Abstract
OBJECTIVES
Alpha-methylacyl-CoA racemase (AMACR) has been used as a diagnostic biomarker for prostate cancer (CaP) and is now a standard biomarker for needle biopsy specimens with ambiguous lesions. In this study, we evaluate the possibility of using AMACR variants to improve the specificity of CaP detection.
METHODS
We used in silico analysis and molecular cloning to discover new AMACR variants and quantitative RT-PCR to measure the transcript levels of AMACR and its variants in four prostate cell lines and 23 pairs of CaP and adjacent normal tissue.
RESULTS
We found four novel variants, IAs, IBL, IBLd, and IBLi. Transcript levels of the majority of AMACR variants were significantly upregulated in CaP as compared with its adjacent normal counterparts. A variants, the functional variants based on bioinformatic analysis, showed levels of transcript expression in CaP in this order: IA≫IAd=IIA≫IIAs>IAs. In contrast, the expression of the B variants, which appear to be nonfunctional due to the absence of exon 3, was lower than that of the A variants. IB was the most abundant form of B variant; and expression of IIB was negligible. More important, the difference between levels of variant IA, IAd, IIA, IIAs, IB, and IBLi in CaP and normal tissue was significantly higher than the difference in levels of total AMACR.
CONCLUSIONS
Our data suggest that AMACR variants have better power than total AMACR in discriminating between CaP and adjacent normal tissue. These findings may be useful for the development of future diagnostic assays.
Keywords: spliced variant, isoforms, cloning, prostate tissue, prostate cancer, quantitative RT-PCR
The high mortality of prostate cancer (CaP) in men and the low specificity of the serum prostate-specific antigen (PSA) assay has generated great interest in finding new biomarkers that could specifically detect CaP at an earlier stage.1 P504S, or alpha-methylacyl-CoA racemase (AMACR), was first discovered in 2000 as a specific marker for CaP.2 Jiang et al. soon validated the aberrant expression of AMACR transcripts in CaP and used the transcripts for detecting CaP antigen in needle biopsy specimens.3,4 Immunohistochemical staining with AMACR antibody alone or in combination with antibody to a basal cell marker is currently a standard adjuvant tool in CaP diagnosis, especially in needle biopsy specimens with ambiguous lesions.5–8 Overexpression of AMACR has also been reported in other human tumors, including those cancers originating from kidneys, liver, lymph nodes, and colon.9,10
AMACR catalyzes the conversion of phytanic and pristanic acid that is essential in the beta-oxidation of branched-chain fatty acids and bile acid intermediates.11 Six different variants of AMACR have been reported as IA, IAd, IIA, IIAs, IB, and IIB.12–14 These transcript variants are considered to be alternatively spliced from a single gene locus located in chromosome 5.15 These alternatively spliced variants of AMACR are grouped as type I and II according to the presence or absence, respectively, of intact exon 5 and are subclassified as A or B, on the basis of the existence or absence, respectively, of exon 3. IA, the most abundant variant, contains all five intact exons and codes for a protein of 382 amino acids. Overexpression of this enzyme may allow CaP cells to switch energy sources, thereby facilitating cancer progression.
AMACR has been studied as a potential early cancer diagnostic biomarker through determining its protein level in western blot assays, its transcript level in urine with quantitative RT-PCR, and the DNA methylation status of promoter in cancer tissue and blood.16–18 We reported that a duplex assay of total AMACR (AMACRt) and prostate cancer antigen 3 (PCA3) transcripts from urine sediment could significantly increase the specificity of CaP detection.19 A small-scale study noted differential expression of A and B forms of AMACR transcripts in CaP tissue.13,14 Here, we report four novel AMACR alternatively spliced variants and evaluate the expression of AMACRt transcript and its ten individual variants in four prostate cell lines and 23 normal-cancer paired prostate tissues.
