Abstract
Cyclic 3′,5′-adenosine monophosphate (cAMP) response-element (CRE) binding protein (CREB) is an important transcription factor that is differentially regulated in cells of various types. We recently reported that RA rapidly activates CREB without using retinoic acid (RA) receptors RAR and RXR in normal human tracheobronchial epithelial (NHTBE) cells. However, little is known about RA’s role in the physiologic regulation of CREB expression in the early mucous differentiation of NHTBE cells. Here, we report that RA upregulated CREB gene expression and that using 5′-serial deletion promoter analysis and mutagenesis analyses, two Sp1-binding sites located at nucleotides −217 and −150, which flank the transcription initiation site, were essential for RA induction of CREB gene transcription. Furthermore, we found that CREs located at nucleotides −119 and −98 contributed to basal promoter activity. Interestingly, RA also upregulated Sp1 in a time- and dose-dependent manner. Knockdown of endogenous Sp1 using small interfering RNA (siRNA) decreased RA-induced CREB gene expression. However, the converse was not true: knockdown of CREB using CREB siRNA did not affect RA-induced Sp1 gene expression. We conclude that RA upregulates CREB gene expression during the early stage of NHTBE cell differentiation and that RA-inducible Sp1 plays a major role in upregulating human CREB gene expression. This result implies that cooperation of these two transcription factors play a crucial role in mediating early events of normal mucous cell differentiation of bronchial epithelial cells.
Keywords: CREB, NHTBE, Retinoic acid, Sp1, Airway, RAR/RXR
Introduction
Retinoic acid (RA) exerts a profound effect on vertebrate development, cellular differentiation, and homeostasis [1–4]. In addition, RA plays a pivotal role in the induction and maintenance of mucociliary differentiation of airway epithelial cells [5–10]. In the respiratory system, RA deficiency causes squamous metaplasia, and RA supplementation restores the normal mucous cell phenotype in cultured primary bronchial epithelial cells [7]. It has been well documented that the physiological effects of RAs are mediated through two families of nuclear receptors (NRs), the RA receptor (RAR) and the retinoid X receptor (RXR), which control transcription initiation by binding to RA-response elements (RAREs) [1, 11, 12]. However, we recently demonstrated that RA rapidly activates CREB without using RAR/RXR in normal human tracheobronchial epithelial (NHTBE) cells [13, 14].
The cyclic 3′,5′-adenosine monophosphate (cAMP) response-element (CRE) binding protein (CREB), a 43-kDa protein, belongs to a family of basic domain-leucine zipper (bZip) transcription factors that contains activating transcription factor 1 (ATF1) and the CRE modulator (CREM) [15, 16]. CREB activates gene transcription by binding to regulatory DNA elements in the promoter regions of genes with the consensus motif; binding has been found to occur at an 8–base pair segment of the canonical CRE motif, 5′-TGACGTCA-3′, and at other sequence variants (called CREs) [17–19]. Phosphorylation of CREB at serine residue 133 (Ser-133) promotes recruitment of the transcriptional co-activator CREB-binding protein (CBP)/p300 and/or the TATA box-binding protein-associated proteins (e.g., TAFII110) of the polymerase II complex to stimulate gene transcription [20–22].
CREB is widely distributed in most tissues and has an essential role in controlling cell growth, differentiation, survival, and cell cycle progression and in determining the fate of many cell types [23–27], as well as being a proto-oncogene in myeloid transformation [28].
The only mechanism reported to be involved in CREB expression is CREB phosphorylation because CREB expression has been considered constitutive [29, 30]. However, there is increasing evidence that CREB gene expression is subject to dynamic regulation that varies depending on the cell type. In primary rat Sertoli cells, activation of the cAMP pathway up-regulates CREB expression via CREs present in the 5′ promoter region of the CREB gene [31]. Regulation of CREB expression has also been demonstrated in neuronal cells in vivo and in vitro [32–35]. However, these findings in Sertoli and neuronal cells are in marked contrast to findings in certain other systems (e.g. CATA.a cells). Widnell et al [35] reported that mRNA of CREB was down-regulated in CATH.a cells (a neural-derived cell line) by activation of the cAMP pathway. Thus, it would appear that perturbation of the cAMP pathway could exert opposite effects on CREB expression depending on the type of cells involved.
Our previous studies showed that RA-activated CREB plays a critical role in the expression of mucin genes, which are typical indicators of bronchial mucous cell differentiation [13, 14]. We also observed that CREB expression is very low in fully differentiated metaplastic squamous bronchial epithelial cells. As it is evidenced that CREB can be positively autoregulated [31], we hypothesized that RA-activated CREB induces CREB gene expression in bronchial epithelial cells. To test the hypothesis, we determined the level of CREB expression after RA treatment in NHTBE cells and analyzed the transcriptional regulation mechanism using the upstream region of the human CREB gene. Interestingly, we found that RA upregulates Sp1 expression and RA-inducible Sp1 contributes to the upregulation of CREB, while basal level of CREB is autoregulated by CREB itself. These data suggest cooperative role of these two transcription factors in regulation of CREB expression during mucous cell differentiation of normal bronchial epithelial cells.
MATERIALS AND METHODS
Cell cultures and chemicals
NHTBE cells (Clonetics, San Diego, CA) were cultured by the air-liquid interface method as described previously [5, 7, 13, 36]. Briefly, second-passage NHTBE cells were seeded at a density of 1 × 105 cells onto 24-mm semipermeable membrane inserts (Transwell-Clear; Corning, Inc., Acton, MA) and grown in serum-free culture medium supplemented with growth factor and hormones. The air-liquid interface was initiated when the cultures were confluent. All-trans RA, cycloheximide (CHX), and actinomycin D (ActD) (Sigma-Aldrich, St. Louis, MO) were dissolved in DMSO. Ro 61-8431 (pan RAR antagonist) and Ro 26-5405 (pan RXR antagonist) were kindly provided by Roche Bioscience (Palo Alto, CA). All cells were maintained at 37°C in an incubator containing 5% CO2.
Quantitative Real-time Polymerase Chain Reaction (QRT-PCR) and Conventional Reverse Transcription PCR (RT-PCR) Analyses
Total RNA was extracted from NHTBE cells cultured in the absence or presence of RA (1 μM) for specific times and concentrated by using RNeasy mini-kits (Qiagen, Valencia, CA). The RT reaction was performed using 1 μg of total RNA that was reverse-transcribed into cDNA using a random hexamer primer (GeneAmp RNA PCR core kit; Applied Biosystems, Foster City, CA) according to the manufacturer’s instructions. All primer sets produced amplicons of expected sizes and sequences. For the quantitative analysis of mRNA expression, the iCycler thermal cycler (Bio-Rad Laboratories, Inc., Rodeo, CA) was used with the DNA binding dye SYBR green (Applied Biosystems-Perkin-Elmer Corp., San Mateo, CA). The PCR cycling conditions for CREB expression were 95°C for 5 min followed by 40 cycles at 95°C for 30 s and 60°C for 1 min. Initial PCR conditions for Sp1 expression were 95°C for 5 min followed by 40 cycles at 95°C, 55°C, and 72°C for 30 s each. PCR for human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) labeled with VIC dye (Applied Biosystems) was done as an endogenous control. Primer sequences were as follows: CREB sense, 5′-ACTGTAACGGTGCCAACTCC-3′ and antisense, 5′-GAATGGTAGTACCCGGCTGA-3′. Human Sp1 primer sets were purchased from SuperArray Biosciences (Frederick, MD). For conventional RT-PCR, total RNA was extracted and then reverse transcribed into cDNA using a random hexamer primer. The conditions for PCR were 30 cycles at 95°C, 55°C, and 68°C for 30 s each. PCR for β-actin (Ambion, Inc., Austin, TX) was done as an internal control. Sp1 primer sequences were used in QRT-PCR. Other primer sequences were as follows: CREB sense, 5′-ACCATGGAATCTGGAGCCGAGAAC-3′ and antisense, 5′-CTGTAGGAAGGCCTCCTTGAAAGA-3′, and RARβ sense, 5′-AGCCTACGTGCCAAAAAAGG-3′ and antisense, 5′-TCTAGGTGTGGAGGCAAATGG-3′. PCR products were then separated in a 2% agarose DNA gel and stained with ethidium bromide.
Cloning of the Human CREB Promoter and Construction of Chimeric Luciferase Reporter Plasmid
The bacterial artificial chromosome (BAC) clone (RPCI-11.C 17I6; Invitrogen Corporation, Carlsbad, CA) was used to clone the human CREB promoter region. PCR was performed to amplify the 5′-flanking regions of the CREB gene by using the human BAC DNA as the template. The primers used to generate different promoter deletion plasmids were as follows: forward, 5′-GAGGAGCTCACGGGTTGAGTTTGAAATGC-3′, 5′-GAGGAGCTCGCCGTCCTGAGAGACCTCTG-3′, 5′-GAGGAGCTCGCATGCTCCGCTGGCG-3′, 5′-GAGGAGCTCCCGCTGCCGGCTTTACC-3′, 5′-GAGGAGCTCAAGGTCTTCGGCAAGTTCC-3′, 5′-GAGGAGCTCTAGCTCGGCTGTTTCCGT-3′, 5′-GAGGAGCTCTGGCTGCGGCTCCTCAGT-3′, 5′-GAGGAGCTCCGGAGTGTTGGTGAGTGACG-3′; and reverse, 5′-GAGGCTAGCTTCTCTCCCCCACGTAACAC-3′. Primers were designed to contain SacI or NheI restriction sites so that the resulting PCR-amplified fragments could be cloned into the multicloning sites upstream of the luciferase reporter gene in the pGL3-basic vector (Promega Corporation, Madison, WI).
