This work examines the localization and functions of two protein disulfide isomerase family oxidoreductases in formation of protein storage bodies in rice endosperm, finding that the two have nonoverlapping localizations, activities, and biological functions.
Abstract
In the rice (Oryza sativa) endosperm, storage proteins are synthesized on the rough endoplasmic reticulum (ER), in which prolamins are sorted to protein bodies (PBs) called type-I PB (PB-I). Protein disulfide isomerase (PDI) family oxidoreductase PDIL2;3, an ortholog of human P5, contains a conserved structural disulfide in the redox-inactive thioredoxin-like (TRX) domain and was efficiently targeted to the surface of PB-I in a redox active site–dependent manner, whereas PDIL1;1, an ortholog of human PDI, was localized in the ER lumen. Complementation analyses using PDIL1;1 knockout esp2 mutant indicated that the a and a′ TRX domains of PDIL1;1 exhibited similar redox activities and that PDIL2;3 was unable to perform the PDIL1;1 functions. PDIL2;3 knockdown inhibited the accumulation of Cys-rich 10-kD prolamin (crP10) in the core of PB-I. Conversely, crP10 knockdown dispersed PDIL2;3 into the ER lumen. Glutathione S-transferase-PDIL2;3 formed a stable tetramer when it was expressed in Escherichia coli, and the recombinant PDIL2;3 tetramer facilitated α-globulin(C79F) mutant protein to form nonnative intermolecular disulfide bonds in vitro. These results indicate that PDIL2;3 and PDIL1;1 are not functionally redundant in sulfhydryl oxidations of structurally diverse storage proteins and play distinct roles in PB development. We discuss PDIL2;3-dependent and PDIL2;3-independent oxidation pathways that sustain disulfide bonds of crP10 in PB-I.
INTRODUCTION
During seed development, rice (Oryza sativa) endosperm cells synthesize large amounts of proteins, including three major groups of storage proteins with distinct solubility profiles (Shewry et al., 1995): acid- and alkaline-soluble glutelins (11S globulin homologs; 60% of the total proteins), alcohol-soluble prolamins (~20%), and saline-soluble α-globulin (~10%). These storage proteins are synthesized on the rough endoplasmic reticulum (ER) and are deposited into two distinct types of protein bodies (PBs): the ER-derived type-I PB (PB-I) and the protein storage vacuole type-II PB (PB-II) (Tanaka et al., 1980; Yamagata et al., 1982). Glutelin precursors (proglutelins) and α-globulin acquire intramolecular disulfide bonds and then exit the ER via the Golgi body to PB-II, in which proglutelins are processed by vacuolar processing enzyme (Wang et al., 2009b; Kumamaru et al., 2010) and form the crystalloid (crystal-like blocks in which lattice structures are often formed), and α-globulin is sequestered to the matrix surrounding the crystalloid (Krishnan et al., 1992; Kawagoe et al., 2005). PB-I contains prolamins, which are classified to Cys-rich (10-, 14-, and 16-kD) and Cys-poor (13-kD) prolamins (Ogawa et al., 1987). The Cys-rich 10-kD prolamin (crP10) and Cys-poor 13-kD prolamin (cpP13) are segregated to the central core and outer layer of PB-I, respectively (Kumamaru et al., 2007). Although these results have suggested that sulfhydryl oxidations of crP10 are involved in the development of PB-I, how crP10 is oxidized in the ER has remained obscure.
Protein disulfide isomerase (PDI), a ubiquitous sulfhydryl oxidoreductase found in all eukaryotic cells, is multifunctional enzyme, which catalyzes a wide range of thiol-disulfide exchange reactions, including oxidation, reduction, and isomerization, and also displays chaperone activity (Hatahet and Ruddock, 2009). The PDI family oxidoreductases contain both redox-active and redox-inactive thioredoxin (TRX)-like domains and show wide variation in the number and arrangement of the TRX-like domains (Ferrari and Söling, 1999). For example, yeast (Saccharomyces cerevisiae) Pdi1p is a U-shaped molecule in which two redox-active TRX-like domains (a and a′, each contains a redox-active Cys-X-X-Cys motif) and two noncatalytic TRX-like domains (b and b′) are arranged in the order abb′a′ (Tian et al., 2006). Human PDI and ERp57 also have the abb′a′ domain organization (Appenzeller-Herzog and Ellgaard, 2008). Another subfamily of PDI, including rice PDIL2;3 and human P5 (hereafter referred to as the P5 subfamily), contains two redox-active TRX-like domains in tandem followed by a single redox-inactive domain (aa′b) (Kikuchi et al., 2002). The genomes of rice, Arabidopsis thaliana, and maize (Zea mays) each encode at least seven members of the PDI family with two redox-active sites (see Supplemental Figure 1 online): PDIL1;1−1;4 (abb′a′) and PDIL2;1−2;3 (aa′b) (Houston et al., 2005; Lu and Christopher, 2008). The essential function and broad substrate specificity of the PDI family oxidoreductases complicate the identification of redundant and specific functions of each member of the PDI family in vivo. Yeast Pdi1p is essential for cell viability, but the functions of Pdi1p can be performed by other PDI family oxidoreductases, such as Mpd1p, when overexpressed (Tachikawa et al., 1995; Nørgaard et al., 2001). In human, extensive studies have elucidated some specific functions of the PDI family oxidoreductases in different cells. For example, PDI prevents neurotoxicity associated with ER stress and with protein misfolding in neurodegenerative diseases, including Parkinson’s disease (Uehara et al., 2006), whereas P5 enables shedding of tumor-associated NKG2D ligands by reducing disulfide bonds on the surface of tumor cells (Kaiser et al., 2007).
Specific functions of plant PDI family oxidoreductases have been postulated with accumulating evidence. In the maize endosperm, the level of PDIL1;1 (an ortholog of rice PDIL1;1; see Supplemental Figure 1 online) is upregulated in the floury-2 mutant, which contains an abnormally processed α-zein (Li and Larkins, 1996). In the soybean (Glycine max) cotyledon, P5 subfamily oxidoreductase PDIM associates noncovalently with proglycinin, 11S globulin-type storage protein (Wadahama et al., 2008). In Arabidopsis, PDI5 (PDIL1;1) regulates the timing of programmed cell death in endothelial cells (Andème Ondzighi et al., 2008), and PDIL2;1 plays a role in embryo sac development (Wang et al., 2008). In Arabidopsis liquid culture, the PDI family oxidoreductases respond differently to various types of stresses (Lu and Christopher, 2008). These results have suggested that some of the PDI family oxidoreductases play distinct role in specific cell types in plants. However, the redundancy or specificity for the PDI family oxidoreductases, when expressed in the same cell, has been difficult to characterize and has thus remained an open question. The rice endosperm provides an ideal system for studying specific functions of the PDI family oxidoreductases. A PDIL1;1 knockout mutant esp2 accumulates large protein aggregates containing proglutelins through intermolecular disulfide bonds in the ER (Takemoto et al., 2002; Onda et al., 2009; Satoh-Cruz et al., 2010). These studies leave little room for doubt that PDIL1;1 facilitates oxidative folding of proglutelins and that its activity is indispensable when the rate of proglutelin synthesis exceeds a critical rate. We revealed that the ER membrane-localized sulfhydryl oxidase Ero1 promotes the formation of native intramolecular disulfide bonds in proglutelins in the peripheral region of endosperm, also known as subaleurone cells, and that ERO1 knockdown results in a markedly decreased production of PDIL1;1, increased production of PDIL2;3, and unaltered production of PDIL1;4 (a paralog of PDIL1;1; see Supplemental Figure 1 online) (Onda et al., 2009), which together suggested that these PDI oxidoreductases play specific roles in the rice endosperm.
In this study, we examined oxidative folding of storage proteins and development of PBs in the rice endosperm, focusing on the role of PDIL1;1 and PDIL2;3. We demonstrated that PDIL1;1 and PDIL2;3 exhibited distinct localizations in the ER lumen of the same cell; PDIL1;1 was uniformly distributed in the ER lumen, whereas PDIL2;3 was localized mainly on the surface of PB-I in the ER. PDIL2;3 knockdown caused aberrant accumulation of prolamins in PB-I, whereas the oxidative folding of vacuole-targeted proteins, such as proglutelins and α-globulin, was hardly affected. Based on these results, we discuss the distinct roles of PDIL1;1 and PDIL2;3 in sulfhydryl oxidations of structurally diverse storage proteins, which are tightly linked to PB development in the rice endosperm.
RESULTS
Localization and Role of PDIL2;3 in the ER of Endosperm Cells
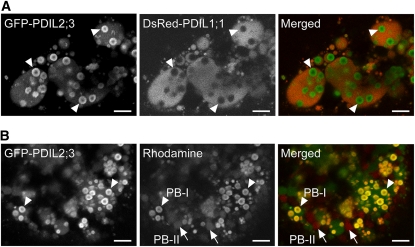
In the endosperm of rice esp2, a knockout mutant of PDIL1;1, the protein level of PDIL2;3 is significantly upregulated, whereas that of PDIL1;4 is relatively constant (Onda et al., 2009). To gain insights into specific roles of PDIL1;1 and PDIL2;3 in the oxidative folding of storage proteins and development of PBs, we compared the subcellular localizations of PDIL1;1 and PDIL2;3 by producing transgenic rice plants coexpressing two chimeric genes encoding PDIL1;1 and PDIL2;3, fused to Discosoma species red fluorescent protein (DsRed-Monomer) and green fluorescent protein (GFP), respectively, which were both modified to contain a signal peptide (sp) at the N terminus. DsRed-PDIL1;1 was uniformly dispersed in the luminal space (Figure 1A, middle panel). Intriguingly, GFP-PDIL2;3 was also detected in the ER lumen, and strong fluorescence signals were detected in rings within the ER (Figure 1A, left panel). Note that the ring-like signals of GFP-PDIL2;3 overlapped with the Rhodamine-stained PB-Is (Figure 1B), indicating that GFP-PDIL2;3 was targeted mainly to the surface of PB-I in the ER lumen.
Figure 1.
PDIL2;3 Localizes on the Surface of PB-I in the Endosperm.
(A) Confocal fluorescence images of the outer region of endosperm (10 d after flowing [DAF]) expressing both spGFP-PDIL2;3 and spDsRed-PDIL1;1. The fluorescence signals of GFP-PDIL2;3 (left panel) and DsRed-PDIL1;1 (middle panel) were converted to green and red, respectively, and merged (right panel). Arrowheads indicate PB-I in the ER lumen.
