Abstract
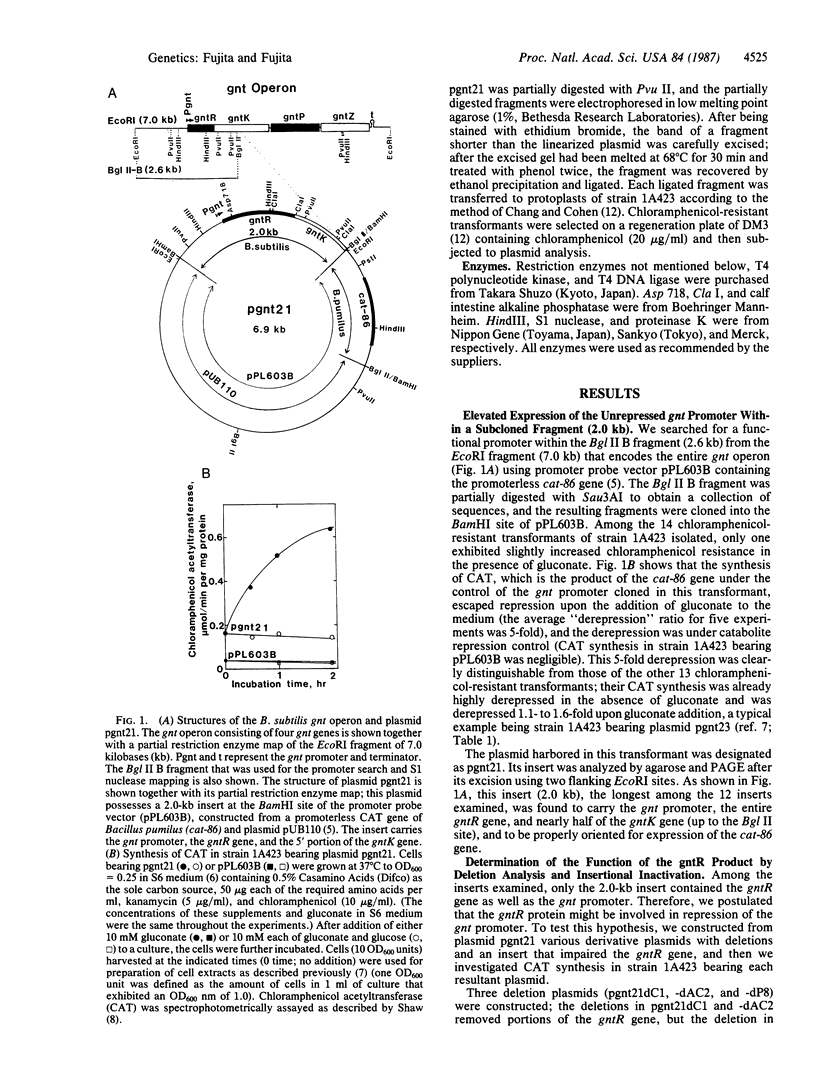
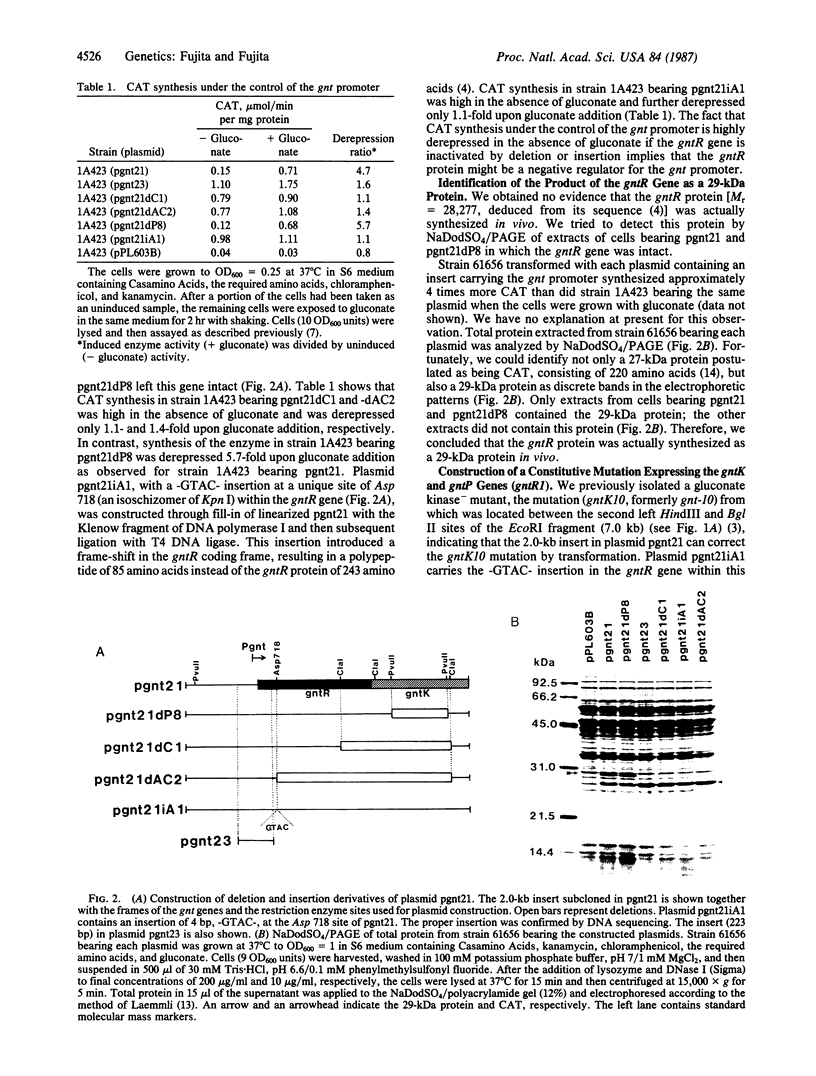
The gluconate (gnt) operon of Bacillus subtilis consists of four gnt genes; the second and third genes code for gluconate kinase (gluconokinase, EC 2.7.1.12) and gluconate permease, respectively. A fragment carrying the promoter of this operon (gnt promoter) and the first gene (gntR) was subcloned into a promoter probe vector (pPL603B). Repression of the expression of cat-86 gene, encoded in the vector portion of a constructed plasmid (pgnt21), that is under the control of the gnt promoter was removed by gluconate. The results of deletion analysis and of insertional inactivation of the gntR gene cloned in pgnt21 suggested that the product of the gntR gene, actually synthesized as a 29-kDa protein in vivo, is involved in repression of the gnt promoter. A 4-base-pair insertional mutation within the gntR gene constructed in vitro was introduced into the B. subtilis chromosomal gnt operon by use of linkage of the 4 base pairs to gntK10 in transformation. The introduced mutation gntR1 caused the constitutive expression of the gluconate kinase and gluconate permease genes. S1 nuclease analysis indicated that the mRNA of this operon is synthesized in the gntR1 strain and amounts of mRNA are not changed very much by gluconate, which acts as an inducer in the wild-type gene. These results strongly indicate that the gntR gene codes for a transcriptional negative regulator for the gnt operon.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Chang S., Cohen S. N. High frequency transformation of Bacillus subtilis protoplasts by plasmid DNA. Mol Gen Genet. 1979 Jan 5;168(1):111–115. doi: 10.1007/BF00267940. [DOI] [PubMed] [Google Scholar]
- Fujita Y., Freese E. Isolation and properties of a Bacillus subtilis mutant unable to produce fructose-bisphosphatase. J Bacteriol. 1981 Feb;145(2):760–767. doi: 10.1128/jb.145.2.760-767.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Fujita T. Genetic analysis of a pleiotropic deletion mutation (delta igf) in Bacillus subtilis. J Bacteriol. 1983 May;154(2):864–869. doi: 10.1128/jb.154.2.864-869.