with two centrioles. In many cell types, the centrioles remain in close proximity, whereas in others (e.g. HeLa), the original mother and daughter centrioles can be widely separate. The 2 o’clock cell is in late G1, and the centrioles have separated slightly and have lost their orthogonal arrangement. Centriole disorientation was once thought to signal the initiation of centriole duplication, but more recent work has revealed that the centrioles become disengaged from each other starting in late mitosis. The cell at 4 o’clock is in early S-phase, and centriole duplication is underway with the assembly of short procentrioles at the proximal ends of the mother centrioles. The procentrioles elongate throughout the rest of interphase, reaching their mature length in mitosis or the following G1. The 6 o’clock cell is in late S or early G2. The procentrioles have become longer. The 8 o’clock cell is in G2, and the mother–daughter centrioles pairs have started to separate as the centrosome is resolving itself into two sister centrosomes. With time, the sister centrosomes continue to separate around the nucleus as the cell cycle approaches mitosis. At mitosis (10 o’clock), the sister centrosomes organize the two poles of the spindle. Each centrosome contains a mother centriole and its daughter. The cell at 12 o’clock is in late telophase, as it is completing cleavage. Centriole duplication is said to be conservative because the procentriole is assembled from subunits in the cytoplasm, not from components of the mother centriole. Centriole distribution to sister centrosomes is said to be semiconservative because parental centrioles are distributed to both centrosomes. Diagram after Wheatley (1982), by permission of Elsevier and the author.
All key events of centriole reproduction are tightly linked to progression through the cell cycle. Centriole duplication is currently thought to start with the disjoining of the mother and daughter centrioles through the action of Plk1 early in mitosis and separase activity at the metaphase–anaphase transition (Tsou and Stearns, 2006a, 2006b; Tsou et al., 2009; Loncarek et al., 2010). This breaking of the association of the two centrioles is said to ‘license’ the centrioles to duplicate in the following interphase. The morphological duplication of the centrioles is first seen during S-phase, with the appearance of short (~50 nm) daughter centrioles (also called procentrioles) near the proximal ends of both centrioles inherited from mitosis as in Figure 1 (Robbins et al., 1968; Kuriyama and Borisy, 1981; Vorobjev and Chentsov, 1982; Wheatley, 1982; Alvey, 1985); the usual sequence is depicted in Figure 3. This event is initiated by a rise in the activity of Cdk2 in association with cyclin E and/or cyclin A, which also initiates DNA replication [reviewed by Sluder (2004)]. The procentrioles elongate during S and G2, reaching mature length in mitosis or the following G1 (Kuriyama and Borisy, 1981; Lange et al., 2000; Figure 3). Daughter centrioles (the elongating procentrioles) do not acquire distal and subdistal appendages until they fully mature in the subsequent cell cycle (Paintrand et al., 1992; Marshall, 2001; Bornens, 2002). When the centrosome splits in G2, a pair of mother–daughter centrioles is found in each sister centrosome.
Since the purpose of mitosis is to divide a mother cell into two genetically identical daughter cells, the cell must ensure that the centrosome inherited from the previous mitosis doubles once and only once. The presence of more than two centrosomes at the onset of mitosis, a condition called centrosome amplification, greatly increases the chances that the cell will assemble a multipolar spindle and distribute the chromosome unequally. Even if such cells eventually assemble bipolar spindles by bundling multiple centrosomes into two poles, the fidelity of chromosome segregation is compromised by the incidence of merotelically attached chromosomes that may segregate to the same spindle poles as their sisters (Heneen, 1970, 1975; Sluder et al., 1997; Ganem et al., 2009, Silkworth et al., 2009). The resulting genomic instability due to whole chromosome losses or gains can lead to loss of normal alleles for tumour-suppressor genes and other genetic imbalances that can promote unregulated growth characteristics and a diminished apoptotic response to cellular damage [reviewed by Orr-Weaver and Weinberg (1998), Brinkley (2001); Nigg (2002)]. Genomic instability is thought to be a major driving force in multistep carcinogenesis (Lengauer et al., 1998; D’Assoro et al., 2002; Goepfert et al., 2002; Pihan et al., 2003).
Two potential abnormalities in the control of centriole duplication cycle will inevitably lead to centrosome amplification. First, centrioles can ‘overduplicate’ by assembling too many procentrioles around the mother centriole (Figure 2d) (Kleylein-Sohn et al., 2007; Rodrigues-Martins et al., 2007; Duensing et al., 2009). Second, mature centrioles may ‘reduplicate’ by assembling procentrioles one at a time, more than once per cell cycle. This is observed in some cell lines, such as CHO and U2OS, when the cells are experimentally arrested in S-phase (Balczon et al., 1995; Stucke et al., 2002).