(B) PB-I (arrowheads) and PB-II (arrows) were visualized by Rhodamine staining. The fluorescence signals of GFP-PDIL2;3 (left panel) and Rhodamine (middle panel) were converted to green and red, respectively, and merged (right panel). The fluorescence signals of DsRed-PDIL1;1 were not visible under the conditions for obtaining Rhodamine signals at the appropriate intensity.
Bars = 5 μm.
Because GFP-PDIL2;3 showed characteristic localizations on the PB-I surface, we hypothesized that PDIL2;3 assists the development of PB-I in the ER lumen. We analyzed the effects of knockdown of PDIL2;3 on the accumulations of prolamins in PB-I. We coexpressed crP10 and cpP13, fused to spGFP and spDsRed, respectively, in the developing endosperm of wild-type or PDIL2;3-knockdown (PDIL2;3-KD) plants (Figure 2A). Lowering the expression level of PDIL2;3 did not upregulate the protein levels of PDIL1;1, PDIL1;4, Ero1, or BiP (Figure 2B). We found that GFP-crP10 was concentrated in the core of PB-I, and DsRed-cpP13 was targeted to the peripheral layers of PB-I in the wild-type endosperm cells (Figure 2C, panel WT), which indicated that GFP or DsRed tagging to crP10 and cpP13, respectively, does not alter their localizations in PB-I. By contrast, in the endosperm of PDIL2;3-KD, GFP-crP10 was not concentrated in the core of PB-I, and the size of PB-I was more heterogeneous compared with the wild type (Figure 2C, panel PDIL2;3-KD). Similar results were obtained by immunocytochemical analyses (Figure 2D). We confirmed the alterations in the distributions of crP10 and cpP13 in PB-I between the wild type and PDIL2;3-KD by immunocytochemical electron microscopy (see Supplemental Figure 2 online).
Figure 2.
Accumulation of crP10 in the Core of PB-I Depends on PDIL2;3.
(A) and (B) Immunoblot analyses of extracts from mature seeds of untransformed wild-type control (lane 1) and transformants expressing both spGFP-crP10 and spDsRed-cpP13 (lanes 2 and 3) in the background of the wild type (+/+) (lane 2) or PDIL2;3-knockdown (KD) (lane 3). The protein levels were compared by immunoblot with indicated antibodies.
(C) Confocal fluorescence images of the outer region of endosperm (10 DAF) expressing both spGFP-crP10 (left panels) and spDsRed-cpP13 (middle panels) in the background of the wild type (top panels) or PDIL2;3-KD (bottom panels). The fluorescence signals of GFP-crP10 and DsRed-cpP13 were converted to green and red, respectively, and merged (right panels).
(D) Immunofluorescence analysis of crP10 and cpP13 in the wild-type and PDIL2;3-KD endosperm (17 DAF). Arrows and arrowheads indicate PB-I and small patches, respectively.
Bars = 5 μm.
Oxidative Folding of Storage Proteins and PB-I Development in the esp2 Seed
The esp2 endosperm cells do not produce typical PBs but form abnormal small particles, which are uniformly stained with Rhodamine (Onda et al., 2009). To see if this is caused in part by the aberrant accumulation of different classes of storage proteins in the small particles, we coexpressed spGFP-crP10 and spDsRed-cpP13 in the esp2 endosperm. As we reported previously (Onda et al., 2009), the protein levels of PDIL2;3, Ero1, and BiP were upregulated in the esp2 seed (Figure 3A, cf. lanes 1 and 2). Expressing spGFP-crP10 and spDsRed-cpP13 in the esp2 seed further upregulated PDIL2;3 (Figure 3A, lane 3). When esp2 seed proteins are extracted and fractionated by centrifugation under nonreducing condition, the pellet (P) contains a large amount of proglutelins in complexes through intermolecular disulfide bonds (Onda et al., 2009). Almost all crP10 was fractionated into the P fraction when proteins were extracted from the wild-type seed, whereas a small amount of crP10 was detected in the S fraction when extracted from the esp2 seed (Figure 3B, panel crP10, lane 3). Other storage proteins, such as α-globulin and RA17, which both contain ABC domains conserved in the 2S albumin superfamily (Nakase et al., 1996; Kawagoe et al., 2005), were mostly in the S fraction when extracted from the wild-type seed (Figure 3B, lane 1), whereas significant amounts of the proteins were fractionated into the P fraction when extracted from the esp2 seed (Figure 3B, lane 4). By contrast, cpP13, which does not contain Cys residue in the mature polypeptide, was fractionated into the S fraction when extracted from the seeds of the wild type and esp2 (Figure 3B, panel cpP13), which supports the notion that the proteins fractionated into the P fraction form large complexes through intermolecular disulfide bonds (Onda et al., 2009). When the expression level of PDIL2;3 was lowered to a level comparable to the wild type, the S/P ratio of proglutelins was not noticeably altered from that of esp2 (Figure 3B, lanes 7 and 8). We then analyzed the localizations of GFP-crP10 and DsRed-cpP13 in the esp2 endosperm. A large fraction of GFP-crP10 accumulated and formed particles of different sizes (Figure 3C, left panel). Interestingly, DsRed-cpP13 did not strongly associate with the particles containing GFP-crP10, but it was spread in the cell, most likely the ER network (Figure 3C, middle panel). When the PDIL2;3 level was significantly lowered in the esp2 seed, GFP-crP10 was mostly localized in PB-I–like particles, but GFP-crP10 was not accumulated in the core of these particles (Figure 3D, left panel). The localization of DsRed-cpP13 in the PDIL2;3-KD endosperm of esp2 showed extensive overlaps with that of GFP-crP10 (Figure 3D, middle panel). These results indicate that the expression level of PDIL2;3 influences the localizations of GFP-crP10 and DsRed-cpP13 in both wild-type and esp2 endosperm.
Figure 3.
Aberrant Localizations of Prolamins in the PDIL1;1-Knockout esp2 Endosperm.
(A) Immunoblot analyses of extracts from mature seeds with indicated antibodies. Total seed proteins were extracted under reducing conditions from untransformed wild-type (+/+) control (lane 1), PDL1;1-knockout (−/−) esp2 (lane 2), esp2 expressing both spGFP-crP10 and spDsRed-cpP13 (lanes 3), and esp2 expressing both spGFP-crP10 and spDsRed-cpP13 under the background of PDIL2;3-knockdown (KD) (lane 4).
(B) Immunoblot analyses of seed proteins. Seed proteins were fractionated into the supernatant (S) and pellet (P) fractions by centrifugation under nonreducing conditions (see Methods for details). pGT, proglutelins; Glb, α-globulin; RA17, α-amylase inhibitor.
(C) Confocal fluorescence images of the outer region of endosperm (10 DAF) expressing both spGFP-crP10 and spDsRed-cpP13 in the background of esp2 (pdil1;1). Arrowheads indicate PB-I–like particles containing GFP-crP10. Note that DsRed-cpP13 did not accumulate in these PB-I-like particles. The fluorescence signals of GFP-crP10 (left panel) and DsRed-cpP13 (middle panel) were converted to green and red, respectively, and merged (right panel). Bars = 5 μm.
(D) Confocal fluorescence images of the PDIL2;3-knockdown (KD) endosperm cells (10 DAF) expressing both spGFP-crP10 and spDsRed-cpP13 in the background of esp2 (pdil1;1). The fluorescence signals of GFP-crP10 (left panel) and DsRed-cpP13 (middle panel) were converted to green and red, respectively, and merged (right panel). Bars = 5 μm.
A Conserved Structural Disulfide in the b Domain of PDIL2;3
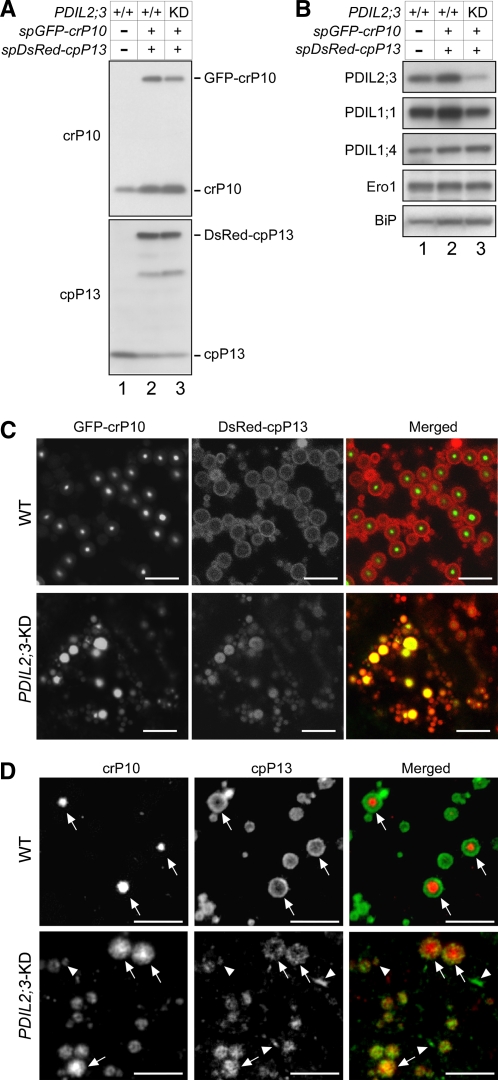
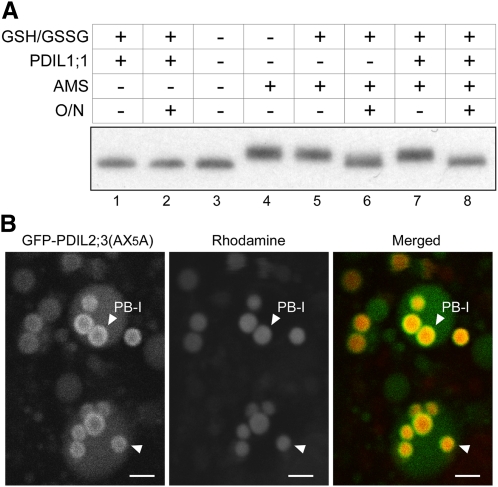
We noted that a significant fraction of PDIL2;3 was fractionated into the P fraction when extracted from esp2 seeds (Figure 3B, panel PDIL2;3, lane 4), which suggested that PDIL2;3 in the P fraction was contained in large complexes through intermolecular disulfide bonds. To gain insights into how PDIL2;3 formed nonnative intermolecular disulfides in the esp2 endosperm, we compared the amino acid sequences of the P5 subfamily oxidoreductases from diverse organisms, including mouse, Xenopus laevis, fruit fry, worm, Arabidopsis, and moss, and noted that the redox-inactive TRX-like b domain contains two absolutely conserved Cys residues separated by five amino acid residues (CX5C) and that the C-terminal tail is rich in acidic amino acid residues (see Supplemental Figures 3 and 4 online). We speculated that the CX5C motif might form a structural disulfide and that the Cys residues in CX5C may have been involved in nonnative intermolecular disulfide bond formation in the esp2 seed. To see whether the CX5C forms a disulfide bond, we conducted protein folding assays with a recombinant PDIL2;3 mutant protein, in which all the four catalytic Cys residues in the a domain (Cys59-X-X-Cys62) and a′ domain (Cys195-X-X-Cys198) were substituted with Ala residues. This PDIL2;3(ama′mb) mutant contains the two Cys residues (Cys-293 and Cys-299) in the CX5C motif and another plant-conserved Cys residue (Cys-223) in the a′ domain (see Supplemental Figure 3 online). Interestingly, size exclusion chromatography (SEC) of soluble glutathione S-transferase (GST)-PDIL2;3(ama′mb), whose calculated molecular mass is 72,396.94 D, revealed that the purified protein migrated at ~316 kD (see Supplemental Figure 5 online), suggesting that GST-PDIL2;3(ama′mb) forms a tetramer in Escherichia coli and that the GST tag attached at the N terminus of PDIL2;3(ama′mb) does not interfere with homotetramerization of the fusion protein. When the GST tag was removed, PDIL2;3(ama′mb) (calculated molecular mass is 45,948.23 D) also migrated as a single peak at ~208 kD (see Supplemental Figure 5 online). We incubated the purified ama′mb protein overnight with recombinant PDIL1;1 in a redox buffer containing reduced glutathione/oxidized glutathione (GSH/GSSG) and compared the mobilities by SDS-PAGE under nonreducing condition. As shown in Figure 4, the mobility of the incubated ama′mb protein was indistinguishable from that without incubation (cf. lanes 1 and 2), indicating that the conformational change upon disulfide bond formation, even if formed, did not significantly change the mobility in the gel. We thus labeled the Cys residues with 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid (AMS), which increases molecular mass by 0.5 kD per Cys residue (Wang et al., 2009a). As expected, AMS labeling of ama′mb protein at the three Cys residues increased the apparent molecular mass by 1.5 kD (Figure 4, lane 4). After incubation of the protein in the refolding buffer, AMS labeling increased the apparent molecular mass only by 0.5 kD (Figure 4, lane 6). Similar results were obtained when ama′mb protein was incubated with recombinant PDIL1;1 (Figure 4, lane 8). These results suggest that PDIL2;3 does not form an intermolecular disulfide bond at these Cys residues in the presence or absence of PDIL1;1. It is thus probable that the conserved CX5C motif in the b domain forms an intramolecular, structural disulfide bond.