MATERIALS AND METHODS
Prostate tissue sample collection and cell lines
We collected 23 pairs of human CaP and matched non-cancerous prostate tissues from the same patient from University of Cincinnati Medical Center and Indiana University Medical Center. These tissues were obtained by radical surgical prostatectomy and verified by urologist (BB) and pathologist (LC) according to the protocol approved by the Institutional Review Board and guidelines at the University of Cincinnati and Indiana University. The characteristics of specimens collected from 23 patients are shown in Table S1. Tissue samples were collected using biopsy punch on surgically removed prostate, and 1mm thick section of the top of the punched sample was discarded and a second 1mm thick section near the top of the larger remaining sample was formalin fixed and hematoxylin & eosin (H&E) stained for determining the percentage of tumor. The rest of the tissues were snap-frozen and stored at −80C prior to RNA extraction. Tumor samples were defined as the tissue with at least 90% tumor based on H&E staining. Human prostate cell lines LNCaP, DU145, and PC-3 were purchased from ATCC (Manassas, VA) and maintained according to the provider’s recommendations. Immortalized normal human prostate epithelial cell line NPrEC was established in our laboratory and maintained as previously described.19
Primer design and bioinformatics
Primers were designed with the aid of Primer3 and compared against each sequence of AMACR variant to ensure that each set of primers amplify only one specific variant (see Table S2). Multiple sequence alignment was carried out using Clustalw from the European Bioinformatics Institute (EBI, http://www.ebi.ac.uk/clustalw). Prediction of the open reading frame of each transcript was carried out with ORF Finder in NCBI (http://www.ncbi.nlm.nih.gov/gorf/gorf.html).
RNA isolation, cDNA synthesis, and quantification of transcripts
RNA isolation, cDNA synthesis, and quantification of transcripts were carried out as previously reported.20 Plasmids containing a variant-specific sequence were applied to real-time PCR reactions for the generation of standard curves for determination of the molecular numbers of transcripts. Expression ratios of AMACR variants in tumor versus non-tumor tissue were calculated and compared.
Molecular modeling
The ProModII algorithm in SWISS-MODEL (http://swissmodel.expasy.org/) was used to generate three-dimensional structures of AMACR isoforms. The crystal structure of AMACR isolated from Mycobacterium tuberculosis (PDB ID: 1X74A) was applied to generate a molecular model for the AMACR IA isoform.21 The molecular model was evaluated with PyMOL v0.98 (http://www.delanoscientific.com).
Statistical analysis
All statistical analyses were performed with statistical software SAS 9.1.3 (SAS Institute) and R€ (http://www.r-project.org). Student’s t-test was used to compare the expression levels of cancer and non-cancer tissues for each variant. A P-value of <0.05 was considered statistically significant. A scoring system ranging from 1 to 9 (low to high) was used to assign to each sample pair according to the expression ratio for individual variant. The total scores from sample pairs for each variant were calculated, and the ability of the variants to distinguish cancer from non-cancer tissues was evaluated by a binomial test.
RESULTS
Identification and cloning of novel alternatively spliced variants of AMACR
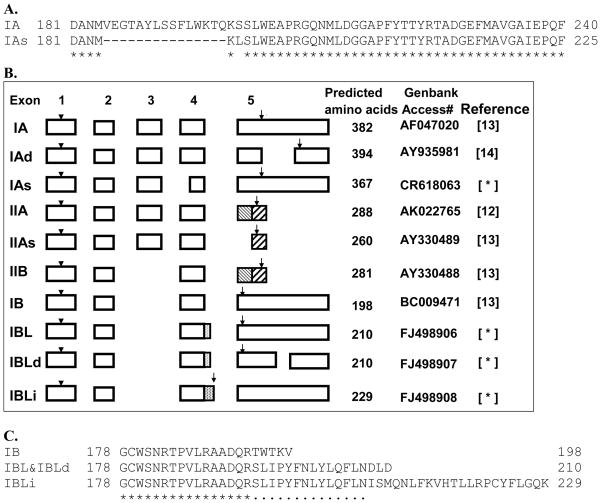
We identified and cloned four novel alternatively spliced AMACR variants in this study. Using the AMACR IA cDNA sequence to perform a nucleotide homolog search via NCBI BLAST, we found a full-length high-throughput cDNA clone isolated from fetal liver that was highly homologous with the IA sequence from the GenBank database but distinct from any known AMACR variant. Sequence alignment with IA revealed that 45 in-frame nucleotides at the 5′ end of exon 4 were missing, resulting in the encoding of a protein comprising 367 amino acid residues (Figure 1A). This variant is now designated as IAs (Genbank accession number: CR618063), a short form of IA. One novel coding SNP (G/A) is found in exon 5 of the AMACR IA mRNA sequence, resulting in a change of lysine to glutamic acid at amino acid residue 277 in the C-terminal tail of the protein. The functional consequence of this change is currently unknown, and the SNP is not yet available in the NCBI SNP database (dbSNP). Figure 1B summarizes the exon utilization of all known and novel AMACR variants, including IAs.