Given the dangers of having too many centrosomes at mitosis, it is of real importance that only one daughter centriole forms at each mother centriole with high fidelity. Although the mechanism by which the cell limits the number of daughter centrioles assembled at each mother centriole is not fully understood, recent studies have started to provide insight into this important issue. In a landmark study, Wong and Stearns (2003) presented evidence that there is an intrinsic block to centrosome reduplication in normal human cells. Their key experiment was one in which they fused a G1 cell with a G2 cell. Consistent with the findings of Johnson and Rao (1971), they found that the heterophasic heterokaryon progressed through interphase, and the G1 nucleus underwent DNA synthesis, while the G2 nucleus did not, confirming a nuclear block to DNA reduplication. The significant new finding of Wong and Stearns (2003) was that the G1 centrosome duplicated, while the G2 centrosome did not, despite passage through the cytoplasmic conditions of S-phase. Loncarek et al. (2008) elegantly demonstrated this phenomenon in a different way. They laser ablated just the daughter centriole of one sister centrosome in S-phase-arrested HeLa and CHO cells. A new procentriole was assembled relatively rapidly, but no more were formed thereafter. Although the conditions of S-phase appear to be constitutively permissive for procentriole assembly, once the mother centriole has duplicated, it will not do so again. This result is seemingly compatible with the idea that the procentriole sterically blocks the site responsible for the assembly of the procentriole on each mother centriole, rendering it incapable of further duplication until the daughter centriole disengages during the ensuing mitosis (Tsou and Stearns, 2006; Nigg, 2007).
Traditional views on the seeding of the procentriole
One of the enduring mysteries in the centrosome field has been how centrioles duplicate so that only one procentriole is assembled at, but slightly separated from, the proximal end of each mother centriole. This numerical and spatial specificity has, in the past, led workers to embrace the possibility that a ‘template’ on the mother centriole provides a unique site for procentriole assembly. Indeed, the notion of ‘templated duplication’ is still part of the contemporary lexicon. There are two key facets to the template hypothesis: first, there is only one unique site on the mother centriole that can seed the assembly of the new procentriole, and second, only one template is formed per mother centriole in each cell cycle. There must also be a reduplication block to prevent a new template from being formed on the daughter centriole in that cell cycle. Together, these characteristics provide the essential numerical and spatial control over procentriole assembly.
Although the term ‘template’ has often been used, the concept(s) behind this term have typically not been clearly spelled out. One of the most comprehensive discussions of the many possible meanings of this term can be found in the thoughtful review of Fulton (1971). The most literal interpretation of the term implies the existence of a preformed structure that directly patterns the nine triplet microtubules and perhaps the cartwheel structure at the proximal end of the procentriole. Such a ‘rubber stamp’ in the parlance of Fulton has derived support from exacting ultrastructural studies of basal body duplication. In Paramecium, there is a plaque next to the parent basal body upon which the barrel of triplet microtubules progressively assembles (Dippell, 1968), and in Chlamydomonas, there is a looped fibre at the mother basal body containing nine densely staining foci that later elaborate into triplet microtubules [Gould, 1975; reviewed by Beisson and Wright (2003)]. However, it has never been certain whether these structures are the proposed template on the mother basal body or alternatively are the early assembly intermediates of the forming daughter basal bodies that were seeded by some other mechanism. Also, careful ultrastructural characterization of gamma-tubulin distribution at centrioles led to the proposal that the centriole duplication starts with the formation of a single focus of gamma tubulin on the wall of the mother centriole, which forms a template from which the microtubules of the procentriole grow (Fuller et al., 1995). Another interesting proposal holds that fibers, which link the proximal ends mother and daughter centrioles, remain attached to both at the time of centriole disjoining in mitosis. When centriole duplication begins in the next cell cycle, these attached fibers form a locus on the proximal end of each parent centriole that serves as the unique site for the self assembly of the initiating cartwheel structure of the procentriole (Salisbury, 2008).
Another version of the concept was outlined by Mazia et al. (1960) who postulated that the centriole contains a self-reproducing seed of molecular dimensions that provides the specific site for the self-assembly of the procentriole. They propose that this self-reproducing seed (a ‘self-replicating entity’ or SRE in the words of Fulton, 1971) organizes a copy of itself at each cell cycle to provide the assembly site for the next generation centriole. In effect, there is a semiautonomous organelle whose duplication is distinct from the visible centriolar structures. Though nobody has found a morphological, biochemical or genetic correlate for this SRE; a recent study has provided functional observations consistent with the existence of such an entity (Collins et al., 2010). These workers report that when microtubule assembly is blocked (not only cytoplasmic microtubules, but also centriolar triplet microtubules), over time there is the accumulation of ‘cryptic’ centriole precursors that soon elaborate into centrioles after the microtubule inhibitor is washed out.