Figure 4.
Structural Disulfide CX5C in the b Domain of PDIL2;3.
(A) Purified PDIL2;3 (ama′mb, Cys59/62/195/198Ala) was incubated with or without PDIL1;1 in a GSH/GSSG redox buffer overnight (O/N) or mixed immediately with the SDS sample buffer after starting the reaction. The denatured proteins were immunoblotted with anti-PDIL2;3 antibody. Alkylation at three Cys residues with AMS shifted the protein by ~1.5 kD (cf. lanes 3 and 4), indicating that the oxidation state of PDIL2;3 protein purified from E. coli was predominantly reduced. By contrast, the oxidized protein, which was presumably alkylated at Cys-223, was shifted by ~0.5 kD (lanes 6 and 8). Note that that PDIL1;1 assisted the oxidation of PDIL2;3 (cf. lanes 7 and 8).
(B) Confocal fluorescence images of the outer region of endosperm (7 DAF) expressing spGFP-PDIL2;3(AX5A). PB-I (arrowheads) were visualized by Rhodamine staining. The fluorescence signals of GFP-PDIL2;3(AX5A) (left panel) and Rhodamine (middle panel) were converted to green and red, respectively, and merged (right panel). Bars = 2 μm.
We asked a question whether the CX5C motif influences the function of PDIL2;3. Mutating the two Cys residues (Cys-293 and Cys-299) in the CX5C structural disulfide to Ala residues (AX5A) did not affect the retention time in SEC (see Supplemental Figure 5 online), indicating that the CX5C motif is not necessary for tetramer formation in E. coli and that the stability of the tetramer in vitro does not depend on the CX5C structural disulfide. We next examined whether the CX5C motif affects the redox activity of PDIL2;3 using reduced, denatured ribonuclease A (rRNase) as substrate. The wild-type PDIL2;3 and mutant PDIL2;3(AX5A), in which Cys-293 and Cys-299 in the CX5C motif were substituted with Ala residues, were preincubated in a redox buffer containing GSH/GSSG to facilitate the formation of the structural disulfide of the CX5C motif and then mixed with the substrate rRNase. The mutant protein exhibited 95.1% ± 1.4% activity (mean ± sd of duplicates) of the wild-type protein, indicating that the CX5C motif in the b domain does not significantly modulate the refolding activity. We then examined the effect of Cys-to-Ala substitution in the CX5C motif on the localization of GFP-PDIL2;3 in the developing endosperm and found that the GFP-PDIL2;3(AX5A) mutant protein was predominantly on the PB-I surface (Figure 4B), indicating that the CX5C structural disulfide is not required for targeting GFP-PDIL2;3 to the PB-I surface.
Characterization of the Redox-Active Sites of PDIL2;3
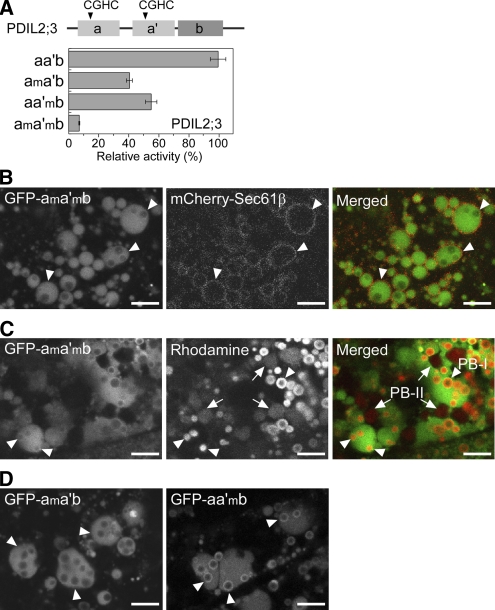
Rice PDIL2;3 contains two putative redox-active sites in the TRX-like domains, one each in the a domain (Cys59-X-X-Cys62) and a′ domain (Cys195-X-X-Cys198) (Figure 5A, top panel; see Supplemental Figure 3 online). To characterize putative redox activities of the a and a′ domains, we expressed a series of GST-PDIL2;3 fusion proteins in E. coli, removed the GST tag, and then examined the redox activities of the a and a′ domains using rRNase as substrate. Cys-to-Ala substitution both in the a and a′ domains (ama′mb, Cys59/62/195/198Ala) resulted in 4% residual activity (Figure 5A, bottom panel). Substitution mutations at the active site in the a domain (ama′b, Cys59/62Ala) or the a′ domain (aa′mb, Cys195/198Ala) resulted in partial reduction of RNase refolding activity (40 and 55% remaining activity for ama′b and aa′mb, respectively; Figure 5A, bottom panel).
Figure 5.
The PB-I-Surface Localization of PDIL2;3 Depends on the Catalytic Active Sites.
(A) Relative rRNase-refolding activities of recombinant PDIL2;3 variants. The arrowheads in the top panel indicate the positions of redox active sites of PDIL2;3: Cys59-X-X-Cys62 (a domain) and Cys195-X-X-Cys198 (a′ domain). The Cys residues in the active sites were substituted with Ala residues: Ala59-X-X-Ala62 (am domain) and Ala195-X-X-Ala198 (a′m domain). The wild type (aa′b) and three mutants (ama′b, aa′mb, and ama′mb) were purified, and their refolding activities were compared. The relative activities are presented as means with standard deviation of three replicates.
(B) Confocal fluorescence images of the endosperm cells (10 DAF) expressing both spGFP-PDIL2;3(ama′mb) and mCherry-Sec61β. The fluorescence signals of GFP-PDIL2;3(ama′mb) (left panel) and mCherry-Sec61β (middle panel) were converted to green and red, respectively, and merged (right panel). Arrowheads indicate PB-I in the ER lumen. Bars = 5 μm.
(C) PB-I (arrowheads) and PB-II (arrows) were visualized by Rhodamine staining. The fluorescence signals of GFP-PDIL2;3(ama′mb) (left panel) and Rhodamine (middle panel) were converted to green and red, respectively, and merged (right panel). Bars = 5 μm.
(D) Confocal fluorescence images of the endosperm (10 DAF) expressing spGFP-PDIL2;3(ama′b) (left panel) or spGFP-PDIL2;3(aa′mb) (right panel). The molecular masses of the fusion proteins were confirmed by immunoblot analysis. Arrowheads indicate PB-I in the ER lumen. Bars = 5 μm.
We next investigated the role of the two active sites of PDIL2;3 in vivo by examining the localizations of mutant proteins in the endosperm, using Sec61β fused to mCherry as an ER membrane marker (Onda et al., 2009). Interestingly, the mutant protein with all the redox-active Cys residues replaced with Ala residues (Cys59/62/195/198Ala; designated GFP-ama′mb) was evenly dispersed within the ER lumen (Figures 5B and 5C). We next analyzed the role of each active site in targeting the protein to the PB-I surface and found that the a domain made a greater contribution to PDIL2;3 localization on the PB-I surface than did the a′ domain (Figure 5D, panels GPF-ama′b and GFP-aa′mb).
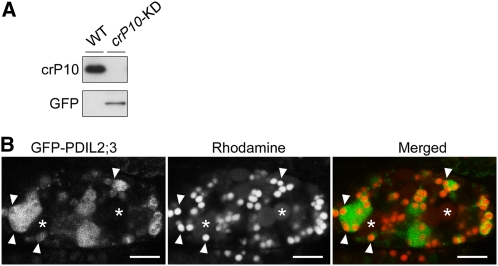
Having demonstrated that the localization of PDIL2;3 on the PB-I surface depends on the redox-active site of the a domain (Figures 5B to 5D) and that the accumulation of crP10 in the core of PB-I depends on PDIL2;3 (Figures 2C and 2D; see Supplemental Figure 2 online), we next analyzed possible effect of crP10 knockdown (crP10-KD) on the localization of GFP-PDIL2;3 in the ER. In knockdown lines with severely reduced levels of crP10 in the seeds, GFP-PDIL2;3 was not localized on the PB-I surface but was dispersed in the ER lumen (Figures 6A and 6B). This aberrant localization of GFP-PDIL2;3 was similar to those of GFP-ama′mb and GFP- ama′b in the ER (Figures 5B to 5D). Taken together, these results indicate that the localizations of PDIL2;3 and crP10 in the ER of endosperm are interdependent and suggest that PDIL2;3 and crP10 play important role in PB-I development.