Site-directed mutations of the human CREB promoter were made in the context of −321/−70 CREB-LUC, using the Quikchange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The oligomers used to introduce multiple point mutations were: CRE1, 5′-GGAGGTGTAGTTTCTCGAGGTGTGTTACGTG-3′; CRE2, 5′-TGTTGGTGAGTCTCGAGGCGGAGGTGTA-3′; CRE1+2, 5′-GTTGGTGAGTCTCGAGGCGGAGGTGTAGTTTCTCGAGGTGTGTTAC-3′; Sp1-1, 5′-TGTGGCCCGGGCCCCTGGGAGAAGCG-3′; and Sp1-2, 5′-CTCGGCACTGGGCCCCGCTGGCTGGCT-3′, where boldface, underlined type indicates a mutation [37]. To generate the Sp1-1+Sp1-2 mutant, the Sp1-1 and Sp1-2 primers were used in combination. All constructs were confirmed by DNA sequencing.
Transfection and Luciferase Assays
For the luciferase assay, NHTBE cells were plated in 12-well culture plates at a density of 1 × 104 cells/well. Cells were grown in BEGM (bronchial epithelial growth medium) without RA. When the cells reached 80% confluence, they were subjected to transient transfection with 0.5 μg of plasmid DNA with FuGENE 6 transfection reagent (Roche Diagnostics, Indianapolis, IN), which was performed according to the manufacturer’s instructions. To normalize the transfection efficiencies of the various luciferase constructs, the phRL-CMV plasmid containing the Renilla luciferase gene was co-transfected into the cells in a molar ratio of 1:50 (phRL-CMV: pGL3). Four hours after transfection, medium was replaced, and the cells were treated with RA. The cultures were maintained for an additional 48 h. CREB promoter activity was determined by measuring firefly luciferase activity, and Renilla luciferase activities were measured sequentially with a dual-luciferase reporter assay system (Promega Corporation) according to the manufacturer’s instructions; the Lumant LB 9507 luminometer (EG & G Berthold, Bad Wildbad, Germany) was used to make the measurements.
EMSA
To assess CREB’s DNA binding activity, we performed EMSA as described previously [13, 38], with necessary modifications. Briefly, nuclear proteins from RA-treated or untreated cells were prepared and stored at −80°C. For EMSA, oligonucleotide-corresponding sequences were as follows: Sp1-1, 5′-TGTGGCCCGGGCGGCTGGGAGAAGCG-3′, Sp1-2, 5′-CTCGGCACTGGGCGGCGCTGGCTGGCT-3′, CRE1, 5′-GGAGGTGTAGTTTGACGCGGTGTGTTACGTG-3′, and CRE2, 5′-TGTTGGTGAGTGACGCGGCGGAGGTGTA-3′. These oligonucleotides were annealed and end-labeled with [γ-32P]ATP using T4 polynucleotidekinase. Ten micrograms of nuclear extract was incubated at room temperature for 30 min with the 32P-labeled CRE probe in binding buffer (20% glycerol, 5 mM MgCl2, 2.5 mM EDTA, 2.5 mM DTT, 250 mM NaCl,50 mM Tris-Cl [pH 7.5], and 0.25 mg/ml poly[dI-dC]). DNA-nuclear protein complexes were separated from the DNA probe by electrophoresis through 5% nondenaturing polyacrylamide gels in 0.5× Tris borate EDTA (TBE) buffer. For supershift analysis, nuclear extracts were incubated with anti-CREB and anti-Sp1 antibodies and then incubated with oligonucleotide probes for 30 min at 37°C. The complex was then analyzed by EMSA. Antibodies against pre-immune serum were included as negative controls. The DNA-protein complexes were resolved by electrophoresis in 5% nondenaturing polyacrylamide gels in0.5× TBE (1× TBE: 25 mM Tris base, 25 mM boric acid, and 0.5 mM EDTA). Gels were dried and autoradiographed at −80°C. All EMSAs were repeated three times, and consistent results were obtained.
ChIP Analyses
ChIP assays were performed as described elsewhere [39, 40]. Briefly, NHTBE cells were activated with RA for 4, 8, 24, or 48 h and then cross-linked by reaction with 1% formaldehyde for 10 min at 37°C. The cells were then washed with cold phosphate-buffered saline (PBS). The cell pellet was resuspended in lysis buffer (1% SDS, 100 mM NaCl, 50 mM Tris-HCl [pH 8.1], and 5 mM EDTA) and then sonicated to an average DNA length of 500 to 1,000 base pairs. Antibodies were added to each of the samples, which were then rotated overnight and a temperature of 4°C. After interaction with protein A beads and incubation overnight at 65°C to reverse the cross-links, the DNA was dissolved in Tris-EDTA buffer and then analyzed by PCR. The anti-CREB, anti-pCREB, and anti-Sp1antibodies (Upstate Biotechnology, Inc., Waltham, MA) or normal rabbit IgG was added separately into the reaction solutions. Primers used for PCR were obtained from CREB promoter sequences: 5′-AAGGTCTTCGGCAAGTTCC-3′ (sense) and 5′-TTCTCTCCCCCACGTAACAC-3′ (antisense).
RNA Interference Analyses
For knockdown by RNA interference, we used SMARTpool siRNA of CREB, RARα, RARβ, RARγ, and RXRα (Upstate Biotechnology, Inc.). Sp1 siRNA purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Briefly, NHTBE cells at 60% to 70% confluence were transfected with a final concentration of 100 nM SMARTpool siRNA or nonspecific control pool (siRNA negative control) using the siIMPORTER siRNA transfection reagent (Upstate Biotechnology, Inc.) according to the manufacturer’s instructions. Sp1 siRNA was also transfected at 100 nM. For luciferase reporter assay, −321/−70 CREB-LUC was co-transfected with siRNA of CREB or Sp1. After 72 h of transfection, when the target protein levels had been reduced by 70% to 80% as assessed by western blot analysis, the cells were treated with or without RA for another 24 h or 48 h and then assessed by the luciferase reporter assay. Then, whole-cell lysate was prepared for western blot, QRT-PCR, RT-PCR, and luciferase assays.
Western Blot Analyses
Western blot analysis was performed as previously described [13]. Briefly, whole-cell extracts were prepared using 2x SDS Laemmli lysis buffer. Equal amounts of total protein (20 μg) were resolved by 10% SDS-polyacrylamide gel electrophoresis (SDS-PAGE). We used mouse monoclonal antibodies against β-actin (clone AC-15; Sigma-Aldrich, St. Louis, MO), rabbit polyclonal antibodies against Sp1 (Upstate Biotechnology, Inc.), and total CREB and phospho-CREB (Upstate Biotechnology, Inc.). Proteins reactive with primary antibody were visualized with a horseradish peroxidase-conjugated secondary antibody and enhanced chemiluminescence reagents (Amersham Bioscience, Arlington Heights, IL).
Statistical Analyses
For all transfection assays and QRT-PCR analysis, data were shown as mean ± SE of triplicate assays in three independent experiments. Statistical analysis was performed using Student’s t test (Prism;GraphPad Software, San Diego, CA), and statistical significance are expressed as * p < 0.05, ** p < 0.01.