Figure 1.
Cloning of three AMACR novel variants. (A) Alignment of the partial predicted amino acid sequences of IA and IAs using CLUSTALW. (B) Block diagrams showing the exon and structure relationship of four variants identified in this study and six known AMACR variants. Detailed transcript sequence information can be obtained from GenBank by the corresponding accession number. Arrowheads indicate a start codon. Arrows indicate a stop codon. Number in parentheses represents the reference number. Novel AMACR variants are marked with an asterisk. (C) Alignment of the partial predicted amino acid sequences at C-termini of three novel variants and IB using CLUSTALW.
We then amplified each variant using specific primer. However, we amplified more than one PCR product when we used the IB variant–specific primer. Sequence analyses of those PCR products revealed three novel cDNA clones with a high homology to IB. We called them IBL, IBLd, and IBLi according to the different features of the sequence (see Figure 1B and 1C and Genbank database with accession number: IBL, FJ498906; IBLd, FJ498907; IBLi, FJ498908). IBL uses a cryptic splice signal in intron 4, gaining 43 bp for exon 4, leading to the formation of a premature stop codon in exon 5 and resulting in a unique amino acid sequence at the C-terminus (Figure 1C). IBLd has the same coding sequence as IBL, but only 749 bp of nucleotide sequences are lost at the 3′ untranslated region of the transcript. Hence, the protein products of IBLd and IBL are the same (Figure 1C). Like IBL, IBLi gains an extra 173 bp from intron 4 because of the existence of another cryptic splice signal. This extra nucleotide sequence contains a stop codon at this extended exon 4, resulting in an alternate 36 amino acid sequence at the C-terminal tail (Figure 1C). We designed more primer pairs flanking the newly discovered sequences with which specific PCR products were successfully amplified from LNCaP cell cDNA, indicating that the three newly cloned AMACR variants are present in this prostate cell line (Figure 3).
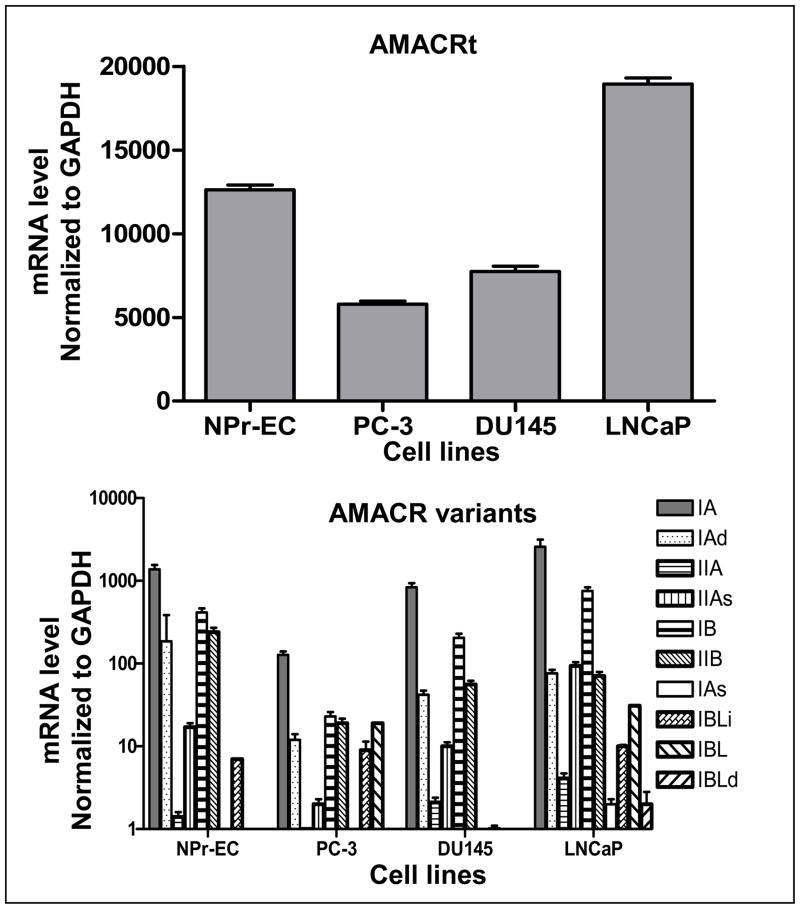
Figure 3.