Figure 6.
PB-I-Surface Localization of GFP-PDIL2;3 Depends on crP10.
(A) The RNAi-mediated knockdown of crP10 in seeds (crP10-KD) was confirmed by immunoblot analysis with anti-crP10 antibody. The molecular mass of GFP-PDIL2;3 (predicted molecular mass = 76 kD) was confirmed by SDS-PAGE, followed by immunoblot analysis with anti-GFP antibody.
(B) Confocal fluorescence images of the crP10-KD endosperm (10 DAF) expressing spGFP-PDIL2;3. PB-I (arrowheads) and PB-II (asterisks) were visualized by Rhodamine staining. The fluorescence signals of GFP-PDIL2;3 (left panel) and Rhodamine (middle panel) were converted to green and red, respectively, and merged (right panel). Bars = 5 μm.
Distinct Functions of PDIL1;1 and PDIL2;3 in the Endosperm
To see whether PDIL2;3 can fulfill the function of PDIL1;1 in the developing endosperm, we performed complementation analyses by expressing spGFP-PDIL1;1 or spGFP-PDIL2;3 in the PDIL1;1-knockout esp2 mutant. Rhodamine staining of the developing endosperm cells showed that spGFP-PDIL1;1 under the control of the rice TIP3 promoter in the esp2 mutants restored the formation of the typical PB-I and PB-II (Figure 7A, compare panels WT and esp2 + PDIL1;1; see Supplemental Figure 6B online). By contrast, small and numerous particles were observed in esp2 expressing spGFP-PDIL2;3 (Figure 7A, panel esp2 + PDIL2;3), which indicated that PDIL2;3 did not complement the function of PDIL1;1 in oxidative folding of storage proteins in the ER.
Figure 7.
PDIL2;3 Does Not Perform the Functions of PDIL1;1 in the Endosperm.
(A) Confocal fluorescence images of the Rhodamine-stained endosperm cells of the wild type, esp2 (pdil1;1), and esp2 expressing spGFP-PDIL1;1 (esp2+PDIL1;1) or spGFP-PDIL2;3 (esp2+PDIL2;3). Typical PB-I (arrowheads) and PB-II (arrows) are indicated. Bars = 5 μm.
(B) Relative rRNase-refolding activities of recombinant PDIL1;1 variants. The arrowheads in the top panel indicate the positions of redox active sites of PDIL1;1: Cys69-X-X-Cys72 (a domain) and Cys414-X-X-Cys417 (a′ domain). The Cys residues in the active sites were substituted with Ala residues: Ala69-X-X-Ala72 (am domain) and Ala414-X-X-Ala417 (a′m domain). The wild type (abb′a′) and three mutants (ambb′a′, abb′a′m, and ambb′a′m) were purified, and their refolding activities were compared. The relative activities are presented as means with standard deviation of three replicates.
(C) Complementation analysis of the esp2 endosperm with wild-type spGFP-PDIL1;1, spGFP-PDIL2;3, or mutated spGFP-PDIL1;1 at the redox active sites. The total proteins extracted from the mature seeds were separated by SDS-PAGE, stained with Coomassie blue (panel CBB) or subjected to immunoblot analysis with antiglutelin antibody (panel pGT). Immunoblot with anti-GFP antibody detected GFP-PDIL1;1 (predicted molecular mass = 84 kD) and GFP-PDIL2;3 (predicted molecular mass = 76 kD) (panel GFP). Seed proteins extracted from untransformed wild-type control (lane 1, WT), PDIL2;3 knockdown in the wild-type background with respect to PDIL1;1 (lane 2, PDIL2;3-KD), and esp2 (lane 3) were also analyzed as controls. pGT, proglutelins; αGT, glutelin acidic subunits; βGT, glutelin basic subunits; crP10, Cys-rich 10-kD prolamin.
We next characterized the functions of the a and a′ domains of PDIL1;1. The a and a′ domains contain putative catalytic sites Cys69-X-X-Cys72 and Cys414-X-X-Cys417, respectively (Figure 7B, top panel). We produced recombinant proteins of the wild-type PDIL1;1 (abb′a′) and three variants, in which Cys residues in the catalytic sites were replaced with Ala residues in the a domain (ambb′a′, Cys69/72Ala), a′ domain (abb′a′m, Cys414/417Ala), or both (ambb′a′m, Cys69/72/414/417Ala), and compared their refolding activities using rRNase as substrate. Mutating a single redox-active site reduced the activity by half (50% residual activity for ambb′a′ and 45% for abb′a′m), whereas substitution in both domains (ambb′a′m) resulted in 16% residual activity (Figure 7B, bottom panel). These results indicate that the a and a′ domains of PDIL1;1 facilitate rRNase refolding with similar efficiency in vitro. To characterize the functions of the a and a′ domains in vivo, we conducted complementation assays by expressing these mutant PDIL1;1 proteins in the esp2 endosperm. The protein profile in the esp2 seeds expressing the ambb′a′m mutant was essentially the same as that of the esp2 seeds (Figure 7C, panels CBB and pGT, lanes 3 and 7). Interestingly, expression of either the ambb′a′ or abb′a′m mutant resulted in partially reduced accumulation of proglutelins (Figure 7C, panel pGT, lanes 5 and 6). Expression of the wild-type PDIL1;1 (abb′a′) resulted in greatly reduced accumulation of proglutelins in the esp2 seeds (Figure 7C, panel pGT, lane 4). Consistent with the results of microscopy analyses (Figure 7A, panel esp2 + PDIL2;3), expression of the wild-type PDIL2;3 did not result in a significant reduction of proglutelins in the esp2 seeds (Figure 7C, panel pGT, lane 8; see Supplemental Figure 6A online). We noted that the relative amount of proglutelins to the processed glutelin subunits in the PDIL2;3-KD seed was indistinguishable from that of the wild type (Figure 7C, cf. lanes 1 and 2). In summary, these gain-of-function and loss-of-function experiments indicate that the a and a′ domains of PDIL1;1 are both functional in the oxidative folding of proglutelins and that PDIL2;3 does not facilitate the oxidative folding of proglutelins.
Redox Activities of PDIL1;1 and PDIL2;3 in Vitro
We previously showed that how sulfhydryls of α-globulin are oxidized plays a critical role in sorting α-globulin in the endosperm (Kawagoe et al., 2005). In this study, we found that almost half of α-globulin was fractionated into the P fraction when proteins were extracted from the esp2 seed under nonreducing conditions, whereas nearly all α-globulin produced in the wild-type seed was fractionated into the S fraction (Figure 3B). These results suggested that PDIL1;1 facilitates the oxidative folding of not only proglutelins but also α-globulin. To investigate whether PDIL2;3 also promotes the oxidative folding of α-globulin, we extracted and fractionated α-globulin from the PDIL2;3-KD seed. We found that α-globulin extracted from the PDIL2;3-KD seed was efficiently fractionated into the S fraction (Figure 8A), suggesting that PDIL2;3 activity is dispensable in the oxidative folding of α-globulin. We next extracted α-globulin from the seeds of esp2 expressing spGFP-PDIL1;1 or spGFP-PDIL2;3. Expressing spGFP-PDIL1;1 in the esp2 seeds reduced α-globulin in the P fraction to an undetectable level (Figure 8B, panel esp2 + PDIL1;1). By contrast, when spGFP-PDIL2;3 was expressed in the esp2 seed, a significantly increased amount of α-globulin was extracted in the P fraction (Figure 8B, panel esp2 + PDIL2;3), suggesting that GFP-PDIL2;3 facilitates intermolecular disulfide bond formation involving α-globulin.
Figure 8.
PDIL2;3 Facilitated Intermolecular Sulfhydryl Oxidation of the α-Globulin(C79F) Mutant.
(A) Proteins extracted from mature seeds of the wild type and PDIL2;3 knockdown (PDIL2;3-KD) were fractionated into the supernatant (fraction S) and pellet (fraction P) (see Methods for detail). The proteins were reduced and separated by SDS-PAGE and subjected to immunoblot analysis with anti-α-globulin (Glb) antibody.
(B) Proteins extracted from mature seeds of the wild type and the esp2 mutants expressing spGFP-PDIL1;1 or spGFP-PDIL2;3 under the control of the rice TIP3 promoter (Onda et al., 2009) were fractionated into the S and P fractions. The protein extracts were reduced and analyzed by SDS-PAGE and subjected to immunoblot analysis with anti-α-globulin antibody. Note that almost a half of α-globulin was fractionated in the P fraction when proteins were extracted from the esp2 seed (Figure 3B, lanes 3 and 4).
(C) Comparison of rRNase-refolding activities of PDIL1;1 and PDIL2;3 in vitro. The reduced, denatured RNase was refolded in the presence of recombinant PDIL1;1 or PDIL2;3 as described in Methods. The relative activities are presented as means with standard deviation of duplicate experiments.
(D) Effects of GSSG concentrations on the rate of rRNase-refolding activities of PDIL1;1 and PDIL2;3 in vitro. All assays were performed at a fixed GSH concentration of 1 mM at 28°C for 15 min. The relative activities are presented as means with standard deviation of duplicate experiments.
(E) and (F) Oxidative folding of reduced, denatured wild-type α-globulin (E) or α-globulin(C79F) mutant (F) in vitro. Protein refolding was performed in the absence (lanes 1 to 3) or presence of recombinant PDIL1;1 (lanes 4 to 6) or PDIL2;3 (lanes 7 to 9) in redox buffers containing a fixed GSH concentration of 1 mM and different GSSG concentrations of 0.05, 0.1, or 0.2 mM (see Methods for detail). The reactions were terminated at 20 min for α-globulin(WT) or 10 min for α-globulin(C79F) by adding 2× SDS sample buffer containing iodoacetamide. The reaction mixtures were centrifuged to separate the supernatant (Sup., top panels) and pellet (bottom panels). The reaction products were separated by SDS-PAGE under nonreducing [Sup. DTT(−)] or reducing condition [Pellet DTT(+)] and stained with Coomassie blue (CBB, left panels) or subjected to immunoblot analysis with anti-α-globulin antibody (right panels). Arrowheads indicate the reduced (red, apparent molecular mass of 26 kD) and oxidized (ox; apparent molecular mass of 22 kD) forms of α-globulin (Glb). The asterisk and dotted line indicate dimers and oligomers of α-globulin, respectively. Arrows indicate the bottom of sample wells.