Students of the centriole have also long recognized the possibility that a new centrioles or basal body simply self-assembles in the vicinity of the mother centriole from subunit pools without a need for some entity to be transferred from the mother to the daughter structure [reviewed by Fulton (1971); Rodrigues-Martins et al, 2007]. Left unanswered in this proposal, however, were the important questions of why the procentriole normally forms only in close spatial proximity to its parent centriole and more puzzling why only one daughter centriole is self-assembled. One possible answer is that the mother centriole provides a special and unique site or singularity that serves as a focus for the assembly of a macromolecular complex that, in time, seeds the progressive assembly of the familiar barrel of triplet microtubules and other structures (Rogers et al., 2009; Guichard et al., 2010). Presumably, this proposed singularity would have to be under tight numerical control to ensure that only one is present at each mother centriole at each cell cycle.
The appeal of the template or singularity hypothesis is, in part, driven by observations that, under normal circumstances, procentriole copy number is under tight control despite abundant and complete cytoplasmic pools of centriolar subunits. For example, early sea urchin and frog zygotes duplicate centrioles in proper copy number at each cell cycle despite the fact that they have complete pools of subunits on hand at fertilization to make many centrioles (Gard et al., 1990; Sluder et al., 1990). Early Drosophila embryos properly duplicate centrioles in the face of subunit pools sufficient to assemble 2 × 1013 centriole pairs (Rodrigues-Martins et al, 2007). Even mammalian somatic cells contain enough centriolar subunits to assemble multiple procentrioles within a single cell cycle. When Plk4 kinase is overexpressed, multiple daughter centrioles assemble around each mother, forming what looks like a rosette around the centrally located mother centriole (Figure 2d) (Habedanck et al., 2005; Kleylein-Sohn et al., 2007). A similar phenotype is induced by overexpression of SAS-6, a central cartwheel protein needed for the initiation of procentriole assembly (Strnad et al., 2007; Duensing et al., 2009). In this case, there are adequate pools of all other centriolar subunits present to match the overexpressed SAS-6 in assembling multiple procentrioles. Given these observations, having the number of procentriole assembly sites (templates or singularities) strictly limited has been an attractive way to explain why only one procentriole is assembled at each mother centriole.
Over the years, the nature and composition of this proposed singularity or SRE has been the subject of much speculation. The seemingly semiconservative nature of centriole duplication and separation of mother–daughter pairs to sister centrosomes has obvious parallels to DNA replication. Naturally, this similarity inspired workers more than 40 years ago to propose that centrioles could be semiautonomous organelles with their own DNA, much like mitochondria. DNA replication is a modern paradigm for a templated reproductive process in which information and copy number are under rigid control. A variant on this theme was the hypothesis that centrioles, like ribosomes, contain RNA that serves a structural role during their assembly. These possibilities inspired numerous studies, the vast majority of which concentrated on trying to demonstrate the existence of centriole-/basal body-specific DNA or RNA under the assumption that presence implies function. All of this work was fraught with serious technical problems and ultimately produced inconclusive observations [reviewed by Fulton (1971); Marshall and Rosenbaum, 2000]. Presently, there is no compelling evidence for the direct involvement of DNA or RNA in the duplication of centrioles or basal bodies.
Life is not that simple
A growing number of studies has produced observations that are incompatible with the template or singularity hypothesis. The first challenge to the need for a template on the mother centriole antedates even the discovery of centrioles! Starting in the late 1800s, a number of investigators reported the parthenogenetic activation of echinoderm egg development [reviewed by Wilson (1925)]. Since centrosome inheritance is strictly paternal in these forms (Sluder et al., 1989a, 1989b), centrosome assembly was induced de novo by the parthenogenetic activation regimes. Later ultrastructural analysis of cytasters induced in sea urchin eggs by parthenogenetic activation confirmed the assembly of centrioles (Kallenbach and Mazia, 1982). Since then, the de novo assembly of centrioles has been directly demonstrated for a wide variety of organisms, including human somatic cells (Figure 2e) (Khodjakov et al., 2002; La Terra et al., 2005; Uetake et al., 2007), Chlamydomonas (Marshall et al., 2001) and various insect eggs when resident centrioles are not present (Riparbelli et al., 2005; Riparbelli et al., 2006; Peel et al., 2007; Rodrigues-Martins et al., 2007). So the presence of a mother centriole and its putative singularity are clearly not needed for the assembly of new centrioles. Nevertheless, these observations have not necessarily spelt the death knell for the singularity hypothesis, as some have said, because the conditions needed for de novo centriole assembly are clearly not normal. De novo centriole assembly certainly demonstrates that self-assembly of centrioles can be forced by unusual conditions that bypass the need for the mother centriole. However, it could be said that under normal circumstances, the system is constrained, so that this special site on the mother centriole acts in a dominant fashion to determine where and in what number procentrioles are self-assembled. The role of the mother centriole in facilitating the assembly of procentrioles is supported by observations that de novo centriole assembly is relatively slow compared with canonical centriole duplication (Khodjakov et al., 2002; La Terra et al., 2005; Rodrigues-Martins et al., 2007).