Expression level of AMACRt (upper panel) and its ten variants (lower panel) in prostate cell lines. Levels of transcript expression of an immortalized normal prostate epithelial cell line (NPrEC) and three prostate cancer cell lines were measured by quantitative real-time PCR. See Materials and Methods for details.
Result of molecular modeling
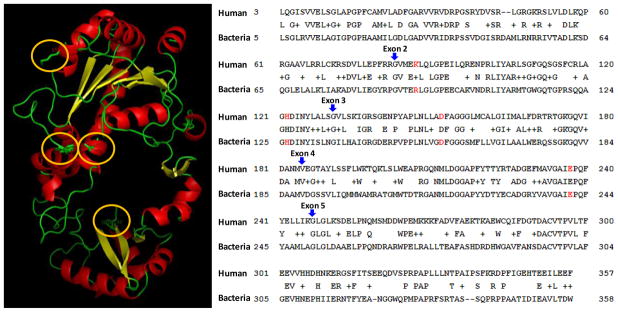
To gain insight onto the functional role of each variant in CaP, we investigated the structure-and-function information for human AMACR. Because this information is not available, we generated, using SWISS-Model, a molecular model for human AMACR IA from a high-resolution crystal of AMACR isolated from Mycobacterium tuberculosis, protein databank ID 1×74 (Figure 2, left panel).21 The structure of human AMACR IA is very similar to that of bacterial AMACR; It forms two domains, a large domain (upper half of the molecule) and a small domain (lower half of the molecule) connected by two linker motifs. Four amino acid residues identified as the major functional sites for AMACR in the bacterial22 were found to be highly conserved in human AMACR IA as lysine 87, histidine 122, aspartic acid 152, and glutamic acid 237 (Figure 2, left panel in orange ovals). The primary sequence positions of these sites are shown in Figure 2 (right panel, highlighted in red). Two sites were located in exon 2, while exon 3 and 4 each has one functionally important amino acid residue, suggesting that all B variants may be nonfunctional because they lack exon 3.
Figure 2.
Molecular model of human AMACR IA isoforms. Similar to bacterial AMACR data published previously, human AMACR (left panel) consists of two major domains, an N-terminal large domain (upper half) and a small domain (lower half). Four amino acid residues of human AMACR IA, lysine 87, histidine 122, aspartic acid 152, and glutamic acid 237, which are highly conserved in bacterial AMACR, are highlighted by orange ovals. The right panel shows the Blast2 result between human AMACR IA protein sequence and the AMACR protein sequence isolated from Mycobacterium tuberculosis. Four highly conserved amino acid residues are highlighted in red. The beginning of each exon is indicated by blue arrow.
Expression of AMACR variant mRNA in human cell lines
Transcripts of IAs and IBLd were detected at very low levels in LNCaP and were almost undetectable in PC3, DU145, and NPrEC. Various levels of mRNA of AMACRt and eight other variants were expressed in four prostate malignant and normal cell lines (Figure 3). IA is the most abundant variant in all prostate cell lines. The expression pattern of AMACRt and the six variants appeared to be very similar in four cell lines. LNCaP presented higher levels of AMACRt and its variants than did the three other cell lines we tested in this study.
Expression of AMACR variant mRNA in paired human prostate tumor tissue and non-tumor tissues
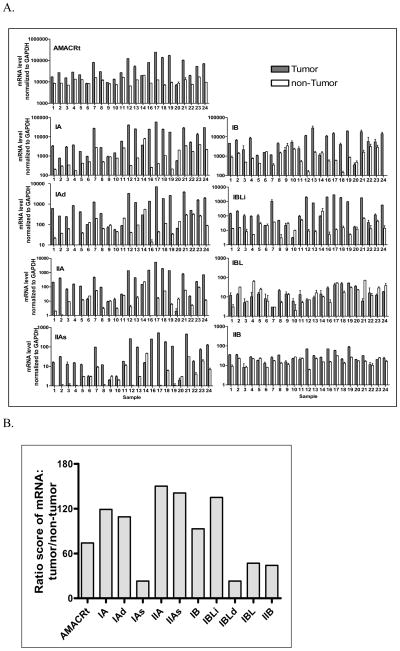
Except for IAs and IBLd, AMACRt and eight other variants were readily detectable in 23 pairs of prostate tumor and non-tumor tissues (Figure 4A). Among all the variants, IA and IB were the most abundant in non-tumor prostate tissues and IAs and IBLd were the least abundant, being almost undetectable in all prostate samples (data not shown). The relative abundance of variant transcripts in non-cancer tissues, in descending order, was IA≥IB>IAd>IIA=IBLi>IBL=IIB>IIAs>IAs=IBLd. However, transcripts of AMACRt and six variants (IA, IAd, IIA, IIAs, IB, IBLi) were significantly upregulated in most of the tumor samples (17–20 out of 23 cases, Figure 4A). Among all the AMACR variants tested in tumor samples, IA was the most abundant variant, followed by IB. These two variants account for most of AMACR transcripts in prostate tumor tissues. Expression of IBL and IIB were either marginally elevated with no statistical significance or no change in most tumor tissues. IAs and IBLd mRNA were only barely detectable in a few tumor samples and undetectable in most tumor tissues. The expression of AMACRt and a particular variant in CaP had no obvious correlation with tumor Gleason scores, stages and serum PSA level. The relative abundance of variant transcripts in tumor tissues (IA>IB>IAd>IIA=IBLi>IIAs>IIB=IBL>IAs=IBLd) was somewhat different from the expression pattern in non-tumor tissues.