RESULTS
RA upregulates CREB expression in time- and dose-dependent manners
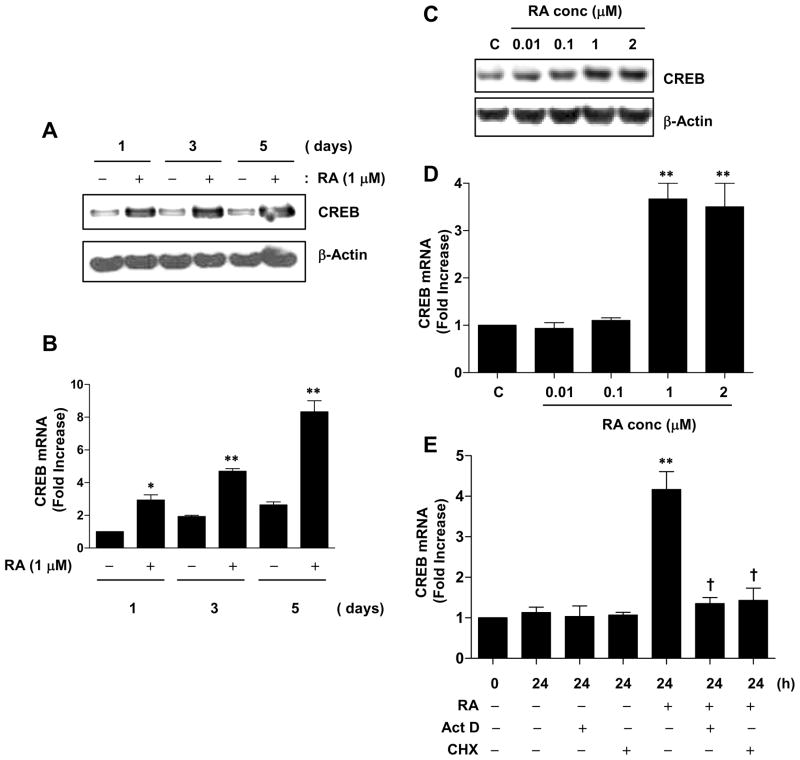
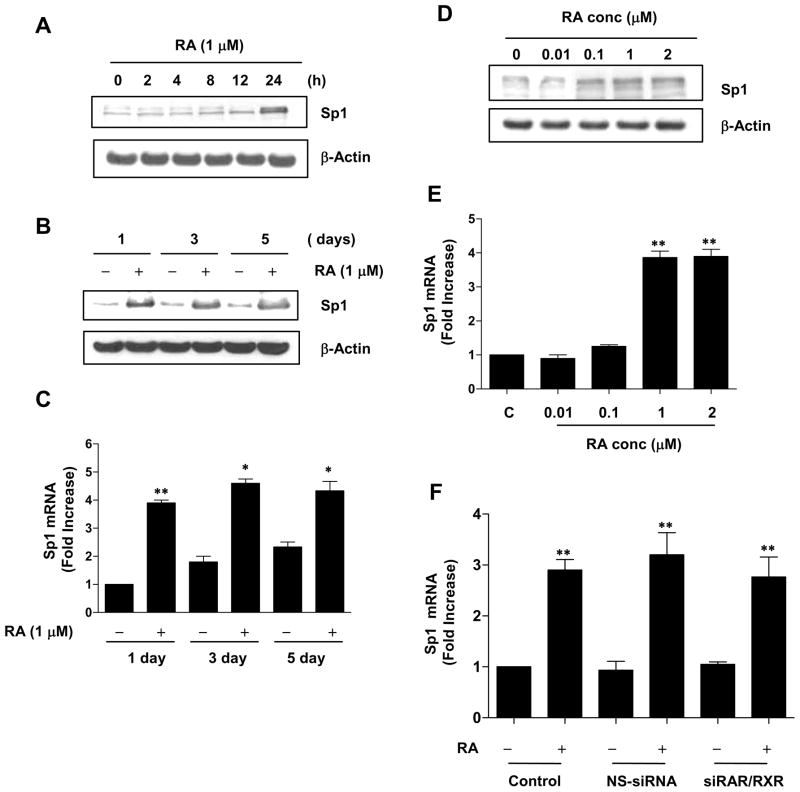
To determine whether RA regulates the CREB gene, we determined the level of CREB gene expression before and after RA treatment. CREB mRNA and protein expression were significantly increased by RA treatment for 1, 3 and 5 days compared to vehicle-treated control (Figure 1, A and B). CREB protein and mRNA also increased in a dose-dependent manner after 24 h of RA treatment; in this analysis (Figure 1, C and D).
Figure 1.
RA upregulates CREB expression. (A–E) NHTBE cells grown in RA-deficient medium for 7 days were incubated with or without 1 μM RA for various periods of time or with various concentrations of RA for 24 h, as indicated on the figure panels. CREB expression was analyzed by western blotting (A and C) and QRT-PCR (B and D). Vehicle (DMSO) was used as negative controls, respectively. (E) ActD and CHX effects on RA-induced CREB expression. NHTBE cells were pre-incubated in the presence or absence of 5 μg/ml ActD or 10 μg/ml CHX for 2 h and were either left untreated or treated with 1 μM RA for 24 h. CREB mRNA expression levels were measured by QRT-PCR. In panels A and C, the western blots are representative of those from three repeat experiments. In panels B, D, and E the data represent the mean ± SE of three independent experiments, each performed in triplicate. * p < 0.05, ** p < 0.01 compared with untreated control. † p < 0.05 compared with the RA-treated group.
To further investigate the effects of RA on CREB gene regulation due to an increase of transcription and translation, we used ActD, a general inhibitor of mRNA transcriptional synthesis, and CHX, a general inhibitor of protein synthesis, to inhibit de novo synthesis of mRNA and protein; NHTBE were then treated with RA. Pretreatment with either ActD or CHX alone did not affect the endogenous level of CREB; however, RA-induced CREB mRNA expression was completely blocked (Figure 1E). Taken together, these findings demonstrate that RA increases CREB gene expression and that this upregulation occurred at transcription level while de novo synthesis of protein is required.
RA-induced upregulation of CREB is independent of retinoic acid receptors
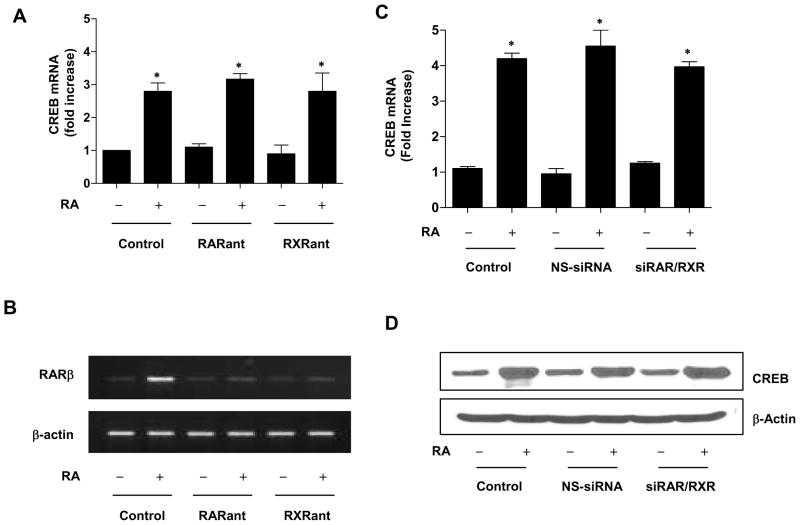
Next, we examined whether RA-induced CREB gene expression is mediated by the conventional RA receptors (RAR and RXR). In cultured NHTBE cells, pretreatment with 10 μM Ro 61-8431 (pan-RAR antagonist) or Ro 26-5405 (pan-RXR antagonist) alone did not affect the endogenous level of CREB. In addition, neither the pan-RAR antagonist nor the pan-RXR antagonist could block RA-induced CREB gene expression (Figure 2A). The activities of pan-RAR and RXR antagonists were confirmed by measuring the mRNA level of the RAR/RXR-mediated RA target gene RARβ (Figure 2B). RA-induced RARβ mRNA expression was almost completely blocked by either antagonist.
Figure 2.
RAR and RXR receptor independent effect on RA-induced CREB regulation. (A and B) NHTBE cells grown in RA-deficient medium for 7 days were preincubated with or without pan-RAR or pan-RXR antagonist (10 μM) for 2 h and then further incubated with RA (1 μM) for 48 h. Vehicle (DMSO) was used as a negative control. (A) QRT-PCR analysis of CREB mRNA expression normalized to GAPDH endogenous control. Data represent the mean ± SE of three independent experiments, each performed in triplicate. * p < 0.05 compared with RA-untreated control. (B) To show the effects of pan-RAR antagonist and pan-RXR antagonist, RARβ mRNA expression was measured by RT-PCR. After treatment, total RNA was extracted and then used to measure the RARβ mRNA level. RT-PCR products were analyzed on a 2% agarose gel using β-actin as a loading control. (C) Effects of siRNA knockdown of RAR/RXR on CREB regulation. NHTBE cells were transiently transfected with siRNAs of RAR and RXR together in combination with either siRNA-Rαβγ/Xα or a nonspecific control pool (NS siRNA). Three days after transfection, the cells were further incubated with or without RA for 24 h. After treatment, CREB mRNA expression was measured by QRT-PCR and the results normalized to those for GAPDH endogenous control. To show the effects of RAR/RXR siRNA, RARβ expression was measured by western blotting. Equal loading was confirmed by stripping the blot and reprobing it for β-actin antibody. Results are representative of those from three independent experiments. * p < 0.01 compared with the untreated control.
To further investigate the effects of RAR/RXR on RA-induced CREB gene expression, we used RAR/RXR siRNA to suppress the endogenous expression of RAR/RXR in NHTBE cells. Cotransfection of the NHTBE cells was done using SMARTpool-sequenced siRNA targeting the human RAR and RXR receptors with a combination of RARα, RARβ, and RARγ/RXRα (Figure 2C). We silenced only the α isotype of RXR because it is known that RXRα plays a critical role in RAR/RXR heterodimers [41, 42]. We observed maximal reduction of endogenous RAR/RXR protein 72 h after transfection. Successful suppression of RAR/RXR has been demonstrated previously and residual RAR/RXR were not sufficient enough to transmit RA signaling [13, 14]. Likewise, the RA-induced expressions of CREB mRNA and protein were not significantly reduced after co-transfection with a combination of RAR and RXR siRNAs (Figure 2, C and D), strongly indicating that RAR and RXR receptors are not involved in RA-induced CREB regulation.