The second challenge comes from observations on cultured cells that overexpression of the Plk4 kinase, the structural centriolar protein SAS-6, or the expression of the high-risk human papilloma virus protein E7, leads to the concurrent assembly of several procentrioles around each mother centriole (Figure 2d) (Kleylein-Sohn et al., 2007; Strnad et al., 2007; Duensing, et al., 2009). These observations argue against the notion that a mother centriole can bear only one singularity or template in any given cell cycle. Importantly, the assembly of multiple procentrioles, under these conditions, occurs only at the mother centrioles, not throughout the cytoplasm, indicating that the centrosome plays a role in providing a favoured location for the formation of multiple sites of procentriole assembly. In addition, some cultured cell types, such as CHO and U2-OS cells, reduplicate centrioles when they are arrested in S-phase with hydroxyurea or aphidicolin. Under this condition, procentrioles mature into daughter centrioles, and after they eventually separate from their mothers, they give birth to a new generation of procentrioles (Loncarek et al., 2008). Thus, progression through mitosis is not required for new sites of procentriole assembly to be formed. Intriguingly, these workers found that the mother centriole sometimes concurrently assembles two procentrioles in hydroxyurea-arrested CHO cells (Figure 2c). Together, these observations argue against the notion that only one singularity or template can be formed at each mother centriole per cell cycle.
The third and the most direct challenge to the existence of centriolar template comes from selective laser ablation of procentrioles in S-arrested HeLa and CHO cells (Loncarek et al., 2008). Ablation of the procentriole in a normal diplosome results in the rapid formation of a new procentriole at the mother centriole. Serial section ultrastructural reconstructions of cells previously followed in vivo reveal that the new procentriole does not always form at the exact location of the previous procentriole. This was most evident in the cases where the laser beam not only ablated the daughter centriole but also took out a bit of the mother centriole, which should have removed the putative template or singularity. The new daughter was sometimes seen to form on the opposite side of the mother. A second line of evidence against the existence of the template came from selective ablation of daughter centrioles in ‘triplosomes’ – complexes consisting of a single mother centriole and two procentrioles (Figure 2c). From the standpoint of the template hypothesis, concurrent formation of multiple procentrioles implies that the mother centriole somehow managed to acquire additional templates. If so, then removal of both procentrioles in laser ablation experiments should result in the formation of two new procentrioles. Analogously, ablation of a single procentriole should release only one presumptive template and thus lead to formation of a single new procentriole. However, the actual results were different. While laser ablation of all procentrioles did induce reduplication of the mother centriole, the number of new procentrioles did not match the number of the original procentrioles (only one new procentriole formed in most cases). Furthermore, ablation of a single procentriole never resulted in centriole reduplication as long as there was a preexisting procentriole attached to the mother. These results suggest that numerical control over new centriole assembly is executed at the level of the centrosome. Consistent with this notion, overexpression of pericentrin, a PCM protein, in S-phase-arrested CHO or HeLa cells leads to the formation of multiple daughter centrioles after the original daughter had been ablated by the laser beam. The simplest interpretation of this result is that the PCM provides a microenvironment that promotes procentriole formation and that extra PCM allows more than one procentriole to assemble at one time. The fact that the extra centrioles assembled at the mother centriole and not throughout the cytoplasm suggests that something about the mother contributes to the establishment of local permissive conditions for procentriole assembly. This raises the interesting, albeit speculative, possibility that normal procentriole formation is a form of regulated and site-specific de novo assembly. That is, canonical centriole duplication is de novo assembly that is spatially restricted to occur within the PCM cloud that surrounds the mother centriole. This notion is supported by the demonstration that increasing the size of the PCM by substantial overexpression of the PCM component pericentrin results in simultaneous assembly of multiple procentrioles (Loncarek et al., 2008). Similarly, more than one procentriole assembles in association with extremely long centrioles that form upon overexpression of CPAP (centrosomal P4.1-associated protein), a protein normally located along the centriolar wall (Kohlmaier et al., 2009). Together, these findings provide additional evidence that the mother centriole is not restricted to having just one singularity that can seed a procentriole, but rather multiple procentrioles can assemble within a cell cycle if there is extra PCM and/or the centriole is longer than normal.
Regulation of analogue conditions produces a digital result
The studies reviewed above signify the demise of the intuitively attractive idea that the mother centriole has a singularity that provides the spatial and numerical control over procentriole assembly. In its place, we are left with a messier notion that the pericentriolar material, perhaps with a contribution from the wall of the mother centriole, provides a local environment that promotes the assembly of a macromolecular complex containing kinases and structural proteins that seeds the daughter centriole. Put differently, the mother centriole provides a platform for the recruitment and/or local activation of a collection of structural and regulatory molecules that lead to the self-assembly of the procentriole (Rodrigues-Martins et al., 2007).