The apparently dissimilar redox activities of PDIL1;1 and PDIL2;3 in vivo prompted us to further characterize their redox activities in vitro. We first compared refolding activities of recombinant PDIL1;1 and PDIL2;3 using rRNase as substrate at a fixed concentration of GSH (1 mM) and GSSG (0.2 mM), making the GSH/GSSG ratio 5:1. The relative refolding activity of PDIL2;3 was significantly lower than that of PDIL1;1 (Figure 8C). The catalyzed refolding of rRNase was then examined as a function of GSSG concentrations (0.05 to 0.2 mM) at a fixed GSH concentration of 1 mM (Figure 8D). Renaturation of rRNase was not significantly facilitated by PDIL1;1 or PDIL2;3 at a GSSG concentration of 0.05 mM, and renaturation of rRNase was enhanced most efficiently at a GSSG concentration of 0.2 mM.
We next used α-globulin as substrate because its reduced and oxidized forms are readily separated in nonreducing SDS-polyacrylamide gel; the oxidized α-globulin, which presumably contains four intramolecular disulfide bonds, migrates at the apparent molecular mass of 22 kD, whereas the reduced, monomeric form migrates at the apparent molecular mass of 26 kD in nonreducing SDS-polyacrylamide gel (Kawagoe et al., 2005). Reduced, denatured recombinant α-globulin was incubated with PDIL1;1 or PDIL2;3 in redox buffer containing a fixed concentration of GSH (1 mM) and different concentrations of GSSG (0.05, 0.1, or 0.2 mM), and the reaction mixtures were centrifuged at 20,000g to separate the supernatant containing monomeric form and relatively small oligomers from the pellet containing large aggregates that were formed in part by forming nonnative intermolecular disulfide bonds. Under these conditions, aggregates fractionating into the pellet were most efficiently produced at GSSG concentration of 0.2 mM both in the absence and presence of PDIL proteins (Figure 8E, panel Pellet, lanes 3, 6, and 9). Oxidation into monomeric form was most efficient in the presence of PDIL1 at GSSG concentration of 0.2 mM (Figure 8E, panel Sup., lane 6). By contrast, refolding α-globulin into monomeric form was not enhanced by PDIL2;3. We noted that PDIL1;1 inhibited redox reactions that create large oligomers at GSSG concentrations of 0.05 and 0.1 mM [Figure 8E, panel Sup, anti-Glb(WT), lanes 4 and 5]. We then used a mutant α-globulin(C79F) as substrate because the Cys-79 residue, which is the second Cys residue in the conserved CCXQL motif, is indispensable for the oxidative folding of α-globulin in vivo (Kawagoe et al., 2005). Interestingly, when α-globulin(C79F) mutant was incubated in the redox buffer containing 0.2 mM GSSG and 1 mM GSH, the pellet fraction did not contain the mutant protein at a detectable level by Coomassie blue staining (Figure 8F, panel Pellet, lane 3), indicating that Cys79 of α-globulin is necessary for the efficient formation of nonnative intermolecular disulfide bonds under these conditions. Immunoblot analysis indicated that PDIL1;1 also inhibited nonnative disulfide bond formation between C79F α-globulin mutant proteins [Figure 8F, panel Sup. anti-Glb(C79F), lanes 4 to 6]. By contrast, a large amount of mutant protein was fractionated into the pellet when incubated with PDIL2;3 at GSSG concentration of 0.2 mM (Figure 8F, panel Pellet, lane 9), suggesting that PDIL2;3 substantially promoted the formation of nonnative disulfide bonds between mutant proteins.
DISCUSSION
Distinct Functions of PDIL1;1 and PDIL2;3 in the Rice Endosperm
The redundancy and specificity of the PDI family oxidoreductases have been difficult to characterize in higher plants. Here, we demonstrated that PDIL1;1 and PDIL2;3 showed distinct localizations and functions in the rice endosperm cells. PDIL1;1, localized in the ER lumen, facilitated the oxidative folding of vacuole-targeted storage proteins, such as proglutelins and α-globulin (Figures 1, 7C, and 8B; see Supplemental Figure 6 online). By contrast, PDIL2;3, localized primarily on the PB-I surface, did not carry out the PDIL1;1 functions, but promoted the specific localization of crP10 in the core of PB-I (Figures 1, 2, 7A, and 7C; see Supplemental Figures 2 and 6A online). Recombinant PDIL1;1 and PDIL2;3 also exhibited distinct catalytic activities in vitro; PDIL1;1 refolded reduced, denatured RNase and α-globulin with a higher efficiency than did PDIL2;3 in a GSH/GSSG ratio-dependent manner, but PDIL2;3 exhibited a significantly higher sulfhydryl oxidase activity to form nonnative intermolecular disulfide bonds when reduced, denatured α-globulin(C79F) mutant was used as substrate (Figure 8F). These in vivo and in vitro results suggest that PDIL1;1 and PDIL2;3 have evolved to acquire distinct redox activities.
The primary substrate binding sites of yeast Pdi1p and human PDI have been mapped to the hydrophobic pocket in the b′ domain (Klappa et al., 1998; Tian et al., 2006), and the substrate binding ability of the b′ domain is likely influenced by the neighboring domains (b and a′) and by the x-linker region between b′ and a′ (Darby et al., 1998; Klappa et al., 1998; Tian et al., 2006; Byrne et al., 2009). Because the P5 subfamily oxidoreductases differ from Pdi1p and human PDI in domain architecture, it is most likely that the P5 subfamily proteins bind substrates in a unique manner. Although the structure of the P5 subfamily proteins has not been elucidated, we revealed that GST-PDIL2;3 formed a tetramer in E. coli and that the purified tetramers with or without GST tag were stable in vitro (see Supplemental Figure 5 online). Because GST attached at the N terminus of PDIL2;3 did not interfere with homotetramerization of the fusion protein in E. coli, we think it likely that GFP-PDIL2;3 also forms a similar tetramer when expressed in the rice endosperm. Further studies are needed to identify critical TRX-like domain(s) and amino acid residues involved in the tetramer formation.
In human culture cells, P5 forms a noncovalent complex with BiP and BiP client proteins (Meunier et al., 2002; Jessop et al., 2009). Although how P5 interacts with BiP remains to be elucidated, Jessop et al. (2009) pointed out the possibility that proteins that are targeted to BiP for their folding can become substrates for P5. In this respect, it is important to note that in the developing rice endosperm BiP is localized on the PB-I surface in an ATP-sensitive manner (Li et al., 1993). Given the high sequence similarity between P5 and PDIL2;3 (see Supplemental Figure 3 online), we speculate that PDIL2;3 may also interact with BiP in the endosperm. If so, BiP may bind nascent prolamins and channel them to PDIL2;3, and the whole complex may be targeted to PB-I in the ER.
The a domain of yeast Pdi1p and Mpd1p contains CX6C and CX8C structural disulfide, respectively (Wilkinson et al., 2005; Vitu et al., 2008). The CX6C structural disulfide in the a domain of Pdi1p destabilizes the active site in the same TRX-like domain (Wilkinson et al., 2005). We revealed that the redox activity of PDIL2;3(AX5A) was not significantly different from that of wild-type PDIL2;3 when the rRNase was used as substrate and that the CX5C motif of PDIL2;3 was not necessary for homotetramerization when produced in E. coli. Furthermore, mutating the CX5C motif to AX5A did not significantly alter the localization of GFP-PDIL2;3 in vivo (Figure 4B). However, it is notable that almost all PDIL2;3 proteins were in the S fraction when proteins were extracted from wild-type seed, whereas a substantial amount of PDIL2;3 was in the P fraction when proteins were extracted from the esp2 seed (Figure 3B, panel PDIL2;3). It is possible that in the esp2 endosperm cells, the Cys residues (Cys-293/Cys-299) in the CX5C motif of PDIL2;3 may have formed nonnative intermolecular disulfide bonds with storage proteins, such as proglutelins and α-globulin, which also accumulated in the P fraction. If so, these results suggest that PDIL1;1 facilitates the oxidative folding of the CX5C structural disulfide; hence, PDIL1;1 may regulate the function of PDIL2;3 in the ER.
Multiple Disulfide Bond Formation Pathways in the Rice Endosperm
Disulfide bond formation plays a dominant role in sorting storage proteins to the two types of PBs in the rice endosperm (Kawagoe et al., 2005), suggesting that sulfhydryl oxidases and the PDI family oxidoreductases are involved in protein sorting in the ER. It is notable that the specific localization of GFP-PDIL2;3 depended primarily on the active site of the a domain (Figures 5B to 5D). Furthermore, crP10 knockdown similarly inhibited the targeting of GFP-PDIL2;3 to the PB-I surface (Figure 6). Conversely, crP10 did not accumulate in the core of PB-I in the PDIL2;3-knockdown seed (Figures 2C and 2D; see Supplemental Figure 2 online). These results together suggest that thiol-disulfide exchange reactions between crP10 and PDIL2;3 occur in PB-I: the accumulation of crP10 in the core of PB-I involves sulfhydryl oxidations of crP10, and the reduced form of PDIL2;3 dissociates from PB-I. Interestingly, in the esp2 endosperm cells, DsRed-cpP13 did not accumulate in PB-I-like particles in the ER but was spread in the cell, most likely in the ER (Figure 3C). It is conceivable that abnormally elevated amounts of unfolded proteins, including proglutelins, α-globulin, and RA17, titrated PDIL2;3 and/or BiP in the ER of esp2 endosperm. We speculate that the aberrant environment in the esp2 ER hindered interactions of PDIL2;3 and/or BiP with otherwise preferred substrates, such as prolamins. These results indicate that PDIL1;1 facilitates the oxidative folding of PB-II–targeted proteins, such as proglutelins and α-globulin, and thereby assists indirectly targeting prolamins to PB-I in the ER.