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Fujita T. Identification and nucleotide sequence of the promoter region of the Bacillus subtilis gluconate operon. Nucleic Acids Res. 1986 Feb 11;14(3):1237–1252. doi: 10.1093/nar/14.3.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Fujita T., Miwa Y., Nihashi J., Aratani Y. Organization and transcription of the gluconate operon, gnt, of Bacillus subtilis. J Biol Chem. 1986 Oct 15;261(29):13744–13753. [PubMed] [Google Scholar]
- Fujita Y., Nihashi J., Fujita T. The characterization and cloning of a gluconate (gnt) operon of Bacillus subtilis. J Gen Microbiol. 1986 Jan;132(1):161–169. doi: 10.1099/00221287-132-1-161. [DOI] [PubMed] [Google Scholar]
- Gryczan T. J., Contente S., Dubnau D. Characterization of Staphylococcus aureus plasmids introduced by transformation into Bacillus subtilis. J Bacteriol. 1978 Apr;134(1):318–329. doi: 10.1128/jb.134.1.318-329.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood C. R., Williams D. M., Lovett P. S. Nucleotide sequence of a Bacillus pumilus gene specifying chloramphenicol acetyltransferase. Gene. 1983 Oct;24(2-3):163–169. doi: 10.1016/0378-1119(83)90076-8. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Maloy S. R., Roth J. R. Regulation of proline utilization in Salmonella typhimurium: characterization of put::Mu d(Ap, lac) operon fusions. J Bacteriol. 1983 May;154(2):561–568. doi: 10.1128/jb.154.2.561-568.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nihashi J., Fujita Y. Catabolite repression of inositol dehydrogenase and gluconate kinase syntheses in Bacillus subtilis. Biochim Biophys Acta. 1984 Mar 22;798(1):88–95. doi: 10.1016/0304-4165(84)90014-x. [DOI] [PubMed] [Google Scholar]
- Shaw W. V. Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol. 1975;43:737–755. doi: 10.1016/0076-6879(75)43141-x. [DOI] [PubMed] [Google Scholar]
- Shibata T., Saito H. Repair of ultraviolet-induced DNA damage in the subcellular systems of Bacillus subtilis. Mutat Res. 1973 Nov;20(2):159–173. doi: 10.1016/0027-5107(73)90186-3. [DOI] [PubMed] [Google Scholar]
- Williams D. M., Duvall E. J., Lovett P. S. Cloning restriction fragments that promote expression of a gene in Bacillus subtilis. J Bacteriol. 1981 Jun;146(3):1162–1165. doi: 10.1128/jb.146.3.1162-1165.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]