This naturally begs the question of how such a permissive microenvironment is precisely controlled under normal circumstances to yield the formation of only one procentriole at each mother with high fidelity in the face of non-limiting pools of centriolar subunits. That is, how can a system, which is capable of making more than one procentriole at the same time, be limited so that only one procentriole self-assembles? Even though there is a centrosome intrinsic block to reduplication (Wong and Stearns, 2003; Tsou and Stearns, 2006b), numerical control must be exercised earlier – at the very first step in the assembly of the generative structure for the procentriole, so that no more than one is made before this reduplication block takes effect.
We suggest that centriole duplication is controlled by a tightly regulated equilibrium of regulatory kinases and structural proteins acting in concert with structural constrains. First, mother centriole length must be limited because multiple daughters are assembled on overly long centrioles in the presence of normal subunit pools for the pericentriolar material and procentriole assembly. Second, the amount of pericentriolar material must be limited in the presence of a centriole of normal length because overexpression of the PCM protein pericentrin leads to the assembly of extra procentrioles. Also, the activity of Plk4 must be limited, both in absolute amount and also temporal duration, when all else is normal. Third, even though the structural molecules involved in the initiation of procentriole formation are not normally limiting, the expression levels of at least some of them must, nonetheless, be controlled. Overexpression of DSAS-6 can drive centriole reduplication in Drosophila embryos and somatic cells and cause unfertilized eggs, which normally do not contain centrioles, to make supernumerary centrosomes de novo (Peel et al., 2007; Rodrigues-Martins et al., 2007). Likewise, excess levels of the structural protein CPAP lead to overly long centrioles that allow the assembly of several procentrioles. The finding that inappropriate levels of any one of these factors can abrogate the proper numerical control of procentriole assembly indicates that there is not one single limit but rather a complex equilibrium. Even though the system normally behaves in a digital fashion to go from zero daughter centrioles to just one per mother, this behaviour appears to be based in the precise, but analog, control of multiple proteins, their activities and the structure provided by the mother centriole. Obviously a fertile ground for future research will be to determine how the cell controls these analogue conditions to produce a digital result.
Acknowledgements
We express our appreciation to Dr Jadranka Loncarek for preparing Figure 2 from her images for this review.
Funding
Work of the authors in this reivew is funded by the National Institutes of Health [grant numbers GM30758 (to G.S.) and GM59363 (to A.K.)].
Abbreviations
- EM
electron microscopy
- MTOC
microtubule-organizing centre
- PCM
pericentriolar material
- SRE
self-replicating entity.
References
- Alvey PL. An investigation of the centriole cycle using 3T3 and CHO cells. J Cell Sci. 1985;78:147–162. doi: 10.1242/jcs.78.1.147. [DOI] [PubMed] [Google Scholar]
- Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- Balczon R, Bao L, Zimmer WE, Brown K, Zinkowski RP, Brinkley BR. Dissociation of centrosome replication events from cycles of DNA synthesis and mitotic division in hydroxyurea-arrested Chinese hamster ovary cells. J Cell Biol. 1995;130:105–115. doi: 10.1083/jcb.130.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisson J, Wright M. Basal body/centriole assembly and continuity. Curr Opin Cell Biol. 2003;15:96–104. doi: 10.1016/s0955-0674(02)00017-0. [DOI] [PubMed] [Google Scholar]
- Bobinnec Y, Khodjakov A, Mir LM, Rieder CL, Edde B, Bornens M. Centriole disassembly in vivo and its effect on centrosome structure and function in vertebrate cells. J Cell Biol. 1998;143:1575–1589. doi: 10.1083/jcb.143.6.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornens M. Centrosome composition and microtubule anchoring mechanisms. Curr Opin Cell Biol. 2002;14:25–34. doi: 10.1016/s0955-0674(01)00290-3. [DOI] [PubMed] [Google Scholar]
- Bornens M, Paintrand M, Berges J, Marty MC, Karsenti E. Structural and chemical characterization of isolated centrosomes. Cell Motil Cytoskeleton. 1987;8:238–249. doi: 10.1002/cm.970080305. [DOI] [PubMed] [Google Scholar]