Figure 4.
Expression of transcript of AMACRt and its eight variants in paired prostate cancer and non-cancer tissues. (A) Expression level of AMACR variants isolated from 23 pair of prostate cancer and non-cancer (from same prostate) tissues were measured by qRT-PCR. Expression of IAs transcript was undetectable or barely detectable in prostate tissues; therefore, data are not shown in this figure. Asterisk indicates that the level of mRNA expression in the tumor is not statistically different from that in non-tumor tissue or is lower than that in non-tumor tissues. (B). Differential expression of AMACR variants in prostate tumor tissues. The AMACR variant that was most strongly related to cancer was determined by calculating and scoring the ratio of relative transcript numbers of each variant in 23 paired prostate tumor and non-tumor tissues. A higher score indicated that the expression of the AMACR variant was highly specific in cancer.
Differential expression of AMACR transcripts in prostate tumor tissues
To determine which of the AMACR variants was most strongly associated with cancer, we calculated and scored the ratio of relative transcript numbers of each variant in 23 paired prostate tumor and non-tumor tissues. We found that IIA mRNA was the variant that was most upregulated in tumor tissue, followed by IIAs, IBLi, IA, IAd, and IB (Figure 4B). Importantly, IIA was about 2 times more capable than AMACRt to distinguish between CaP and non-CaP. Measuring other variants, including IA, IBLi, IAd, IIAs and IB, was also at least 1 to 2 times better than simply measuring total AMACR level in detecting CaP. Our results suggest that by using variant-specific primers, we will be able to detect minute changes in AMACR level, offering a more sensitive way to detect CaP.
COMMENT
We reported four novel AMACR variants and investigated the expression profile of AMACR and its ten variants in four prostate cell lines and 23 paired prostate tumor and matched non-tumor counterparts. The new variant IAs differs from variant IA only by being 15 amino acid residues shorter. IAs also contains a polypeptide-KASL at the C-terminal end, which is thought to function as type 1 carboxyl-terminal peroxisomal targeting signal (PTS1). Thus, like IA, IAs presumably may reside in both peroxisome and mitochondria. Although the amino acid composition of IA and IAs are highly homologous (94.7%) and have a similar acidic pI (6.1) and subcellular localization (peroxisome and mitochondria), their abundance in prostate is completely different. IA seems to be the main form of AMACR, whereas IAs is barely detectable, suggesting that this unconventional splicing event in prostate is rare. The other three novel variants, IBL, IBLi, and IBLd, were cloned and found to be almost completely homologous to IB in protein sequence except for a difference in 5 to 36 amino acids in the C-terminal tail.
It is unclear if these novel variants function differently from the wild type. A recent study of AMACR isolated from M. tuberculosis identified four amino acid residues critical for AMACR function.21 Using bioinformatic analyses, we also found these sites highly conserved in the human AMACR IA isoform. The lysine 87 in exon 2 is speculated to be crucial for substrate (acyl CoA) binding whereas the other amino acid residues, histidine 122 in exon 2, aspartic acid 152 in exon 3 and glutamic acid 237 in exon 4, are vital in defining the catalytic domain of human AMACR. Theoretically, all variants containing intact exons 1 to 4 should be functional. The function of exon 5 remains unknown. Therefore, variants IA, IAd, IIA IIAs, and even IAs with shortened exon 4 (A variant) may be fully capable of functioning as a racemase. On the other hand, those variants that lack exons 3 (B variant) and 4, may not be functional in a cellular environment. However, cancer cells may take advantage of this to gain function to increase their chance of survival.23. There are now ten known variants of AMACR, including five A and five B forms (seven I types and three II types).