Sp1 binding sites are required for CREB promoter transactivation
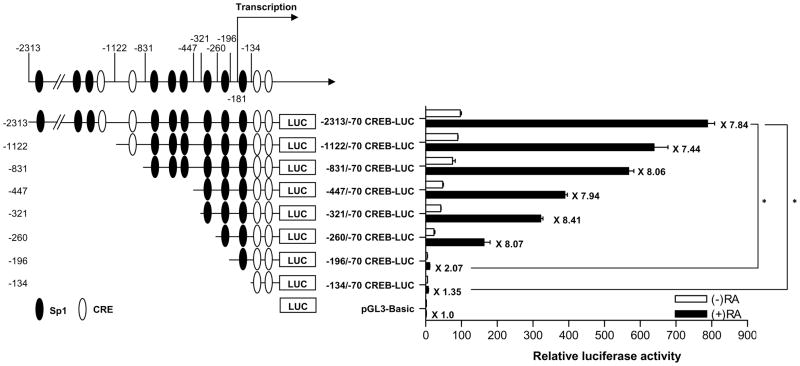
To determine the key transcription factor involved in RA-induced CREB regulation, we first generated a series of 5′ deletion constructs of human CREB promoter by subcloning appropriate restriction fragments upstream of the luciferase gene in the pGL3-Basic luciferase reporter vector. Then we transiently transfected them into NHTBE cells to determine their RA-induced luciferase activities. In the current analysis (Figure 3), we found that RA caused a 7.84-fold increase in the luciferase activity of reporter construct containing the CREB promoter (nucleotides −2313 to −70), relative to untreated control. In addition, RA increased luciferase activity even further, to 8.07-fold over control, in tests using a deletion mutant of CREB promoter that contained nucleotides −260 to −70. However, the deletion mutants that contained nucleotides from the −196 to −70 and −134 to −70 regions of the CREB promoter significantly decreased RA-induced luciferase activity compared with the 2.2-kb CREB promoter (nucleotides −2313 to −70); the −196/−70 and −134/−70 CREB-LUC mutants yielded increases of only 2.07-fold and 1.35-fold over control, whereas that yielded by the −2313/−70 CREB-LUC mutant was 7.84-fold over control (p < 0.01) (Figure 3). These results show that the region between nucleotides −260 and −134 upstream of the CREB coding sequence, which includes two Sp1 motifs, is critical for transactivation of the CREB promoter by RA.
Figure 3.
Functional deletion analysis of the human CREB promoter region. The diagram represents the locations of the Sp1 and CRE motifs and the numbers on the left side represent the upstream start site of the CREB promoter region. NHTBE cells were transiently transfected with serially deleted CREB promoter containing luciferase reporter constructs and then treated with RA. After 48 h of transfection, cell extracts were assayed for luciferase activity, which was normalized to renilla luciferase activity, as described in Materials and Methods. The data are expressed relative to the basal activity of the CREB promoter (indicated as −2313/−70 CREB-LUC = 100%) and represent the mean± SE of three independent experiments, each performed in triplicate. The asterisk indicates the difference from the RA-stimulated −2313/−70 CREB-LUC construct (* p < 0.01). The numbers on the left side of the diagram represent the start position of the 5′ location on the CREB promoter region.
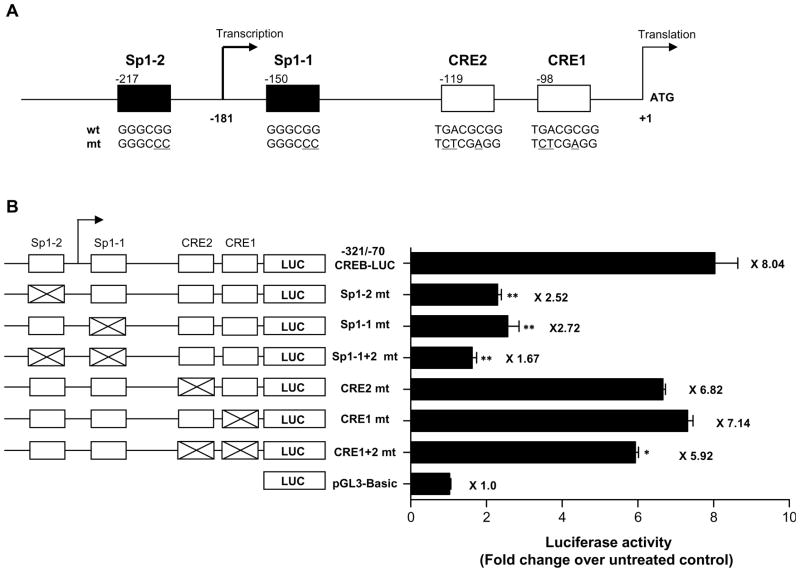
By analyzing the CREB promoter sequences in the TFSEARCH database (http://mbs.cbrc.jp/research/db/TFSEARCH.html) at a threshold score of 74.0, we identified two putative Sp1-binding sites (Sp1 motifs) and two putative CREB binding sites (CRE motifs) in the −260 to −70 CREB promoter region, as shown in Figure 4A. To further determine the role of Sp1 and CRE motifs in CREB expression, we next generated point-mutated Sp1 and CRE motifs in the CREB promoter luciferase reporter vector and tested their transactivation by RA (Figure 4A). We found that Sp1 mutants, as both single mutants (Sp1-1 at −150 or Sp1-2 at −217) and as a double mutant (Sp1-1 + Sp1-2), significantly reduced promoter activity compared with the RA-stimulated wild-type CREB promoter (nucleotides −321 to −70)/luciferase reporter construct as a template to make a construct with site-specific mutations; Sp1-1 alone increased activity 2.72-fold over control, Sp1-2 alone increased it 2.52-fold, and Sp1-1 + Sp1-2 increased it 1.67-fold, whereas the RA-stimulated CREB promoter (nucleotides −321 to −70)/luciferase reporter construct increased it 8.04-fold. Whereas neither CRE mutant alone (CRE1 mt nor CRE2 mt) affected promoter activity, the double CRE mutant (CRE1 + CRE2 mt) slightly decreased the promoter activity compared with the RA-stimulated CREB promoter (nucleotides −321 to −70)/luciferase reporter construct. CRE1 alone increased promoter activity 7.14-fold over control, CRE2 alone increased it 6.82-fold, and CRE1 + CRE2 increased it 5.92-fold. These data clearly demonstrate that the Sp1 motifs, which surround the transcription initiation start site, are required for RA-induced CREB promoter transactivation.
Figure 4.
Analysis of mutated Sp1 and CRE motifs in the CREB promoter. (A) The schematic diagram represents the locations and sequences of the wild-type (wt) and mutated (mt) Sp1 and CRE motifs. (B) NHTBE cells were transiently transfected with various mutants of Sp1 and CRE in the CREB promoter–luciferase reporter constructs. The diagrams show the relative locations of the Sp1 and CRE motifs in the −321 to −70 CREB-LUC construct. An X in a motif designates a mutation. After transfection, the cells were further incubated with 1 μM RA or vehicle control for 48 h. The data are expressed relative to the fold increase in luciferase acitivity over untreated control and represent the means ± SE of three independent experiments, each performed in triplicate. The asterisk indicates the difference from the RA-stimulated wild-type −321/−70 CREB-LUC construct (* p < 0.05, ** p < 0.01).
Phylogenetic analysis of CREB promoter shows conservation of CRE, Sp1 motifs in several species
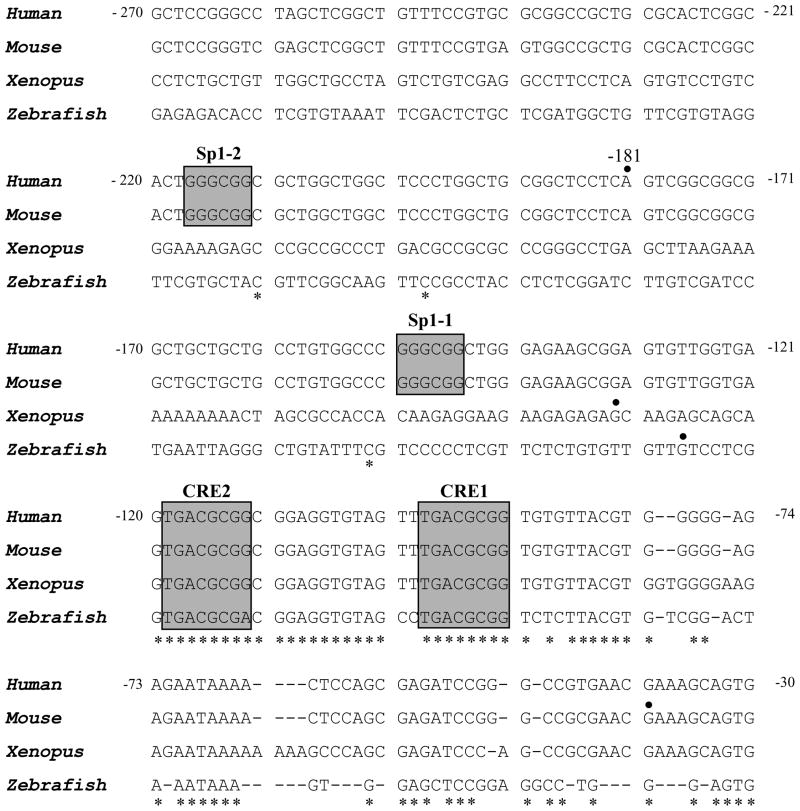
To determine the evolutionary conserved transcription factor that may play a critical role on the regulation of the CREB gene, we analyzed the human CREB gene promoter around the transcription start site and compared it with mouse, xenopus, and zebrafish CREB gene promoters (Figure 5). Genomic DNA sequences of the CREB promoter region from four different species were obtained (Ensembl genome browser, http://www.ensembl.org/) and aligned using the NCBI blast two-sequence alignment program (bl2seq). The human CREB promoter is enriched in the nucleotides guanosine and cytosine (65% G+C) and contains no CAAT or TATA box motifs [43]; however, it does contain the 5′-TCCTCAGTCGG-3′ sequence, which resembles the initiator sequence 5′-PyCCTAPyTCTG-3′ that has been shown in TATA-less promoters to initiate gene transcription at the conserved A nucleotide [44]. The numbering was based on the translation start site, defined as +1. Because of the lack of a TATA box, the CREB promoter is numbered relative to the translation start site, placing the major transcription initiation start site at position −181. As Figure 5 shows, two Sp1 motifs and CRE motifs are present within the 191-base pair promoter fragment; they are located between nucleotides −260 and −70. Sp1 sites surround the major transcription initiation site (at positions −217 and −150), and the two CREs are located −119 and −98 base pairs downstream of the transcription start site. The two CRE motifs are highly conserved among the four species, but Sp1 motifs are conserved only in human and mouse.