- Brinkley BR. Managing the centrosome numbers game: from chaos to stability in cancer cell division. Trends Cell Biol. 2001;11:18–21. doi: 10.1016/s0962-8924(00)01872-9. [DOI] [PubMed] [Google Scholar]
- Brinkley BR, Stubblefield E, Hsu TC. The effects of colcemid inhibition and reversal on the fine structure of the mitotic apparatus of Chinese hamster cells in vitro. J Ultrastruct Res. 1967;19:1–18. doi: 10.1016/s0022-5320(67)80057-1. [DOI] [PubMed] [Google Scholar]
- Collins ES, Hornick JE, Durcan TM, Collins NS, Archer W, Karanjeet KB, et al. Centrosome biogenesis continues in the absence of microtubules during prolonged S-phase arrest. J Cell Physiol. 2010;225:454–465. doi: 10.1002/jcp.22222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Assoro AB, Lingle WL, Salisbury JL. Centrosome amplification and the development of cancer. Oncogene. 2002;21:6146–6153. doi: 10.1038/sj.onc.1205772. [DOI] [PubMed] [Google Scholar]
- Dippell RV. The development of basal bodies in paramecium. Proc Natl Acad Sci U S A. 1968;61:461–468. doi: 10.1073/pnas.61.2.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duensing A, Spardy N, Chatterjee P, Zheng L, Parry J, Cuevas R, et al. Centrosome overduplication, chromosomal instability, and human papillomavirus oncoproteins. Environ Mol Mutagen. 2009;50:741–747. doi: 10.1002/em.20478. [DOI] [PubMed] [Google Scholar]
- Fuller SD, Gowen BE, Reinsch S, Sawyer A, Buendia B, Wepf R, Karsenti E. The core of the mammalian centriole contains gamma tubulin. Curr Biol. 1995;5:1384–1393. doi: 10.1016/s0960-9822(95)00276-4. [DOI] [PubMed] [Google Scholar]
- Fulton C. Centrioles. In: Reinert J, Ursprung H, Baxter R, editors. Origin and continuity of cell organelles. New York: Springer; 1971. pp. 170–213. [Google Scholar]
- Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gard DL, Hafezi S, Zhang T, Doxsey SJ. Centrosome duplication continues in cycloheximide-treated Xenopus blastulae in the absence of a detectable cell cycle. J Cell Biol. 1990;110:2033–2042. doi: 10.1083/jcb.110.6.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goepfert TM, Adigun YE, Zhong L, Gay J, Medina D, Brinkley WR. Centrosome amplification and overexpression of aurora A are early events in rat mammary carcinogenesis. Cancer Res. 2002;62:4115–4122. [PubMed] [Google Scholar]
- Gould RR, Borisy GG. The pericentriolar material in Chinese hamster ovary cells nucleates microtubule formation. J Cell Biol. 1977;73:601–615. doi: 10.1083/jcb.73.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould RR. The basal bodies of Chlamydomonas reinhardtii. Formation from probasal bodies, isolation, and partial characterization. J Cell Biol. 1975;65:65–74. doi: 10.1083/jcb.65.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichard P, Chretien D, Marco S, Tassin AM. Procentriole assembly revealed by cryo-electron tomography. EMBO J. 2010;29:1565–1572. doi: 10.1038/emboj.2010.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA. The polo kinase Plk4 functions in centriole duplication. Nat Cell Biol. 2005;7:1140–1146. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- Heneen WK. Kinetochores and microtubules in multipolar mitosis and chromosome orientation. Exp Cell Res. 1975;91:57–62. doi: 10.1016/0014-4827(75)90140-8. [DOI] [PubMed] [Google Scholar]
- Heneen WK. In situ analysis of normal and abnormal patterns of the mitotic apparatus in cultured rat-kangaroo cells. Chromosoma. 1970;29:88–117. doi: 10.1007/BF01183663. [DOI] [PubMed] [Google Scholar]
- Johnson RT, Rao PN. Nucleo-cytoplasmic interactions in the achievement of nuclear synchrony in DNA synthesis and mitosis in multinucleate cells. Biol Rev Camb Philos Soc. 1971;46:97–155. doi: 10.1111/j.1469-185x.1971.tb01180.x. [DOI] [PubMed] [Google Scholar]
- Kallenbach RJ, Mazia D. Origin and maturation of centrioles in association with the nuclear envelope in hypertonic-stressed sea urchin eggs. Eur J Cell Biol. 1982;28:68–76. [PubMed] [Google Scholar]
