This work demonstrated that conjugation of glutathione with indole-3-acetonitrile is catalyzed by GSTF6 in Arabidopsis. Comprehensive feeding experiments of MKK9DD/pad2 and pad2 plants suggested that glutathione-indole-3-acetonitrile and its catabolites are intermediates within the camalexin biosynthetic pathway.
Abstract
Camalexin, a major phytoalexin in Arabidopsis thaliana, consists of an indole ring and a thiazole ring. The indole ring is produced from Trp, which is converted to indole-3-acetonitrile (IAN) by CYP79B2/CYP79B3 and CYP71A13. Conversion of Cys(IAN) to dihydrocamalexic acid and subsequently to camalexin is catalyzed by CYP71B15. Recent studies proposed that Cys derivative, not Cys itself, is the precursor of the thiazole ring that conjugates with IAN. The nature of the Cys derivative and how it conjugates to IAN and subsequently forms Cys(IAN) remain obscure. We found that protein accumulation of multiple glutathione S-transferases (GSTs), elevation of GST activity, and consumption of glutathione (GSH) coincided with camalexin production. GSTF6 overexpression increased and GSTF6-knockout reduced camalexin production. Arabidopsis GSTF6 expressed in yeast cells catalyzed GSH(IAN) formation. GSH(IAN), (IAN)CysGly, and γGluCys(IAN) were determined to be intermediates within the camalexin biosynthetic pathway. Inhibitor treatments and mutant analyses revealed the involvement of γ-glutamyl transpeptidases (GGTs) and phytochelatin synthase (PCS) in the catabolism of GSH(IAN). The expression of GSTF6, GGT1, GGT2, and PCS1 was coordinately upregulated during camalexin biosynthesis. These results suggest that GSH is the Cys derivative used during camalexin biosynthesis, that the conjugation of GSH with IAN is catalyzed by GSTF6, and that GGTs and PCS are involved in camalexin biosynthesis.
INTRODUCTION
Phytoalexins are defined as low molecular weight antimicrobial compounds that are produced by plants after infection or stress (Hammerschmidt, 1999). Camalexin (3-thiazol-2’-yl-indole) is the major phytoalexin that accumulates in Arabidopsis thaliana plants after infection with microorganisms (Tsuji et al., 1992; Thomma et al., 1999; Glawischnig, 2007) and following treatment with abiotic factors (Tsuji et al., 1993; Zhao et al., 1998; Mert-Turk et al., 2003; Bouizgarne et al., 2006; Kishimoto et al., 2006). Several intermediates and key enzymes for camalexin biosynthesis in Arabidopsis have been identified through analysis of camalexin-deficient mutants and incorporation of radiolabeled compounds. Many phytoalexins produced by cruciferous plants are indoles with sulfur-containing moieties. The sulfur-containing moiety for camalexin is a thiazole ring at the 3-position of the indole (Devys et al., 1990; Monde et al., 1990; Tsuji et al., 1992). The indole ring of camalexin originates from Trp (Zook, 1998). Trp is converted to indole-3-acetaldoxime (IAOx) by the cytochrome P450 enzymes CYP79B2/CYP79B3 (Hull et al., 2000; Mikkelsen et al., 2000; Glawischnig et al., 2004). IAOx is a key metabolic intermediate for camalexin, indole glucosinolates, and auxin biosynthesis (Glawischnig et al., 2004). The dehydration of IAOx to indole-3-acetonitrile (IAN), catalyzed by CYP71A13, is required for IAOx flux through camalexin biosynthesis (Nafisi et al., 2007). The final two steps of camalexin biosynthesis, the catalysis of Cys(IAN) into dihydrocamalexic acid (DHCA) and DHCA into camalexin, are catalyzed by CYP71B15 (PAD3) (Schuhegger et al., 2006; Böttcher et al., 2009).