Figure 5.
Alignment of the nucleotide sequences around the transcription initiation sites of CREB genes from four different species. The respective transcription initiation sites are indicated by the solid dot (●). Gaps in the sequence are indicated by dashes, and nucleotide numbers are shown for the human sequence. The positions of sequence identity are also indicated (*). The previously known sequence elements, including Sp1 and CRE sequences, are boxed.
Transcription factor Sp1 and CREB bind specifically to the CREB promoter
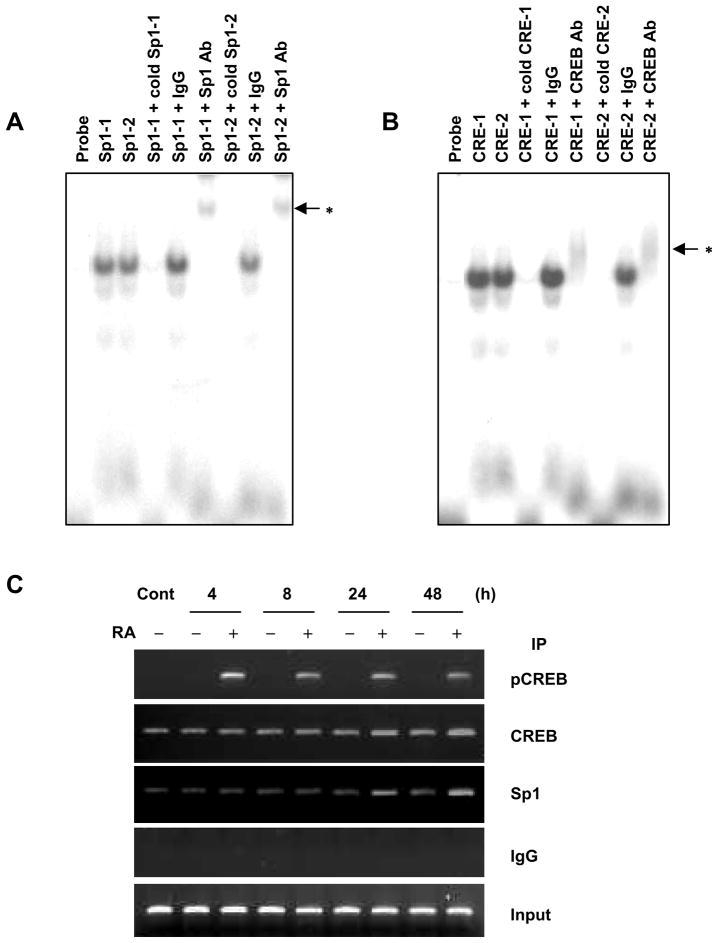
To determine whether Sp1 and CREB specifically bind to their binding motifs (boxes in Figure 5) in the CREB promoter, we performed competition EMSA and ChIP assays with anti-CREB, anti-pCREB, and anti-Sp1 antibodies. EMSA data showed that the radiolabeled oligonucleotides of Sp1-1, Sp1-2, CRE1, and CRE2 formed DNA protein. In addition, pretreatment with these cold oligonucleotides abolished nuclear protein binding to radiolabeled oligonucleotides. To further confirm Sp1 and CREB binding to putative Sp1 and CRE motifs after RA treatment, supershift assays were preformed in the presence of anti-Sp1 or anti-CREB antibodies. After incubation of either of these antibodies, attenuation and band shift were seen in the DNA-protein bindings (Figure 6, A and B, lanes 6 and 9). However, normal rabbit IgG did not induce attenuation or band shift of these DNA-protein bindings (Figure 6, A and B, lanes 5 and 8). These results indicate that Sp1 and CREB specifically bind to Sp1- and CRE-binding motifs, respectively (Figure 6, A and B).
Figure 6.
The effect of RA on CREB promoter binding of Sp1 and CREB. (A and B) EMSA was performed using NHTBE cells that were cultured for 7 days with or without RA and then treated with RA for 24 h. Ten micrograms of nuclear extract was prepared, and DNA-protein binding activity was measured by EMSA using various [γ-32P]ATP-labeled oligonucleotides corresponding to Sp1-1, Sp1-2, CRE1, and CRE2, as described in Materials and Methods. The DNA-binding reaction was performed in the absence or presence of a 100-fold excess of each oligonucleotide unlabeled (that is, an unlabeled competitor). Supershift assays were performed using 2 μg of the anti-CREB and anti-Sp1 antibodies, as indicated above each lane in panels A and B. The labeled nucleotide and nuclear protein complexes were separated by electrophoresis on 5% polyacrylamide gels, and the gels were dried and autoradiographed at −80°C. The asterisks (*) indicate shifted bands. (C) ChIP analysis was performed using NHTBE cells grown in RA-deficient medium for 7 days. The cells then were incubated with or without 1 μM RA for the indicated time. ChIP assays were performed using anti-phospho-CREB (pCREB), anti-CREB, or anti-Sp1 antibodies to precipitate CREB promoter that corresponded to the position −321/−70 region. One percent of the chromatin was assayed to verify equal loading (Input), and nonspecific IgG was assayed under otherwise identical conditions as a negative control. Results are representative of those from three independent experiments.
ChIP assay confirmed that endogeneous Sp1 and CREB bind to this promoter region (Figure 6C). One pair of primers was used to amplify the CREB promoter regions from −321 to −70 (252 base pairs), which contained the Sp1 and CRE sites. Sp1 and CREB were recruited to the CREB promoter with or without RA treatment. However, the level of recruited CREB and Sp1 protein was increased after 24 and 48 h of RA treatment. These results indicate that RA increased Sp1 and phosphorylated CREB binding to the CREB promoter in vivo.
RA induces Sp1 expression
Because ChIP analysis (Figure 6C) showed that RA increases Sp1 binding to the CREB promoter, we examined the possibility that RA is also involved in the regulation of endogenous Sp1 in NHTBE cells. As shown in Figure 7, A and B, Sp1 protein expression levels remained unchanged until 4 h of RA treatment and then gradually increased thereafter. In addition, QRT-PCR revealed that Sp1 mRNA was upregulated 3.91-fold within 1 day of RA treatment and remained elevated thereafter (Figure 7C). These results indicate that RA treatment significantly increases Sp1 mRNA and protein compared with control treatment. In addition, Sp1 protein and mRNA were increased in a dose-dependent manner at 24 h of RA treatment (Figure 7, D and E).
Figure 7.
RA increases Sp1 expression. NHTBE cells were grown in RA-deficient medium for 7 days and then incubated with or without 1 μM RA for the indicated periods of time or with various concentrations of RA for 24 h. Vehicle (DMSO) was used as a negative control. Sp1 gene expression was analyzed using western blotting (A, B, and D) and QRT-PCR (C and E), as described in Figure 1. (F) NHTBE cells were transiently transfected with siRNAs of RARs and RXR together or with nonspecific siRNA as described in Figure 2. Three days after transfection, the cells were further incubated with or without RA for 24 h. After treatment, Sp1 mRNA expression was measured by QRT-PCR. In panels C, E, and F, the data represent the mean ± SE of three independent experiments, each performed in triplicate. * p < 0.05, ** p < 0.01 compared with untreated control. † p < 0.01 compared with the RA-treated group. In panels A, B, and D, western blot results are representative of those from three repeat experiments.
Next, to determine whether RAR/RXR is involved in RA-induced Sp1 gene regulation, we used the siRNA approach to deplete expression of RARα, RARβ, RARγ, and RXRα in NHTBE cells. RA-induced expression of Sp1 mRNA was not affected after cells were co-transfected with the combination of RAR and RXR siRNAs, implying that RAR and RXR receptors are not involved in RA-induced Sp1 gene regulation (Figure 7F).