- Khodjakov A, Rieder CL, Sluder G, Cassels G, Sibon O, Wang CL. De novo formation of centrosomes in vertebrate cells arrested during S phase. J Cell Biol. 2002;158:1171–1181. doi: 10.1083/jcb.200205102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleylein-Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof YD, Nigg EA. Plk4-induced centriole biogenesis in human cells. Dev Cell. 2007;13:190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Kohlmaier G, Loncarek J, Meng X, McEwen BF, Mogensen MM, Spektor A, et al. Overly long centrioles and defective cell division upon excess of the SAS-4-related protein CPAP. Curr Biol. 2009;19:1012–1018. doi: 10.1016/j.cub.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriyama R, Borisy GG. Centriole cycle in Chinese hamster ovary cells as determined by whole-mount electron microscopy. J Cell Biol. 1981;91:814–821. doi: 10.1083/jcb.91.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Terra S, English CN, Hergert P, McEwen BF, Sluder G, Khodjakov A. The de novo centriole assembly pathway in HeLa cells: cell cycle progression and centriole assembly/maturation. J Cell Biol. 2005;168:713–722. doi: 10.1083/jcb.200411126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange BM, Faragher AJ, March P, Gull K. Centriole duplication and maturation in animal cells. Curr Top Dev Biol. 2000;49:235–249. doi: 10.1016/s0070-2153(99)49011-8. [DOI] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- Loncarek J, Hergert P, Magidson V, Khodjakov A. Control of daughter centriole formation by the pericentriolar material. Nat Cell Biol. 2008;10:322–328. doi: 10.1038/ncb1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loncarek J, Hergert P, Khodjakov A. Curr Biol. 2010 Jul 27;20:1277–1282. doi: 10.1016/j.cub.2010.05.050. Epub 2010 Jun 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF. What is the function of centrioles? J Cell Biochem. 2007;100:916–922. doi: 10.1002/jcb.21117. [DOI] [PubMed] [Google Scholar]
- Marshall WF. Centrioles take center stage. Curr Biol. 2001;11:R487–R496. doi: 10.1016/s0960-9822(01)00289-5. [DOI] [PubMed] [Google Scholar]
- Marshall WF, Rosenbaum JL. Are there nucleic acids in the centrosome? Curr Top Dev Biol. 2000;49:187–205. doi: 10.1016/s0070-2153(99)49009-x. [DOI] [PubMed] [Google Scholar]
- Mazia D, Harris PJ, Bibring T. The multiplicity of the mitotic centers and the time-course of their duplication and separation. J Biophys Biochem Cytol. 1960;7:1–20. doi: 10.1083/jcb.7.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA. Centrosome duplication: of rules and licenses. Trends Cell Biol. 2007;17:215–221. doi: 10.1016/j.tcb.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Centrosome aberrations: cause or consequence of cancer progression? Nat Rev Cancer. 2002;2:815–825. doi: 10.1038/nrc924. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver TL, Weinberg RA. A checkpoint on the road to cancer. Nature. 1998;392:223–224. doi: 10.1038/32520. [DOI] [PubMed] [Google Scholar]
- Paintrand M, Moudjou M, Delacroix H, Bornens M. Centrosome organization and centriole architecture: their sensitivity to divalent cations. J Struct Biol. 1992;108:107–128. doi: 10.1016/1047-8477(92)90011-x. [DOI] [PubMed] [Google Scholar]
- Palazzo RE, Vogel JM, Schnackenberg BJ, Hull DR, Wu X. Centrosome maturation. Curr Top Dev Biol. 2000;49:449–470. doi: 10.1016/s0070-2153(99)49021-0. [DOI] [PubMed] [Google Scholar]
- Peel N, Stevens NR, Basto R, Raff JW. Overexpressing centriole-replication proteins in vivo induces centriole overduplication and de novo formation. Curr Biol. 2007;170:834–843. doi: 10.1016/j.cub.2007.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel M, Meyer P, Khodjakov A, Rieder CL, Bornens M. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J Cell Biol. 2000;149:317–330. doi: 10.1083/jcb.149.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pihan GA, Wallace J, Zhou Y, Doxsey SJ. Centrosome abnormalities and chromosome instability occur together in pre-invasive carcinomas. Cancer Res. 2003;63:1398–1404. [PubMed] [Google Scholar]
- Preble AM, Giddings TM, Jr, Dutcher SK. Basal bodies and centrioles: their function and structure. Curr Top Dev Biol. 2000;49:207–233. doi: 10.1016/s0070-2153(99)49010-6. [DOI] [PubMed] [Google Scholar]
- Rieder CL, Faruki S, Khodjakov A. The centrosome in vertebrates: more than a microtubule-organizing center. Trends Cell Biol. 2001;11:413–419. doi: 10.1016/s0962-8924(01)02085-2. [DOI] [PubMed] [Google Scholar]
- Rieder CL, Borisy GG. The centrosome cycle in PtK2 cells: asymmetric distribution and structural changes in the pericentriolar material. Biol Cell. 1982;44:117–132. [Google Scholar]
- Riparbelli MG, Giordano R, Callaini G. Centrosome inheritance in the parthenogenetic egg of the collembolan Folsomia candida. Cell Tissue Res. 2006;326:861–872. doi: 10.1007/s00441-006-0253-x. [DOI] [PubMed] [Google Scholar]
- Riparbelli MG, Tagu D, Bonhomme J, Callaini G. Aster self-organization at meiosis: a conserved mechanism in insect parthenogenesis? Dev Biol. 2005;278(1):220–230. doi: 10.1016/j.ydbio.2004.11.009. [DOI] [PubMed] [Google Scholar]