Double staining with AMACR antibody and a basal cell marker-specific antibody is now widely used to help in diagnosis of CaP from prostate needle biopsy specimens that are considered to be “atypical small acinar proliferations suspicious for, but not diagnostic of malignancy,” based on H&E criteria.24 Application of AMACR to a urine assay as a complement to a serum PSA assay has been studied in our20,25 and other laboratories.16,17 Existence of multiple alternatively spliced variants in CaP prompted us to investigate whether those variants play an important role in the development of CaP and could be used to improve the non-invasive assay for CaP detection. We first performed a study of the expression of ten individual AMACR variants at the mRNA level in four prostate cell lines and 23 pairs of tumor and non-tumor prostate tissues. We found that AMACR and its five variants (IA, IAd, IIA, IIAs, and IB) shared similar mRNA expression patterns in all cell lines and paired prostate tissues (Figures 3 and 4). Levels of mRNA of variants among prostate tissues showed greater variability, with some tumor samples displaying more than a 100-fold increase over matched a non-tumor tissue control and other variants showing much less variation between tumor and non-tumor tissues. These differences may reflect the heterogeneity of CaP development and differentiations in prostate carcinomas.12–14 Four pairs of samples showed no difference of the mRNA level between tumor and non-tumor prostate tissues for AMACRt and most variants, perhaps caused by (1) an insufficient amount of tumor present in the samples; (2) a discrepancy between pathological analysis and biochemical analysis: “normal tissue cells” may have already gone through biochemical changes that precede morphologic evidence of neoplastic transformation; (3) contamination of “normal tissues” mixed with a small amount of high grade prostate intraepithelial neoplasia (HGPIN) or cancerous cells. In daily clinic practice, CaP on needle biopsies showed negative racemase immunostaining at about 18% and also, benign glands can occasionally be stained positively for AMACR with majority being weakly positive, which is consistent with the data found in our study.26,27
Because HGPIN was frequently immunostained with AMACRt, it will be helpful to determine if any variant(s) can distinguish between CaP and HGPIN by antibodies to individual variants or qPCR coupled with laser capture microdissection. By comparing the ratio of mRNA expression between tumor and matched non-tumor tissues, we found consistently high levels of expression of the IIA variant in prostate tumors, indicating that IIA may be the best variant for differentiating CaP from non-tumor tissues (Figure 4B). However, it may not be feasible to measure the IIA variant transcript in CaP cells collected from body fluid such as urine because of the limited availability of CaP cells. With the improvement of the sample collection protocol and the development of a sensitive assay for mRNA measurement, IIA may be a better molecular marker than AMACRt for a non-invasive CaP detection assay. Alternatively, measurement of IA or of multiple variants with high ratio scores could further improve the sensitivity of the assay.
Mubiru et al. reported different subcellular locations presents in A and B variants by immunostaining of CaP tissues.13,14 Our data suggest that two variants (IA and IAs) may be found in both peroxisome and mitochondria whereas the other eight variants may only be present in mitochondria. Kunju et al. described that both commercial AMACR monoclonal and polyclonal antibodies showed moderate to strong staining in CaP (90% sensitivity) and weak focal and non-circumferential staining in morphologically benign glands.28 Therefore, development of novel antibodies to AMACR variants with high ratios of mRNA level (tumor/non-tumor) and the combination of several of these variants to form a cocktail kit may dramatically increase sensitivity and reduce background in the immunostaining of prostate needle biopsies for CaP detection.
CONCLUSIONS
AMACR variants show more differentiation than AMACRt between the levels of expression in normal and cancerous prostate tissue, perhaps improving the specificity of the AMACR-based CaP detection assay and antibody development.