Both Sp1 and CREB are required for RA-induced CREB gene expression
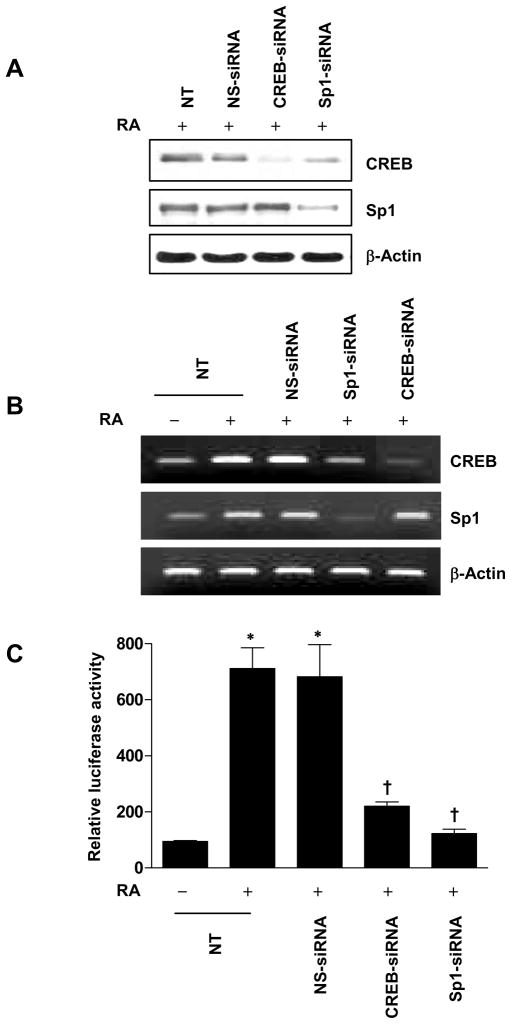
To further investigate the roles of Sp1 and CREB on CREB gene regulation and transactivation, we used an siRNA approach to suppress the endogenous expression of Sp1 and CREB in NHTBE cells and determined the level of RA-inducible Sp1 and CREB protein expression. We found that RA-induced CREB protein expression was reduced after transfection with Sp1 siRNA, whereas CREB siRNA did not affect RA-induced Sp1 expression (Figure 8A). Furthermore, RT-PCR analysis demonstrated that the decrease in endogenous CREB mRNA was dependent on Sp1 silencing, whereas CREB siRNA did not affect RA-induced Sp1 mRNA expression (Figure 8B). This result shows that Sp1 is important for CREB gene expression.
Figure 8.
Effect of siRNA knockdown of Sp1 on CREB gene expression. (A and B) NHTBE cells were transiently transfected with CREB siRNA or Sp1 siRNA. A nonspecific siRNA pool (NS siRNA) was used as the control. Three days after transfection, the cells were further incubated with or without RA for 24 h. After treatment, CREB expression was analyzed by western blotting (A) and QRT-PCR (B). (C) NHTBE cells were cotransfected with −321/−70 CREB-LUC construct plasmid and CREB siRNA or Sp1 siRNA. Three days after transfection, the cells were further incubated with or without RA for 48 h. The luciferase activity was determined as described in Figure 3. The data are expressed relative to the basal activity of the CREB promoter (indicated as −321/−70 CREB-LUC = 100%) and as means± SE of three independent experiments, each performed in triplicate. * p < 0.01 compared with untreated control. † p < 0.01 compared with the RA-treated group.
Next, to further determine the role of Sp1 and CREB on RA-induced transactivation of CREB promoter activity, we co-transfected pGL3-Basic luciferase reporter vector containing CREB promoter (nucleotides −321 to −70) region and Sp1 siRNA or CREB siRNA. Using a luciferase activity assay, we found that knockdown of either endogenous Sp1 or endogenous CREB significantly decreased RA-inducible CREB gene transactivation (Figure 8C). Sp1 knockdown decreased the CREB promoter activity significantly more than CREB knockdown did, (82.9% versus 69.1%), compared with RA-treated nontransfected control. These results strongly support both Sp1 and CREB are required for RA-induced transactivation of CREB promoter activity.
Moreover, these data suggest that Sp1 has a more important role than CREB in RA-inducible CREB gene regulation and that Sp1 and CREB may interact and/or that these protein interactions dependently or independently support binding to the imperfect CRE or Sp1 motifs to facilitate transcription from the CREB promoter.
DISCUSSION
Earlier studies have shown that RA deficiency induced metaplastic squamous differentiation in airway mucosa and normal mucociliary phenotype can be restored by RA treatment [5, 9]. Our recent study showed that one of the earliest event occurred during this restoration of normal mucous phenotype from squamous metaplasia involved in CREB activated by RA. In the present study, we found that RA not only activated CREB but also induced expression of CREB gene in bronchial epithelial cell and that Sp1 played an important role in the upregulation of the CREB gene expression during mucous differentiation of normal human bronchial epithelial cells. By conducting a detailed analysis of the promoter of CREB in order to gain an understanding of the molecular mechanisms associated with the upregulation of CREB transcription in response to RA, we found that Sp1 and CRE motifs in the essential promoter region of the CREB were required for full transcriptional activity in the RA-induced transcription of the human CREB gene. These RA-inducible CREB gene up-regulation mechanisms appeared to be mainly controlled by binding of the two Sp1 binding motifs in the essential promoter region to Sp1, which is also upregulated by RA.
Our studies showed that the two Sp1-binding motifs near the upstream of the transcription start site have a major role in CREB gene regulation. Other investigators have also shown that transient overexpression of Sp1 significantly induces CREB promoter activity in Drosophila SL2 cells, suggesting that Sp1 is a potent activator of CREB gene expression [45]. Mutation of the downstream Sp1 motif (Sp1-1 mt) in the CREB promoter resulted in a 50% decrease in basal activity in Sertoli cells. However, mutation of the Sp1 site, which is located upstream of the transcription initiation site (Sp1-2 mt), did not affect CREB promoter activity [37]. However, Coven et al. [46] report that only one of the Sp1 site (Sp1-2) were required for the normal upregulation of CREB promoter activity in C6 cells. In this study, we also showed that the RA-inducible essential promoter region was located in the nucleotide −260 to −70 segment of the CREB promoter region (Figure 3). Tests for mutagenesis in two putative Sp1-binding motifs and two CRE motifs showed that alone, neither the CRE-1 mt nor the CRE-2 mt affected promoter activity; however, the double CRE mutant (CRE1 mt + CRE2 mt) slightly decreased RA-inducible CREB promoter activity. The Sp1-1 and Sp1-2 mutants alone and combined (Sp1-1 mt + Sp1-2 mt) significantly reduced RA-inducible promoter activity compared with wild-type CREB promoter (Figure 4). Our results strongly suggest that two CRE motifs bind CREB, which contributes to basal promoter activity, and that two Sp1-binding motifs bind Sp1, which is essential for RA upregulation of CREB gene transcription. .
In contrast to our findings, Walker et al. [37] reported that the presence of three CRE-like sequences in the first 1,240 base pairs upstream of the translation start site in the CREB promoter region suggested regulation of the cAMP-inducible CREB gene expression pathway. They showed that cAMP-inducible CREB gene expression required the two CREs located downstream of the major transcription start site in the CREB gene promoter. They also showed that only Sp1-1 binding sites contributed to basal promoter activity. In neuronal cells, up- or down-regulation of CREB expression by activation of the cAMP pathway has been reported both in vivo and in vitro [32, 34, 35]. Activation of the cAMP pathway resulted in downregulation of CREB mRNA and protein in CATA.a cells (a neural-derived cell line) [47], whereas the same treatments did not alter CREB expression in PC12 pheochromocytoma cells [34]. These findings imply that regulation of CREB expression by the cAMP pathway depends on specific cell type.
The results of our EMSA and ChIP analyses further support the belief that Sp1 binds specifically to the Sp1-binding motif and that CREB binds specifically to CRE motifs, in vitro and in vivo (Figure 6). These results also suggest that the Sp1-1, Sp1-2, CRE1, and CRE2-binding motifs in this promoter region can function in vivo. Our finding that Sp1 and CREB binding on the CREB promoter region was increased after 24 h of RA treatment suggests that both the RA-induced upregulated Sp1 and phosphorylated CREB may have a role in upregulating the CREB gene expression during the early stages of epithelial cell differentiation (Figure 6C). It has been reported that the promoter region of human and mouse CREB has three variant CREs spread over nearly 800 base pairs [31, 48]. These CREs are highly conserved in organisms ranging from zebrafish to humans; however, Sp1-binding motifs are conserved only in mice and humans (Figure 5). Thus, both Sp1- and CRE-binding motifs might have important roles in CREB gene regulation in many organisms. As shown in Figure 5, the transcription start sites are located in different regions of the promoter region in humans and mice, implying that different transcription regulation mechanisms exist in the two species.
It has also been reported that nuclear factor (NF)-κB and tumor necrosis factor (TNF)-α are regulators of CREB expression. Delfino and Walker [49] showed that overexpressed NF-κB interacts with the NF-κB-binding site in the CREB promoter and increases promoter activity in Sertoli and NIH 3T3 cells. These results suggest that NF-κB may be an important regulator of the genes required for spermatogenesis and a general regulator of CREB gene expression in nontestis cells. However, our reporter assay indicates that RA-induced CREB activity was retained by luciferase vector containing CREB promoter region from nucleotides −447 to −70, from which the putative NF-κB-binding sites had been deleted (Figure 3). We therefore postulate that NF-κB is not required for RA-inducible CREB expression in NHTBE cells.
RA reportedly has a profound effect on the regulation of cell growth and differentiation, primarily through two families of nuclear receptors: RARs and RXRs. Several studies have shown that RARs and RXRs, upon binding to ligands, promote transcription through interaction with co-activators [50, 51]. However, our data suggest that RA upregulates CREB independently of RAR and RXR because NHTBE cells depleted of RARs and RXRα by means of siRNAs targeting RARα, RARβ, RARγ, and RXRα could still mediate RA-induced transactivation of CREB promoter activity. In addition, in our study, selective pan-RAR and pan-RXR antagonists did not block RA activation of CREB (Figure 2). We previously reported that RA rapidly induced CREB activation via a nonclassical RA-signaling mechanism without using its canonical RARs and RXR [13]. All these results strongly suggest that the activation and regulation of the CREB gene by RA are both RAR and RXR independent.