In a recent study, we demonstrated that ER membrane-localized sulfhydryl oxidase Ero1 facilitates the formation of native disulfide bonds in proglutelins and that Ero1, PDIL2;3, and BiP are produced at markedly higher levels in the esp2 seed (Onda et al., 2009). Considering that crP10 forms polymers in the core of PB-I and that PDIL2;3 was concentrated on the PB-I surface (Figures 1 and 2), we speculate that either the a or a′ domain of PDIL2;3 is oxidized by Ero1 or other sulfhydryl oxidases, such as QSOX (Fass, 2008; Sevier, 2010), and that the oxidized form of PDIL2;3 then transfers oxidizing equivalents to prolamins in PB-I. After having said so, we point out that crP10 was predominantly fractionated into the P fraction when proteins were extracted from the PDIL2;3-knockdown seed (Figure 3B), suggesting that crP10 forms large protein complexes through intermolecular disulfide bonds. We thus speculate that crP10 oxidation in PB-I is sustained by PDIL2:3-dependent and PDIL2;3-independent oxidation pathways.
A recent study has revealed the existence of Ero1-independent mechanisms to generate disulfide bonds in mammalian cells (Zito et al., 2010). In this respect, it is important to note that ERO1 knockdown in the rice endosperm inhibits the oxidative folding of proglutelins, but proglutelins nonetheless form nonnative intermolecular disulfide bonds (Onda et al., 2009), suggesting that sulfhydryl oxidases other than Ero1 accept electrons from the PDI family oxidoreductases. Comprehensive gene expression analyses indeed revealed that rice QSOX (accession number AK121660) is expressed in the developing endosperm (Sato et al., 2010); thus, QSOX may also play a role in sulfhydryl oxidation of storage proteins in rice. Further studies are needed to elucidate relative contributions of Ero1 and other sulfhydryl oxidases, such as QSOX, in oxidation of prolamins in PB-I and oxidative folding of PB-II–targeted proteins in the ER lumen. Interestingly, human Ero1-α preferentially oxidizes the redox-active Cys pair in the a′ domain of PDI (Wang et al., 2009a), whereas it is the a domain of Pdi1p that yeast Ero1p predominantly oxidizes (Vitu et al., 2010). These results indicate that oxidizing equivalents flow from Ero1 to structurally diverse substrates through a subset of redox-active TRX-like domains of the PDI family oxidoreductases. Although it is unknown which redox-active TRX-like domains of the PDI family are preferentially oxidized by Ero1 in plants, it is surprising that expressing PDIL1;1(ambb′a′) or PDIL1;1(abb′a′m) in the esp2 endosperm significantly reduced the level of proglutelins to a similar extent (Figure 7C). These results strongly suggest that oxidoreductases other than PDIL1;1 supply oxidizing equivalents to nascent proglutelins and that the a and a′ redox-active TRX-like domains of PDIL1;1 assist independently the oxidative folding of proglutelins by reduction and/or isomerization reactions. Consistently, the a and a′ redox-active TRX-like domains of PDIL1;1 refolded rRNase at a similar rate in vitro (Figure 7B). In summary, we revealed that the PDI family oxidoreductases play distinct roles in multiple pathways of sulfhydryl oxidations of structurally diverse storage proteins in rice.
METHODS
Materials
Oryza sativa (cv Kinmaze) and an N-methyl-N-nitrosourea–induced mutant, esp2 (cv Kinmaze), were grown in the field. The full-length cDNA clones for PDIL2;3 (AK062254), PDIL1;1 (AK068268), crP10 (AK108254), and cpP13 (AK242260) were obtained from the National Institute of Agrobiological Sciences (Tsukuba, Japan). Antibodies against PDIL1;1, glutelins, α-globulin, and BiP were prepared as described previously (Takemoto et al., 2002; Kawagoe et al., 2005). Anti-GFP antibody was purchased from MBL. Anti-PDIL2;3 antibody was raised in rabbit by injecting purified PDIL2;3 (ama′b) used for the in vitro rRNase refolding assay. Anti-crP10, anti-cpP13, and anti-RA17 antibodies used for immunoblot analysis were prepared as follows. A PCR fragment encoding crP10 (Ile25–Cys134) was amplified from the full-length cDNA (AK108254) with primers 5′-ATCACCACTATGCAGTAT-3′ and 5′-TATCTCGAGACAACAACCACAGGAAGA-3′. A PCR fragment encoding cpP13 (Ala19–Leu150) was similarly amplified from the full-length cDNA (AK242260) with primers 5′-ATACCATGGCGCAGTTTGATGTTTTA-3′ and 5′-ATACTCGAGCAAGACACCGCCAAGGGT-3′. The PCR fragments were cloned in appropriate sites in pET23d (Novagen). A PCR fragment encoding RA17 (Asp28-His163) was amplified from the genomic DNA isolated from the seedling (Nipponbare) with primers 5′-ATACATATGGACCACCACCAAGTCTAC-3′ and 5′-TATCTCGAGGTGACCGGTTCTTGGGGT-3′ and inserted into pET22b (Novagen) at the appropriate sites. The plasmids were transferred to Rosetta2(DE3) (Novagen), and protein production was induced with 0.4 mM isopropyl-β-d(−)-thiogalactopyranoside (IPTG) at 37°C for 3 h. The recombinant proteins in inclusion bodies were solubilized in a denaturing buffer (8 M urea, 100 mM NaH2PO4, 20 mM 2-mercaptoethanol, and 10 mM Tris·HCl, pH 8.0) and were purified with a Ni-NTA agarose column (Qiagen) according to the manufacturer’s instructions. The purified protein was injected into rat (crP10 and RA17) and rabbit (cpP13) for antibody production.
Vector Construction and Rice Transformation
The binary vectors for rice transformation were constructed from modified plasmids derived from the Gateway Entry and Destination vectors (Invitrogen). For the construction of spGFP-PDIL2;3, a fragment encoding Ser19–Leu441 of PDIL2;3 was amplified from the full-length cDNA (AK062254) with primers 5′-CGCGGATCCTCGCCGGTTTCC-3′ and 5′-CCGCTCGAGCGGTCACAACTCGTCATTTACTGGA-3′ and inserted into PTIP3-spGFP-TGT1 (Onda et al., 2009) at the appropriate sites. For the construction of spDsRed-PDIL1;1, a fragment encoding Glu26–Leu512 of PDIL1;1 was amplified from the full-length cDNA (AK068268) with primers 5′-CGCGGATCCGAGGAGGCGGCGGCT-3′ and 5′-CCGCTCGAGCGGTTAGAGCTCATCCTTGAGAGGC-3′ and inserted into PTIP3-spDsRed(monomer)-TGT1 (Onda et al., 2009) at the appropriate sites. The vector containing spGFP-PDIL1;1 was described previously (Onda et al., 2009). For the construction of spGFP-crP10, a fragment encoding Ile25–Cys134 of crP10 was amplified from the full-length cDNA (AK108254) with primers 5′-CATCACCACTATGCAGTAT-3′ and 5′-TATCTCGAGTCAACAACAACCACAGG-3′ and inserted into the spGFP under the control of maize (Zea mays) Ubiquitin1 promoter (Onda et al., 2009). A fragment encoding Gln20–Leu150 of cpP13 was similarly amplified from the full-length cDNA (AK242260) with primers 5′-CCAGTTTGATGTTTTAGGT-3′ and 5′-TATCTCGAGTTTCACATGTCACATA-3′ and inserted into spDsRed-Monomer (Onda et al., 2009) to generate spDsRed-cpP13 under the control of maize Ubiquitin1 promoter. The PDIL2;3 RNA interference (RNAi) construct contained a 461-bp linker of the chloramphenicol resistance gene (CmR), flanked by inverted repeats of the 682-bp PDIL2;3 fragment (amplified with primers 5′-TATGGTACCATAATAGCAGCTCAAGGTAC-3′ and 5′-TATGAATTCGCAGGAATATGGAATAAGAG-3′), under the control of the APS2b promoter and the GT1 terminator (Onda et al., 2009). The crP10 RNAi construct contained the CmR linker flanked by inverted repeats of the 429-bp BamHI/XhoI fragment from the full-length cDNA (AK108254), under the control of the α-globulin promoter and the NOS terminator (Onda et al., 2009). Cys-to-Ala substitutions at the active sites of the a and a′ domains of PDIL1;1 and PDIL2;3 and at the CX5C motif of PDIL2;3 were performed using mutagenic primers and the QuikChange Lightning site-directed mutagenesis kit (Stratagene) according to the manufacturer’s instructions. The mutations were confirmed by DNA sequencing. The primer sequences and procedures are described in Supplemental Table 1 online.
The binary vectors were transferred into Agrobacterium tumefaciens strain EHA105 by electroporation and were used to transform rice (cv Yukihikari and Kinmaze) as described previously (Kawagoe et al., 2005). For complementation analysis of esp2 mutants, calli were induced from the esp2 seeds and used for Agrobacterium-mediated transformation.
Protein Extraction from Mature Rice Seeds
Total seed proteins were extracted by the following method unless otherwise indicated. Mature seeds were powdered in liquid nitrogen, and total seed proteins were extracted in buffer (10% [v/v] glycerol, 4% [w/v] SDS, 6 M urea, 100 mM dithiothreitol (DTT), and 50 mM Tris·HCl, pH 6.8; 200 μL per 10 mg of mature seed) by vigorous shaking for 2.5 h at 25°C. The homogenate was centrifuged at 20,400g for 5 min at 25°C, and the resulting supernatant was subjected to SDS-PAGE.
Protein fractionation under nonreducing conditions was performed as described previously (Onda et al., 2009). For protein fractionation into the S and P fractions, seed proteins were first extracted from the fine powders of mature seeds in nonreducing buffer (20% [v/v] glycerol, 4% [w/v] SDS, 8 M urea, and 50 mM Tris·HCl, pH 6.8; 320 μL per 10 mg of seed] by shaking for 3 h at room temperature. The homogenate was centrifuged at 10,000g for 5 min, and the resulting supernatant was reduced with 100 mM DTT (fraction S). The pellet was washed twice with the nonreducing buffer and then homogenized in reducing buffer (20% [v/v] glycerol, 4% [w/v] SDS, 8 M urea, 100 mM DTT, and 50 mM Tris·HCl, pH 6.8) at the same final volume as the S fractions overnight at room temperature. The soluble fractions were collected by centrifugation at 10,000g for 5 min (fraction P).