Supplementary Material
Acknowledgments
We are grateful to University of Cincinnati Medical Center and Indiana University Medical Center for providing prostate tumor tissues; Barbara Burke and Barbara Bracken for assistance in patient information enlistment and sample collection, Winnie Tang and Xiang Zhang for technique assistance, and Chensi Ouyang for critical reading of the manuscript. Research was supported in part by an award from the Department of Defense Prostate Cancer Research Program #PC051110 (SMH) and a NIH grant ES006096 (SMH)
Footnotes
Financial disclosure
All authors have no direct or indirect commercial financial incentive associated with publishing the article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Etzioni R. Statistical issues in the evaluation of screening and early detection modalities. Urol Oncol. 2008;26:308–315. doi: 10.1016/j.urolonc.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 2.Xu J, Stolk JA, Zhang X, et al. Identification of differentially expressed genes in human prostate cancer using subtraction and microarray. Cancer Res. 2000;60:1677–1682. [PubMed] [Google Scholar]
- 3.Jiang Z, Woda BA, Rock KL, et al. P504S: a new molecular marker for the detection of prostate carcinoma. Am J Surg Pathol. 2001;25:1397–1404. doi: 10.1097/00000478-200111000-00007. [DOI] [PubMed] [Google Scholar]
- 4.Luo J, Zha S, Gage WR, et al. Alpha-methylacyl-CoA racemase: a new molecular marker for prostate cancer. Cancer Res. 2002;62:2220–2226. [PubMed] [Google Scholar]
- 5.Molinie V, Fromont G, Sibony M, et al. Diagnostic utility of a p63/alpha-methyl-CoA-racemase (p504s) cocktail in atypical foci in the prostate. Mod Pathol. 2004;17:1180–1190. doi: 10.1038/modpathol.3800197. [DOI] [PubMed] [Google Scholar]
- 6.Trpkov K, Bartczak-McKay J, Yilmaz A. Usefulness of cytokeratin 5/6 and AMACR applied as double sequential immunostains for diagnostic assessment of problematic prostate specimens. Am J Clin Pathol. 2009;132:211–220. doi: 10.1309/AJCPGFJP83IXZEUR. [DOI] [PubMed] [Google Scholar]
- 7.Herawi M, Epstein JI. Immunohistochemical antibody cocktail staining (p63/HMWCK/AMACR) of ductal adenocarcinoma and Gleason pattern 4 cribriform and noncribriform acinar adenocarcinomas of the prostate. Am J Surg Pathol. 2007;31:889–894. doi: 10.1097/01.pas.0000213447.16526.7f. [DOI] [PubMed] [Google Scholar]
- 8.Wang W, Sun X, Epstein JI. Partial atrophy on prostate needle biopsy cores: a morphologic and immunohistochemical study. Am J Surg Pathol. 2008;32:851–857. doi: 10.1097/PAS.0b013e31815a0508. [DOI] [PubMed] [Google Scholar]
- 9.Jiang Z, Fanger GR, Woda BA, et al. Expression of alpha-methylacyl-CoA racemase (P504s) in various malignant neoplasms and normal tissues: astudy of 761 cases. Hum Pathol. 2003;34:792–796. doi: 10.1016/s0046-8177(03)00268-5. [DOI] [PubMed] [Google Scholar]
- 10.Shilo K, Dracheva T, Mani H, et al. Alpha-methylacyl CoA racemase in pulmonary adenocarcinoma, squamous cell carcinoma, and neuroendocrine tumors: expression and survival analysis. Arch Pathol Lab Med. 2007;131:1555–1560. doi: 10.5858/2007-131-1555-MCRIPA. [DOI] [PubMed] [Google Scholar]
- 11.Ferdinandusse S, Denis S, IJlst L, et al. Subcellular localization and physiological role of alpha-methylacyl-CoA racemase. J Lipid Res. 2000;41:1890–1896. [PubMed] [Google Scholar]
- 12.Shen-Ong GL, Feng Y, Troyer DA. Expression profiling identifies a novel alpha-methylacyl-CoA racemase exon with fumarate hydratase homology. Cancer Res. 2003;63:3296–3301. [PubMed] [Google Scholar]
- 13.Mubiru JN, Shen-Ong GL, Valente AJ, et al. Alternative spliced variants of the alpha-methylacyl-CoA racemase gene and their expression in prostate cancer. Gene. 2004;327:89–98. doi: 10.1016/j.gene.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 14.Mubiru JN, Valente AJ, Troyer DA. A variant of the alpha-methyl-acyl-CoA racemase gene created by a deletion in exon 5 and its expression in prostate cancer. Prostate. 2005;65:117–123. doi: 10.1002/pros.20277. [DOI] [PubMed] [Google Scholar]