We also showed that RA upregulated Sp1 gene expression in time- and dose-dependent manners (Figure 7A–7E). Interestingly, our data suggest that RA-inducible Sp1 gene upregulation is independent of RAR and RXR function: depletion of RARs/RXRα using siRNA did not affect the RA-induced transactivation of Sp1 promoter activity. However, further extensive studies are required to determine how RA upregulates Sp1 expression without using its conventional receptors, RAR/RXR. Notably, our data showed that endogenous depletion of Sp1 significantly reduced the RA-inducible CREB gene but that CREB depletion did not affect Sp1 gene upregulation (Figure 8). In addition, Sp1 depletion completely abolished RA-inducible CREB promoter activity (to a level only 1.2-fold higher than that of untreated controls), and CREB depletion significantly abolished the CREB promoter activity. Our results strongly suggest that Sp1 has a more critical role than CREB in RA-inducible CREB gene regulation and that these two proteins facilitate the binding of imperfect CRE and Sp1 motifs to each other. It is plausible that a basal level of CREB is positively auto-regulated by RA-activated CREB. In addition, Sp1 protein, which is increased by RA, binds to two Sp1-binding sites in the CREB promoter region to further facilitate transactivation of CREB genes in early stage of NHTBE cells differentiation.
In summary, our findings further demonstrates that RA upregulates CREB gene expression during the early stage of NHTBE cell differentiation and that RA-inducible Sp1 plays a major role in upregulating human CREB gene expression. This finding and our earlier findings suggest that several transcription factors, including CREB, Sp1, and RAR/RXR may coordinately regulate differentiation program of bronchial epithelial cells. CREB and Sp1 may play an important role in preparing environment for mucous cell differentiation when the level of RAR/RXR is not sufficient in the cells, such as in squamous metaplasia state.
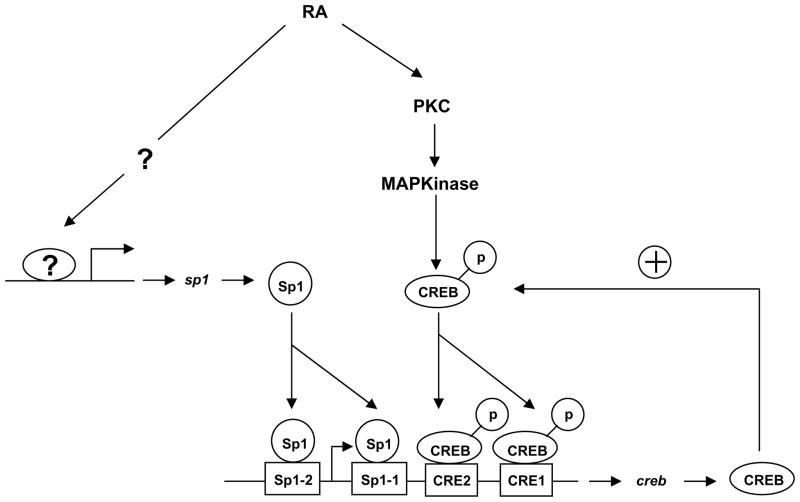
Figure 9.
A proposed model explaining upregulation of CREB expression by RA in NHTBE cells. In this model, RA rapidly activates protein kinase C and transmits an activation signal to phosphorylate nuclear CREB via the Ras/ERK/Rsk pathway, thereby increasing its transactivation activity. A basal CREB regulation of the positive feedback loop is constituted by induction of CREB, which is consequently activated by RA binding of CRE motifs. In addition, Sp1 protein, which is increased by RA, binds to Sp1-binding sites in the CREB promoter region to further activate transcription of CREB genes.
Acknowledgments
The authors thank Dr. Seung-Hee Ryu for technical assistance. This work was supported by National Heart, Lung, and Blood Institute Grant No. R01-HL-077556 (to J.S.K.), and National Cancer Institute Cancer Center Support Grant CA-16672 (to The University of Texas M. D. Anderson Cancer Center).
Abbreviations used
- ActD
actinomycin D
- ATF1
activating transcription factor 1
- BAC
bacterial artificial chromosome
- bZip
basic domain-leucine zipper
- cAMP
Cyclic 3′,5′-adenosine monophosphate
- ChIP
chromatin immunoprecipitation
- CHX
cycloheximide
- CBP
CREB-binding protein
- CRE
cAMP response-element
- CREB
CRE binding protein
- CREM
CRE modulator
- EMSA
electrophoretic mobility shift assays
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- NHTBE
normal human tracheobronchial epithelial
- NF-κB
nuclear factor-κB
- NRs
nuclear receptors
- RA
retinoic acid
- RAR
RA receptor
- RXR
retinoid X receptor
- siRNA
small interfering RNA
- TBE
Tris borateEDTA
- TNF-α
tumor necrosis factor-α
References
- 1.Chambon P. A decade of molecular biology of retinoic acid receptors. FASEB J. 1996;10:940–954. [PubMed] [Google Scholar]
- 2.Chytil F. Retinoids in lung development. FASEB J. 1996;10:986–992. doi: 10.1096/fasebj.10.9.8801181. [DOI] [PubMed] [Google Scholar]
- 3.Gudas LJ. Retinoids and vertebrate development. J Biol Chem. 1994;269:15399–15402. [PubMed] [Google Scholar]
- 4.Okuno M, Kojima S, Matsushima-Nishiwaki R, Tsurumi H, Muto Y, Friedman SL, Moriwaki H. Retinoids in cancer chemoprevention. Curr Cancer Drug Targets. 2004;4:285–298. doi: 10.2174/1568009043333023. [DOI] [PubMed] [Google Scholar]
- 5.Gray TE, Guzman K, Davis CW, Abdullah LH, Nettesheim P. Mucociliary differentiation of serially passaged normal human tracheobronchial epithelial cells. Am J Respir Cell Mol Biol. 1996;14:104–112. doi: 10.1165/ajrcmb.14.1.8534481. [DOI] [PubMed] [Google Scholar]
- 6.Koo JS, Jetten AM, Belloni P, Yoon JH, Kim YD, Nettesheim P. Role of retinoid receptors in the regulation of mucin gene expression by retinoic acid in human tracheobronchial epithelial cells. Biochem J. 1999;338 (Pt 2):351–357. [PMC free article] [PubMed] [Google Scholar]
- 7.Koo JS, Yoon JH, Gray T, Norford D, Jetten AM, Nettesheim P. Restoration of the mucous phenotype by retinoic acid in retinoid-deficient human bronchial cell cultures: changes in mucin gene expression. Am J Respir Cell Mol Biol. 1999;20:43–52. doi: 10.1165/ajrcmb.20.1.3310. [DOI] [PubMed] [Google Scholar]
- 8.McDowell EM, Keenan KP, Huang M. Effects of vitamin A-deprivation on hamster tracheal epithelium. A quantitative morphologic study. Virchows Arch B Cell Pathol Incl Mol Pathol. 1984;45:197–219. doi: 10.1007/BF02889865. [DOI] [PubMed] [Google Scholar]
- 9.McDowell EM, Keenan KP, Huang M. Restoration of mucociliary tracheal epithelium following deprivation of vitamin A. A quantitative morphologic study. Virchows Arch B Cell Pathol Incl Mol Pathol. 1984;45:221–240. doi: 10.1007/BF02889866. [DOI] [PubMed] [Google Scholar]
- 10.Thornton DJ, Carlstedt I, Howard M, Devine PL, Price MR, Sheehan JK. Respiratory mucins: identification of core proteins and glycoforms. Biochem J. 1996;316 (Pt 3):967–975. doi: 10.1042/bj3160967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gronemeyer H, Gustafsson JA, Laudet V. Principles for modulation of the nuclear receptor superfamily. Nat Rev Drug Discov. 2004;3:950–964. doi: 10.1038/nrd1551. [DOI] [PubMed] [Google Scholar]
- 12.Leid M, Kastner P, Chambon P. Multiplicity generates diversity in the retinoic acid signalling pathways. Trends Biochem Sci. 1992;17:427–433. doi: 10.1016/0968-0004(92)90014-z. [DOI] [PubMed] [Google Scholar]
- 13.Aggarwal S, Kim SW, Cheon K, Tabassam FH, Yoon JH, Koo JS. Nonclassical action of retinoic acid on the activation of the cAMP response element-binding protein in normal human bronchial epithelial cells. Mol Biol Cell. 2006;17:566–575. doi: 10.1091/mbc.E05-06-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim SW, HJ, Ryu SH, Chung WC, Yoon JH, Koo JS. Regulation of Mucin Gene Expression by CREB via a Nonclassical RA Signaling Pathway. Mol Cell Biol. 2007 doi: 10.1128/MCB.02385-06. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Radhakrishnan I, Perez-Alvarado GC, Parker D, Dyson HJ, Montminy MR, Wright PE. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- 16.Dwarki VJ, Montminy M, Verma IM. Both the basic region and the ‘leucine zipper’ domain of the cyclic AMP response element binding (CREB) protein are essential for transcriptional activation. EMBO J. 1990;9:225–232. doi: 10.1002/j.1460-2075.1990.tb08099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Comb M, Birnberg NC, Seasholtz A, Herbert E, Goodman HM. A cyclic AMP- and phorbol ester-inducible DNA element. Nature. 1986;323:353–356. doi: 10.1038/323353a0. [DOI] [PubMed] [Google Scholar]
- 18.Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- 19.Montminy MR, Sevarino KA, Wagner JA, Mandel G, Goodman RH. Identification of a cyclic-AMP-responsive element within the rat somatostatin gene. Proc Natl Acad Sci U S A. 1986;83:6682–6686. doi: 10.1073/pnas.83.18.6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chrivia JC, Kwok RP, Lamb N, Hagiwara M, Montminy MR, Goodman RH. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 21.Ferreri K, Gill G, Montminy M. The cAMP-regulated transcription factor CREB interacts with a component of the TFIID complex. Proc Natl Acad Sci U S A. 1994;91:1210–1213. doi: 10.1073/pnas.91.4.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jun DY, Kim MK, Kim IG, Kim YH. Characterization of the murine cyclin D2 gene: exon/intron organization and promoter activity. Mol Cells. 1997;7:537–543. [PubMed] [Google Scholar]