Alkylation of Recombinant PDIL2;3 with AMS
The oxidation state of PDIL2;3 in vitro was examined with the purified recombinant protein (ama′mb, Cys59/62/195/198Ala), which contains three Cys residues, by alkylation of free thiols with AMS (Jessop and Bulleid, 2004). The protein (0.5 μM) was incubated at 28°C in a redox buffer (0.1 M Tris·HCl, pH 8.0, 2 mM EDTA, 1 mM GSH, and 0.2 mM GSSG) with or without purified recombinant PDIL1;1 (0.5 μM). Disulfide exchange reactions were stopped either immediately or after overnight incubation by adding trichloroacetic acid to a final concentration of 10% (w/v). The proteins were precipitated at −30°C for 30 min, followed by centrifugation at 15,000g for 20 min at 4°C. The pellet was washed with ethanol/ether (1:1 v/v), air-dried, and suspended in SDS sample buffer consisting of 62.5 mM Tris·HCl, pH 6.8, 10% (v/v) glycerol, and 2% (w/v) SDS with or without 100 mM AMS (Invitrogen). Alkylation of free thiols with AMS was conducted on ice overnight in the dark. The denatured proteins were separated on SDS-polyacrylamide gel (5 to 20% acrylamide gradient; ATTO) under nonreducing condition, blotted onto a polyvinylidene difluoride membrane, and immunodetected with anti-PDIL2;3 antibody and Immobilon Western (Millipore).
Refolding Assays of Reduced, Denatured RNase in Vitro
Recombinant GST proteins fused with rice PDIL1;1 (Glu26–Leu512), rice PDIL2;3 (Ser19–Leu441), or their variants were constructed using pGEX-2TK or pGEX-6P-3 (GE Healthcare) and expressed in Rosetta2(DE3) (Novagen) at 37°C for 3 h with 0.4 mM IPTG induction. Cells were harvested and lysed with lysozyme and DNase, and the lysate was centrifuged at 11,000g for 10 min at 4°C. The fusion proteins in the supernatant were purified with Glutathione Sepharose 4B (MicroSpin GST Purification Module; GE Healthcare) and cleaved with protease according to the manufacturer’s instructions. SEC of GST-PDIL2;3(ama′mb), PDIL2;3(ama′mb), and PDIL2;3(AX5A) was performed using a Superdex 200 column with 20 mM Tris·HCl buffer, pH 7.0, containing 200 mM NaCl at a flow rate of 1 mL/min. Ribonuclease A (Sigma-Aldrich R5125) was reduced and denatured by incubation of the native enzyme overnight at room temperature in reducing/denaturing buffer (6 M guanidine hydrochloride, 140 mM DTT, 100 mM Tris·HCl, pH 8.0, and 2 mM EDTA). The reduced and denatured RNase A was then separated from excess DTT and guanidine hydrochloride by centrifugal gel filtration in a Micro Bio-Spin 6 column (Bio-Rad), which was prewashed with 0.1% acetic acid in water for buffer exchange. RNase A refolding assays were essentially conducted by the method of Lyles and Gilbert (1991) in buffer (0.1 M Tris·HCl, pH 8.0, 4.5 mM cyclic CMP [cCMP], 2 mM EDTA, 1 mM GSH, 0.2 mM GSSG, 10 μM reduced, denatured RNase A [rRNase], and 1 μM PDIL) at 37°C for 13 min for PDIL1;1 and 30 min for PDIL2;3. Kinetic assays and effects of GSSG concentrations on rRNase refolding activities were conducted by following the method of Rancy and Thorpe (2008). Briefly, rRNase was incubated at 28°C in buffer (0.1 M Tris·HCl, pH 8.0, 2 mM EDTA, 1 mM GSH, 0.2 mM or otherwise indicated concentrations of GSSG, 10 μM rRNase, and 1 μM PDIL). The reactions were stopped by mixing with water containing N-ethylmaleimide to give a final concentration of 2 mM N-ethylmaleimide. The samples were then diluted with an equal volume of Tris buffer containing 2 mM cCMP. As for the oxidation analysis of the CX5C motif of PDIL2;3, the wild-type PDIL2;3 and PDIL2;3(AX5A) mutant were preincubated in the reaction buffer without rRNase and cCMP at 28°C for 3 h prior to the refolding assay. The hydrolysis of cCMP was tested by measuring the absorbance at 296 nm. The data were converted to a percentage of the activity of the wild-type PDIL proteins.
Refolding Assays of Reduced, Denatured α-Globulin in Vitro
The sequences encoding the wild-type and C79F α-globulin (Glu23–Tyr186), which lacks the signal peptide, were amplified by PCR with primers 5′-ATACCATGGAGTCGGAGATGAGGTTCAGG-3′ and 5′-TATGAGCTCTCAGTGGTGGTGGTGGTGGTGGTACTGGCCGGCGGCGAAGAT-3′ (the His tag is underlined) and cloned into appropriate sites in pET23d vector (Novagen). Construction of α-globulin C79F mutant was described previously (Kawagoe et al., 2005). The wild-type and C79F α-globulin were expressed in Rosetta2(DE3) (Novagen) with IPTG induction, yielding inclusion bodies. The polypeptides in the inclusion bodies were purified with a Ni column under denaturing conditions and eluted in elution buffer (8 M urea, pH 4.5, 100 mM NaH2PO4, and 10 mM Tris·HCl). The purified polypeptides were reduced in the presence of DTT (final concentration 140 mM) at room temperature for 3 h. The reduced polypeptides were separated from excess DTT and urea by centrifugal gel filtration in a Micro Bio-Spin 6 column (Bio-Rad), which was prewashed with 0.1% acetic acid in water. The refolding reaction was initiated by adding the wild type or C79F α-globulin (final concentration of 10 μM) and recombinant PDIL proteins (final concentration of 2.2 μM) to the refolding buffer (150 mM NaCl [50 mM NaCl for C79F α-globulin], 100 mM Tris·HCl, pH 8.0, 2 mM EDTA, 1 mM GSH, and three concentrations [0.05, 0.1, or 0.2 mM] of GSSG). The samples were incubated at 28°C for 20 min for the wild type and 10 min for C79F α-globulin. The reactions were stopped by adding equal volume of nonreducing 2× SDS sample buffer (20% [v/v] glycerol, 4% [w/v] SDS, and 125 mM Tris·HCl, pH 6.8) containing 70 mM iodoacetamide. The samples were incubated at 4°C overnight and centrifuged at 20,000g for 5 min. The supernatant was directly subjected to nonreducing SDS-PAGE (10 to 20% acrylamide gradient). The pellets were washed with nonreducing 1× SDS sample buffer and were then resuspended in 1× SDS sample buffer containing 100 mM DTT. The resulting samples were centrifuged and the supernatant was subjected to SDS-PAGE (10 to 20% acrylamide gradient). The gels were either stained with Coomassie Brilliant Blue or subjected to immunoblot analysis using anti-α-globulin antibody as described previously (Kawagoe et al., 2005).
Immunofluorescence Microscopy and Immunocytochemical Analysis
Rice prolamins were purified from mature seeds as described elsewhere (Takemoto et al., 2002). cpP13 (pI 6.65) was purified by isoelectric focusing from Cys-poor prolamins that have been extracted into 60% (v/v) n-propanol containing 1 mM EDTA; the purified cpP13 was subjected to preparative SDS-PAGE. crP10 was purified by preparative SDS-PAGE. Antibodies against crP10 and cpP13 were raised in rabbit and mouse, respectively. Rhodamine-conjugated anti-rabbit and FITC-conjugated anti-mouse antibodies were purchased from Funakoshi Chemical. The fluorescence images of the developing endosperm were analyzed with a fluorescence microscope (AX80; Olympus) as described previously (Takemoto et al., 2002).
Confocal Laser Scanning Microscopy
Sections (100 μm) of the endosperm were prepared from developing seeds of rice plants. The seed sections were incubated in a Rhodamine solution to stain protein bodies PB-I and PB-II at different intensities as described previously (Onda et al., 2009). The fluorescence images of the peripheral region of endosperm, also called subaleurone cells, were analyzed with a confocal laser scanning microscope (TCS SP2 AOBS; Leica) at 488 and 543 nm as described previously (Onda et al., 2009). The data were processed in Adobe Photoshop CS3.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ERO1 (CB654132), α-Globulin (Glb) (D50643), PDIL1;1 (AK068268), PDIL1;4 (AK071514), PDIL2;3 (AK062254), crP10 (AK108254), cpP13 (AK242260), and RA17 (AK242340).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phylogenetic Tree of PDI Proteins.
Supplemental Figure 2. Immunolabeling of crP10 and cpP13 in PB-I of the Wild-Type and PDIL2;3-KD Endosperm Cells.
Supplemental Figure 3. Sequence Alignment of Human P5 and Rice PDIL2;3.
Supplemental Figure 4. Sequence Alignment of the b Domain of the P5 Subfamily Oxidoreductases.
Supplemental Figure 5. Size Exclusion Chromatography of GST-PDIL2;3(ama′mb), PDIL2;3(ama′mb), and PDIL2;3(AX5A).
Supplemental Figure 6. Complementation Analysis of esp2, a Null Mutant of PDIL1;1.
Supplemental Table 1. List of PCR Primers for PDIL1;1 and PDIL2;3 Mutagenesis.
Supplemental Data Set 1. Text File of Alignment Corresponding to Phylogenetic Analysis in Supplemental Figure 1.
Supplementary Material
Acknowledgments
We thank Z. Fujimoto for assisting SEC analysis and Chiemi Takaboshi and Mariko Shibahara for technical assistance. This work was supported by the Research and Development Program for New Bio-Industry Initiatives from the Bio-Oriented Technology Research Advanced Institution, by a grant from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Genomics for Agricultural Innovation, IPG-0023), and by a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (21380008).