- 15.Rubin MA, Zhou M, Dhanasekaran SM, et al. alpha-Methylacyl coenzyme A racemase as a tissue biomarker for prostate cancer. JAMA. 2002;287:1662–1670. doi: 10.1001/jama.287.13.1662. [DOI] [PubMed] [Google Scholar]
- 16.Zehentner BK, Secrist H, Zhang X, et al. Detection of alpha-methylacyl-coenzyme-A racemase transcripts in blood and urine samples of prostate cancer patients. Mol Diagn Ther. 2006;10:397–403. doi: 10.1007/BF03256217. [DOI] [PubMed] [Google Scholar]
- 17.Rogers CG, Yan G, Zha S, et al. Prostate cancer detection on urinalysis for alpha methylacyl coenzyme a racemase protein. J Urol. 2004;172:1501–1503. doi: 10.1097/01.ju.0000137659.53129.14. [DOI] [PubMed] [Google Scholar]
- 18.Zhang X, Leav I, Revelo MP, et al. Deletion hotspots in AMACR promoter CpG island are cis-regulatory elements controlling the gene expression in the colon. PLoS Genet. 2009;5:e1000334. doi: 10.1371/journal.pgen.1000334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mobley JA, Leav I, Zielie P, et al. Branched fatty acids in dairy and beef products markedly enhance alpha-methylacyl-CoA racemase expression in prostate cancer cells in vitro. Cancer Epidemiol Biomarkers Prev. 2003;12:775–783. [PubMed] [Google Scholar]
- 20.Ouyang B, Bracken B, Burke B, et al. A duplex quantitative polymerase chain reaction assay based on quantification of alpha-methylacyl-CoA racemase transcripts and prostate cancer antigen 3 in urine sediments improved diagnostic accuracy for prostate cancer. J Urol. 2009;181:2508–2513. doi: 10.1016/j.juro.2009.01.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhaumik P, Kursula P, Ratas V, et al. Crystallization and preliminary X-ray diffraction studies of an alpha-methylacyl-CoA racemase from Mycobacterium tuberculosis. Acta Crystallogr D Biol Crystallogr. 2003;59:353–355. doi: 10.1107/s0907444902020735. [DOI] [PubMed] [Google Scholar]
- 22.Savolainen K, Bhaumik P, Schmitz W, et al. Alpha-methylacyl-CoA racemase from mycobacterium tuberculosis: Mutational and structural characterization of the active site and the fold. J Biol Chem. 2005;280:12611–12620. doi: 10.1074/jbc.M409704200. [DOI] [PubMed] [Google Scholar]
- 23.Leung YK, Lau KM, Mobley J, et al. Overexpression of cytochrome P450 1A1 and its novel spliced variant in ovarian cancer cells: alternative subcellular enzyme compartmentation may contribute to carcinogenesis. Cancer Res. 2005;65:3726–3734. doi: 10.1158/0008-5472.CAN-04-3771. [DOI] [PubMed] [Google Scholar]
- 24.Iczkowski KA. Current prostate biopsy interpretation: criteria for cancer, atypical small acinar proliferation, high-grade prostatic intraepithelial neoplasia, and use of immunostains. Arch Pathol Lab Med. 2006;130:835–843. doi: 10.5858/2006-130-835-CPBICF. [DOI] [PubMed] [Google Scholar]
- 25.Zielie PJ, Mobley JA, Ebb RG, et al. A novel diagnostic test for prostate cancer emerges from the determination of alpha-methylacyl-coenzyme a racemase in prostatic secretions. J Urol. 2004;172:1130–1133. doi: 10.1097/01.ju.0000133560.87118.4d. [DOI] [PubMed] [Google Scholar]
- 26.Evans AJ. Alpha-methylacyl CoA racemase (P504S): overview and potential uses in diagnostic pathology as applied to prostate needle biopsies. J Clin Pathol. 2003;56:892–897. doi: 10.1136/jcp.56.12.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou M, Aydin H, Kanane H, et al. How often does alpha-methylacyl-CoA-racemase contribute to resolving an atypical diagnosis on prostate needle biopsy beyond that provided by basal cell markers? Am J Surg Pathol. 2004;28:239–243. doi: 10.1097/00000478-200402000-00012. [DOI] [PubMed] [Google Scholar]
- 28.Kunju LP, Chinnaiyan AM, Shah RB. Comparison of monoclonal antibody (P504S) and polyclonal antibody to alpha methylacyl-CoA racemase (AMACR) in the work-up of prostate cancer. Histopathology. 2005;47:587–596. doi: 10.1111/j.1365-2559.2005.02281.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.