- Robbins E, Jentzsch G, Micali A. The centriole cycle in synchronized HeLa cells. J Cell Biol. 1968;36:329–339. doi: 10.1083/jcb.36.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues-Martins A, Riparbelli M, Callaini G, Glover DM, Bettencourt-Dias M. Revisiting the role of the mother centriole in centriole biogenesis. Science. 2007;316:1046–1050. doi: 10.1126/science.1142950. [DOI] [PubMed] [Google Scholar]
- Rogers GC, Rusan NM, Roberts DM, Peifer M, Rogers SL. The SCF Slimb ubiquitin ligase regulates Plk4/Sak levels to block centriole reduplication. J Cell Biol. 2009;184:225–239. doi: 10.1083/jcb.200808049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury JL. Breaking the ties that bind centriole numbers. Nat Cell Biol. 2008;10:255–257. doi: 10.1038/ncb0308-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silkworth WT, Nardi IK, Scholl LM, Cimini D. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS One. 2009;4:e6564. doi: 10.1371/journal.pone.0006564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluder G. Centrosome duplication and its regulation in the higher animal cell. In: Nigg EA, editor. Centrosomes in development and disease. Weinheim: Wiley-VCH; 2004. pp. 167–189. [Google Scholar]
- Sluder G, Thompson EA, Miller FJ, Hayes J, Rieder CL. The checkpoint control for anaphase onset does not monitor excess numbers of spindle poles or bipolar spindle symmetry. J Cell Sci. 1997;110:421–429. doi: 10.1242/jcs.110.4.421. [DOI] [PubMed] [Google Scholar]
- Sluder G, Miller FJ, Cole R, Rieder CL. Protein synthesis and the cell cycle: centrosome reproduction in sea urchin eggs is not under translational control. J Cell Biol. 1990;110:2025–2032. doi: 10.1083/jcb.110.6.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluder G, Miller FJ, Lewis K, Davison ED, Rieder CL. Centrosome inheritance in starfish zygotes: selective loss of the maternal centrosome after fertilization. Dev Biol. 1989a;131:567–579. doi: 10.1016/s0012-1606(89)80027-2. [DOI] [PubMed] [Google Scholar]
- Sluder G, Miller FJ, Rieder CL. Reproductive capacity of sea urchin centrosomes without centrioles. Cell Motil Cytoskeleton. 1989b;13:264–273. doi: 10.1002/cm.970130405. [DOI] [PubMed] [Google Scholar]
- Sluder G, Rieder CL. Centriole number and the reproductive capacity of spindle poles. J Cell Biol. 1985;100:887–896. doi: 10.1083/jcb.100.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad P, Leidel S, Vinogradova T, Euteneuer U, Khodjakov A, Gonczy P. Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev Cell. 2007;13:203–213. doi: 10.1016/j.devcel.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucke VM, Sillje HH, Arnaud L, Nigg EA. Human Mps1 kinase is required for the spindle assembly checkpoint but not for centrosome duplication. EMBO J. 2002;21:1723–1732. doi: 10.1093/emboj/21.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou MF, Wang WJ, George KA, Uryu K, Stearns T, Jallepalli PV. Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Dev Cell. 2009;17:344–354. doi: 10.1016/j.devcel.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou MF, Stearns T. Mechanism limiting centrosome duplication to once per cell cycle. Nature. 2006a;442:947–951. doi: 10.1038/nature04985. [DOI] [PubMed] [Google Scholar]
- Tsou MF, Stearns T. Controlling centrosome number: licenses and blocks. Curr Opin Cell Biol. 2006b;18:74–78. doi: 10.1016/j.ceb.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Uetake Y, Loncarek J, Nordberg JJ, English CN, La Terra S, Khodjakov A, et al. Cell cycle progression and de novo centriole assembly after centrosomal removal in untransformed human cells. J Cell Biol. 2007;176:173–182. doi: 10.1083/jcb.200607073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobjev IA, Chentsov Yu S. Centrioles in the cell cycle. I. epithelial cells. J Cell Biol. 1982;93:938–949. doi: 10.1083/jcb.93.3.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley DN. The centriole: a central enigma of cell biology. Amsterdam: Elsevier Biomedical Press; 1982. [Google Scholar]
- Wilson EB. The cell in development and heredity. New York: Macmillan; 1925. [Google Scholar]
- Wong C, Stearns T. Centrosome number is controlled by a centrosome-intrinsic block to reduplication. Nat Cell Biol. 2003;5:539–544. doi: 10.1038/ncb993. [DOI] [PubMed] [Google Scholar]
- Zimmerman W, Sparks CA, Doxsey SJ. Amorphous no longer: the centrosome comes into focus. Curr Opin Cell Biol. 1999;11:122–128. doi: 10.1016/s0955-0674(99)80015-5. [DOI] [PubMed] [Google Scholar]