References
- Andème Ondzighi C., Christopher D.A., Cho E.J., Chang S.C., Staehelin L.A. (2008). Arabidopsis protein disulfide isomerase-5 inhibits cysteine proteases during trafficking to vacuoles before programmed cell death of the endothelium in developing seeds. Plant Cell 20: 2205–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appenzeller-Herzog C., Ellgaard L. (2008). The human PDI family: Versatility packed into a single fold. Biochim. Biophys. Acta 1783: 535–548 [DOI] [PubMed] [Google Scholar]
- Byrne L.J., Sidhu A., Wallis A.K., Ruddock L.W., Freedman R.B., Howard M.J., Williamson R.A. (2009). Mapping of the ligand-binding site on the b’ domain of human PDI: Interaction with peptide ligands and the x-linker region. Biochem. J. 423: 209–217 [DOI] [PubMed] [Google Scholar]
- Darby N.J., Penka E., Vincentelli R. (1998). The multi-domain structure of protein disulfide isomerase is essential for high catalytic efficiency. J. Mol. Biol. 276: 239–247 [DOI] [PubMed] [Google Scholar]
- Fass D. (2008). The Erv family of sulfhydryl oxidases. Biochim. Biophys. Acta 1783: 557–566 [DOI] [PubMed] [Google Scholar]
- Ferrari D.M., Söling H.D. (1999). The protein disulphide-isomerase family: Unravelling a string of folds. Biochem. J. 339: 1–10 [PMC free article] [PubMed] [Google Scholar]
- Hatahet F., Ruddock L.W. (2009). Protein disulfide isomerase: A critical evaluation of its function in disulfide bond formation. Antioxid. Redox Signal. 11: 2807–2850Erratum. Antioxid. Redox Signal. 12: 322 [DOI] [PubMed] [Google Scholar]
- Houston N.L., Fan C.Z., Xiang J.Q., Schulze J.M., Jung R., Boston R.S. (2005). Phylogenetic analyses identify 10 classes of the protein disulfide isomerase family in plants, including single-domain protein disulfide isomerase-related proteins. Plant Physiol. 137: 762–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessop C.E., Bulleid N.J. (2004). Glutathione directly reduces an oxidoreductase in the endoplasmic reticulum of mammalian cells. J. Biol. Chem. 279: 55341–55347 [DOI] [PubMed] [Google Scholar]
- Jessop C.E., Watkins R.H., Simmons J.J., Tasab M., Bulleid N.J. (2009). Protein disulphide isomerase family members show distinct substrate specificity: P5 is targeted to BiP client proteins. J. Cell Sci. 122: 4287–4295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser B.K., Yim D., Chow I.T., Gonzalez S., Dai Z., Mann H.H., Strong R.K., Groh V., Spies T. (2007). Disulphide-isomerase-enabled shedding of tumour-associated NKG2D ligands. Nature 447: 482–486 [DOI] [PubMed] [Google Scholar]
- Kawagoe Y., Suzuki K., Tasaki M., Yasuda H., Akagi K., Katoh E., Nishizawa N.K., Ogawa M., Takaiwa F. (2005). The critical role of disulfide bond formation in protein sorting in the endosperm of rice. Plant Cell 17: 1141–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi M., Doi E., Tsujimoto I., Horibe T., Tsujimoto Y. (2002). Functional analysis of human P5, a protein disulfide isomerase homologue. J. Biochem. 132: 451–455 [DOI] [PubMed] [Google Scholar]
- Klappa P., Ruddock L.W., Darby N.J., Freedman R.B. (1998). The b’ domain provides the principal peptide-binding site of protein disulfide isomerase but all domains contribute to binding of misfolded proteins. EMBO J. 17: 927–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan H.B., White J.A., Pueppke S.G. (1992). Characterization and localization of rice (Oryza sativa L) seed globulins. Plant Sci. 81: 1–11 [Google Scholar]
- Kumamaru T., Ogawa M., Satoh H., Okita T.W. (2007). Protein body biogenesis in cereal endosperm. Endosperm: Plant Cell Monographs (8), Olsen O.A., (Heidelberg, Germany: Springer; ), pp. 141–158 [Google Scholar]
- Kumamaru T., Uemura Y., Inoue Y., Takemoto Y., Siddiqui S.U., Ogawa M., Hara-Nishimura I., Satoh H. (2010). Vacuolar processing enzyme plays an essential role in the crystalline structure of glutelin in rice seed. Plant Cell Physiol. 51: 38–46 [DOI] [PubMed] [Google Scholar]
- Li C.P., Larkins B.A. (1996). Expression of protein disulfide isomerase is elevated in the endosperm of the maize floury-2 mutant. Plant Mol. Biol. 30: 873–882 [DOI] [PubMed] [Google Scholar]
- Li X.X., Wu Y.J., Zhang D.Z., Gillikin J.W., Boston R.S., Franceschi V.R., Okita T.W. (1993). Rice prolamine protein body biogenesis: a BiP-mediated process. Science 262: 1054–1056 [DOI] [PubMed] [Google Scholar]
- Lu D.P., Christopher D.A. (2008). Endoplasmic reticulum stress activates the expression of a sub-group of protein disulfide isomerase genes and AtbZIP60 modulates the response in Arabidopsis thaliana. Mol. Genet. Genomics 280: 199–210 [DOI] [PubMed] [Google Scholar]
- Lyles M.M., Gilbert H.F. (1991). Catalysis of the oxidative folding of ribonuclease A by protein disulfide isomerase: Dependence of the rate on the composition of the redox buffer. Biochemistry 30: 613–619 [DOI] [PubMed] [Google Scholar]
- Meunier L., Usherwood Y.K., Chung K.T., Hendershot L.M. (2002). A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol. Biol. Cell 13: 4456–4469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakase M., Adachi T., Urisu A., Miyashita T., Alvarez A.M., Nagasaka S., Aoki N., Nakamura R., Matsuda T. (1996). Rice (Oryza sativa L) α-amylase inhibitors of 14–16 kDa are potential allergens and products of a multigene family. J. Agric. Food Chem. 44: 2624–2628 [Google Scholar]
- Nørgaard P., Westphal V., Tachibana C., Alsøe L., Holst B., Winther J.R. (2001). Functional differences in yeast protein disulfide isomerases. J. Cell Biol. 152: 553–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M., Kumamaru T., Satoh H., Iwata N., Omura T., Kasai Z., Tanaka K. (1987). Purification of protein body-I of rice seed and its polypeptide composition. Plant Cell Physiol. 28: 1517–1527 [Google Scholar]
- Onda Y., Kumamaru T., Kawagoe Y. (2009). ER membrane-localized oxidoreductase Ero1 is required for disulfide bond formation in the rice endosperm. Proc. Natl. Acad. Sci. USA 106: 14156–14161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rancy P.C., Thorpe C. (2008). Oxidative protein folding in vitro: a study of the cooperation between quiescin-sulfhydryl oxidase and protein disulfide isomerase. Biochemistry 47: 12047–12056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y., Antonio B.A., Namiki N., Takehisa H., Minami H., Kamatsuki K., Sugimoto K., Shimizu Y., Hirochika H., Nagamura Y. (2010). RiceXPro: A platform for monitoring gene expression in japonica rice grown under natural field conditions. Nucleic Acids Res. 39: D1141–D1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh-Cruz M., Crofts A.J., Takemoto-Kuno Y., Sugino A., Washida H., Crofts N., Okita T.W., Ogawa M., Satoh H., Kumamaru T. (2010). Protein disulfide isomerase like 1-1 participates in the maturation of proglutelin within the endoplasmic reticulum in rice endosperm. Plant Cell Physiol. 51: 1581–1593 [DOI] [PubMed] [Google Scholar]
- Sevier C.S. (2010). New insights into oxidative folding. J. Cell Biol. 188: 757–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewry P.R., Napier J.A., Tatham A.S. (1995). Seed storage proteins: Structures and biosynthesis. Plant Cell 7: 945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachikawa H., Takeuchi Y., Funahashi W., Miura T., Gao X.D., Fujimoto D., Mizunaga T., Onodera K. (1995). Isolation and characterization of a yeast gene, MPD1, the overexpression of which suppresses inviability caused by protein disulfide isomerase depletion. FEBS Lett. 369: 212–216 [DOI] [PubMed] [Google Scholar]
- Takemoto Y., Coughlan S.J., Okita T.W., Satoh H., Ogawa M., Kumamaru T. (2002). The rice mutant esp2 greatly accumulates the glutelin precursor and deletes the protein disulfide isomerase. Plant Physiol. 128: 1212–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Sugimoto T., Ogawa M., Kasai Z. (1980). Isolation and characterization of 2-types of protein bodies in the rice endosperm. Agric. Biol. Chem. 44: 1633–1639 [Google Scholar]
- Tian G., Xiang S., Noiva R., Lennarz W.J., Schindelin H. (2006). The crystal structure of yeast protein disulfide isomerase suggests cooperativity between its active sites. Cell 124: 61–73 [DOI] [PubMed] [Google Scholar]
- Uehara T., Nakamura T., Yao D.D., Shi Z.Q., Gu Z.Z., Ma Y.L., Masliah E., Nomura Y., Lipton S.A. (2006). S-nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration. Nature 441: 513–517 [DOI] [PubMed] [Google Scholar]
- Vitu E., Gross E., Greenblatt H.M., Sevier C.S., Kaiser C.A., Fass D. (2008). Yeast Mpd1p reveals the structural diversity of the protein disulfide isomerase family. J. Mol. Biol. 384: 631–640 [DOI] [PubMed] [Google Scholar]
- Vitu E., Kim S., Sevier C.S., Lutzky O., Heldman N., Bentzur M., Unger T., Yona M., Kaiser C.A., Fass D. (2010). Oxidative activity of yeast Ero1p on protein disulfide isomerase and related oxidoreductases of the endoplasmic reticulum. J. Biol. Chem. 285: 18155–18165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadahama H., Kamauchi S., Nakamoto Y., Nishizawa K., Ishimoto M., Kawada T., Urade R. (2008). A novel plant protein disulfide isomerase family homologous to animal P5 - molecular cloning and characterization as a functional protein for folding of soybean seed-storage proteins. FEBS J. 275: 399–410 [DOI] [PubMed] [Google Scholar]
- Wang H.Z., Boavida L.C., Ron M., McCormick S. (2008). Truncation of a protein disulfide isomerase, PDIL2-1, delays embryo sac maturation and disrupts pollen tube guidance in Arabidopsis thaliana. Plant Cell 20: 3300–3311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Li S.J., Sidhu A., Zhu L., Liang Y., Freedman R.B., Wang C.C. (2009a). Reconstitution of human Ero1-Lalpha/protein-disulfide isomerase oxidative folding pathway in vitro. Position-dependent differences in role between the a and a’ domains of protein-disulfide isomerase. J. Biol. Chem. 284: 199–206 [DOI] [PubMed] [Google Scholar]
- Wang Y.H., Zhu S.S., Liu S.J., Jiang L., Chen L.M., Ren Y.L., Han X.H., Liu F., Ji S.L., Liu X., Wan J.M. (2009b). The vacuolar processing enzyme OsVPE1 is required for efficient glutelin processing in rice. Plant J. 58: 606–617 [DOI] [PubMed] [Google Scholar]
- Wilkinson B., Xiao R.Y., Gilbert H.F. (2005). A structural disulfide of yeast protein-disulfide isomerase destabilizes the active site disulfide of the N-terminal thioredoxin domain. J. Biol. Chem. 280: 11483–11487 [DOI] [PubMed] [Google Scholar]
- Yamagata H., Sugimoto T., Tanaka K., Kasai Z. (1982). Biosynthesis of storage proteins in developing rice seeds. Plant Physiol. 70: 1094–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zito E., Chin K.T., Blais J., Harding H.P., Ron D. (2010). ERO1-β, a pancreas-specific disulfide oxidase, promotes insulin biogenesis and glucose homeostasis. J. Cell Biol. 188: 821–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.