- 23.Reusch JE, Colton LA, Klemm DJ. CREB activation induces adipogenesis in 3T3-L1 cells. Mol Cell Biol. 2000;20:1008–1020. doi: 10.1128/mcb.20.3.1008-1020.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nguyen LQ, Kopp P, Martinson F, Stanfield K, Roth SI, Jameson JL. A dominant negative CREB (cAMP response element-binding protein) isoform inhibits thyrocyte growth, thyroid-specific gene expression, differentiation, and function. Mol Endocrinol. 2000;14:1448–1461. doi: 10.1210/mend.14.9.0516. [DOI] [PubMed] [Google Scholar]
- 25.Scobey M, Bertera S, Somers J, Watkins S, Zeleznik A, Walker W. Delivery of a cyclic adenosine 3′,5′-monophosphate response element-binding protein (creb) mutant to seminiferous tubules results in impaired spermatogenesis. Endocrinology. 2001;142:948–954. doi: 10.1210/endo.142.2.7948. [DOI] [PubMed] [Google Scholar]
- 26.Klemm DJ, Watson PA, Frid MG, Dempsey EC, Schaack J, Colton LA, Nesterova A, Stenmark KR, Reusch JE. cAMP response element-binding protein content is a molecular determinant of smooth muscle cell proliferation and migration. J Biol Chem. 2001;276:46132–46141. doi: 10.1074/jbc.M104769200. [DOI] [PubMed] [Google Scholar]
- 27.Sung JY, Shin SW, Ahn YS, Chung KC. Basic fibroblast growth factor-induced activation of novel CREB kinase during the differentiation of immortalized hippocampal cells. J Biol Chem. 2001;276:13858–13866. doi: 10.1074/jbc.M010610200. [DOI] [PubMed] [Google Scholar]
- 28.Shankar DB, Cheng JC, Kinjo K, Federman N, Moore TB, Gill A, Rao NP, Landaw EM, Sakamoto KM. The role of CREB as a proto-oncogene in hematopoiesis and in acute myeloid leukemia. Cancer Cell. 2005;7:351–362. doi: 10.1016/j.ccr.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 29.Gonzalez GA, Montminy MR. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989;59:675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- 30.Molina CA, Foulkes NS, Lalli E, Sassone-Corsi P. Inducibility and negative autoregulation of CREM: an alternative promoter directs the expression of ICER, an early response repressor. Cell. 1993;75:875–886. doi: 10.1016/0092-8674(93)90532-u. [DOI] [PubMed] [Google Scholar]
- 31.Meyer TE, Waeber G, Lin J, Beckmann W, Habener JF. The promoter of the gene encoding 3′,5′-cyclic adenosine monophosphate (cAMP) response element binding protein contains cAMP response elements: evidence for positive autoregulation of gene transcription. Endocrinology. 1993;132:770–780. doi: 10.1210/endo.132.2.8381074. [DOI] [PubMed] [Google Scholar]
- 32.Brecht S, Gass P, Anton F, Bravo R, Zimmermann M, Herdegen T. Induction of c-Jun and suppression of CREB transcription factor proteins in axotomized neurons of substantia nigra and covariation with tyrosine hydroxylase. Mol Cell Neurosci. 1994;5:431–441. doi: 10.1006/mcne.1994.1053. [DOI] [PubMed] [Google Scholar]
- 33.Nibuya M, Nestler EJ, Duman RS. Chronic antidepressant administration increases the expression of cAMP response element binding protein (CREB) in rat hippocampus. J Neurosci. 1996;16:2365–2372. doi: 10.1523/JNEUROSCI.16-07-02365.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Widnell KL, Russell DS, Nestler EJ. Regulation of expression of cAMP response element-binding protein in the locus coeruleus in vivo and in a locus coeruleus-like cell line in vitro. Proc Natl Acad Sci U S A. 1994;91:10947–10951. doi: 10.1073/pnas.91.23.10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Widnell KL, Self DW, Lane SB, Russell DS, Vaidya VA, Miserendino MJ, Rubin CS, Duman RS, Nestler EJ. Regulation of CREB expression: in vivo evidence for a functional role in morphine action in the nucleus accumbens. J Pharmacol Exp Ther. 1996;276:306–315. [PubMed] [Google Scholar]
- 36.Kolodziejski PJ, Musial A, Koo JS, Eissa NT. Ubiquitination of inducible nitric oxide synthase is required for its degradation. Proc Natl Acad Sci U S A. 2002;99:12315–12320. doi: 10.1073/pnas.192345199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walker WH, Fucci L, Habener JF. Expression of the gene encoding transcription factor cyclic adenosine 3′,5′-monophosphate (cAMP) response element-binding protein (CREB): regulation by follicle-stimulating hormone-induced cAMP signaling in primary rat Sertoli cells. Endocrinology. 1995;136:3534–3545. doi: 10.1210/endo.136.8.7628390. [DOI] [PubMed] [Google Scholar]
- 38.Song KS, Lee WJ, Chung KC, Koo JS, Yang EJ, Choi JY, Yoon JH. Interleukin-1 beta and tumor necrosis factor-alpha induce MUC5AC overexpression through a mechanism involving ERK/p38 mitogen-activated protein kinases-MSK1-CREB activation in human airway epithelial cells. J Biol Chem. 2003;278:23243–23250. doi: 10.1074/jbc.M300096200. [DOI] [PubMed] [Google Scholar]
- 39.Braunstein M, Sobel RE, Allis CD, Turner BM, Broach JR. Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol Cell Biol. 1996;16:4349–4356. doi: 10.1128/mcb.16.8.4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo RX, Postigo AA, Dean DC. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 41.Brown NS, Smart A, Sharma V, Brinkmeier ML, Greenlee L, Camper SA, Jensen DR, Eckel RH, Krezel W, Chambon P, Haugen BR. Thyroid hormone resistance and increased metabolic rate in the RXR-gamma-deficient mouse. J Clin Invest. 2000;106:73–79. doi: 10.1172/JCI9422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Durand B, Saunders M, Leroy P, Leid M, Chambon P. All-trans and 9-cis retinoic acid induction of CRABPII transcription is mediated by RAR-RXR heterodimers bound to DR1 and DR2 repeated motifs. Cell. 1992;71:73–85. doi: 10.1016/0092-8674(92)90267-g. [DOI] [PubMed] [Google Scholar]
- 43.Meyer TE, Habener JF. Cyclic adenosine 3′,5′-monophosphate response element binding protein (CREB) and related transcription-activating deoxyribonucleic acid-binding proteins. Endocr Rev. 1993;14:269–290. doi: 10.1210/edrv-14-3-269. [DOI] [PubMed] [Google Scholar]
- 44.Smale ST, Baltimore D. The “initiator” as a transcription control element. Cell. 1989;57:103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- 45.Shell SA, Fix C, Olejniczak D, Gram-Humphrey N, Walker WH. Regulation of cyclic adenosine 3′,5′-monophosphate response element binding protein (CREB) expression by Sp1 in the mammalian testis. Biol Reprod. 2002;66:659–666. doi: 10.1095/biolreprod66.3.659. [DOI] [PubMed] [Google Scholar]
- 46.Coven E, Ni Y, Widnell KL, Chen J, Walker WH, Habener JF, Nestler EJ. Cell type-specific regulation of CREB gene expression: mutational analysis of CREB promoter activity. J Neurochem. 1998;71:1865–1874. doi: 10.1046/j.1471-4159.1998.71051865.x. [DOI] [PubMed] [Google Scholar]
- 47.Suri C, Fung BP, Tischler AS, Chikaraishi DM. Catecholaminergic cell lines from the brain and adrenal glands of tyrosine hydroxylase-SV40 T antigen transgenic mice. J Neurosci. 1993;13:1280–1291. doi: 10.1523/JNEUROSCI.13-03-01280.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cole TJ, Copeland NG, Gilbert DJ, Jenkins NA, Schutz G, Ruppert S. The mouse CREB (cAMP responsive element binding protein) gene: structure, promoter analysis, and chromosomal localization. Genomics. 1992;13:974–982. doi: 10.1016/0888-7543(92)90010-p. [DOI] [PubMed] [Google Scholar]
- 49.Delfino FJ, Walker WH. NF-kappaB induces cAMP-response element-binding protein gene transcription in sertoli cells. J Biol Chem. 1999;274:35607–35613. doi: 10.1074/jbc.274.50.35607. [DOI] [PubMed] [Google Scholar]
- 50.Lin RJ, Nagy L, Inoue S, Shao W, Miller WH, Jr, Evans RM. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature. 1998;391:811–814. doi: 10.1038/35895. [DOI] [PubMed] [Google Scholar]
- 51.Malpel S, Mendelsohn C, Cardoso WV. Regulation of retinoic acid signaling during lung morphogenesis. Development. 2000;127:3057–3067. doi: 10.1242/dev.127.14.3057. [DOI] [PubMed] [Google Scholar]