Early reports showed that the thiazole ring of camalexin originates from Cys (Zook and Hammerschmidt, 1997). It is unknown whether Cys itself or a Cys derivative conjugates to IAN to subsequently form Cys(IAN). However, recent studies indicate that a Cys derivative, but not Cys itself, conjugates to IAN. The first evidence of this is from the analysis of phytoalexin-deficient2 (pad2) mutant plants. Camalexin accumulation in the pad2 mutant was reduced by ~90% compared with wild-type plants following infection with Pseudomonas syringae pv maculicola strain ES4326 (psm ES4326) (Glazebrook and Ausubel, 1994). Gene cloning indicated that PAD2 encodes glutamylcysteine synthetase 1 (GSH1) (Parisy et al., 2007). However, the pad2 mutant contained about five times more Cys and 22% of glutathione (GSH) compared with wild-type plants. An exogenous supply of GSH restored pathogen-induced camalexin accumulation. Therefore, GSH or GSH derivatives, but not Cys itself, may function as the Cys donors in camalexin biosynthesis. The second point of evidence that a Cys derivative conjugates to IAN comes from IAN feeding and intermediates identification experiments. In plants fed with IAN and challenged with silver nitrate to induce camalexin biosynthesis, Böttcher et al. (2009) identified GSH(IAN), γGluCys(IAN), and Cys(IAN). These results suggest that GSH or GSH catabolites, such as γGluCys, may directly conjugate to IAN through a certain enzyme(s) to form GSH(IAN) or γGluCys(IAN). These conjugates may be further hydrolyzed to Cys(IAN) by specific, currently unidentified, peptidase(s).
Plant glutathione S-transferases (GSTs; EC 2.5.1.18), a diverse group of enzymes, catalyze the conjugation of GSH with a range of electrophilic substrates (Edwards et al., 2000; Sheehan et al., 2001; Dixon et al., 2002a; Frova, 2003). GSTs are involved in responses to biotic and abiotic stresses, hormones, and developmental changes (Dixon et al., 2002a; Moons, 2005; Frova, 2006). Therefore, GSTs are candidate enzymes for catalyzing the conjugation of GSH or GSH derivatives to IAN in camalexin biosynthesis.
Mitogen-activated protein kinase (MAPK) cascades are highly conserved signaling modules in eukaryotes and are composed of three kinase modules, MAPKKK, MAPKK, and MAPK (MKKK, MKK, and MPK, respectively, in Arabidopsis according to systemic nomenclature) (MAPK Group, 2002). By linking to upstream receptors and downstream targets in various ways, MAPK cascades play important roles in regulating plant stress responses, growth, and development (Nakagami et al., 2005; Mishra et al., 2006; Colcombet and Hirt, 2008; Pitzschke et al., 2009; Rodriguez et al., 2010). Activation of MPK3 and MPK6 by upstream MKKs (such as Arabidopsis MKK4, MKK5, and MKK9 and tobacco homolog Nt MEK2) strongly induced camalexin biosynthesis in transgenic Arabidopsis plants through coordinately upregulating the expression of genes in the pathway (Ren et al., 2008; Xu et al., 2008). In our previous studies, we developed transgenic plants with MKK mutant genes under the control of an inducible promoter (Ren et al., 2008; Xu et al., 2008). These transgenic plants are ideal tools for further analysis of the enzymes and intermediates in the camalexin biosynthesis pathway using either genetic or pharmacological methods.
In this study, we found that multiple GSTF proteins were strongly induced by the activation of MKK9. Among the GSTFs, GSTF6 was shown to play an important role in camalexin biosynthesis. Expression of Arabidopsis GSTF6 catalyzed GSH conjugation with IAN in yeast. GSH(IAN), γGluCys(IAN), and (IAN)CysGly were proven to be intermediates in the camalexin biosynthetic pathway. Our results suggest that GSH is the Cys derivative that donates Cys in camalexin biosynthesis and that GSTF6 catalyzes the conjugation of GSH with IAN. Although the catalytic activities have not yet been analyzed, inhibitor treatments and mutant analyses revealed that GGT1, GGT2, and PCS1 could possibly function in further catabolism of GSH(IAN) during camalexin biosynthesis.
RESULTS
Multiple Members of the phi Family of GSTs (GSTF) Are Induced in Transgenic Plants Expressing an Active Form of MKK9
Previously, we demonstrated that activation of MKK9 induced camalexin biosynthesis, possibly through coordinated activation of genes in the biosynthetic pathway (Xu et al., 2008). To identify protein(s) that may be involved in MKK9-MPK3/MPK6 cascade-regulated pathways, whole-cell protein extracts from MKK9 mutants transgenic plants were resolved by isoelectric focusing (IEF)/SDS-PAGE. We performed the experiments using IEF gel strips with a pI range of 4 to 7. Figure 1 shows Coomassie blue–stained two-dimensional (2D) gel images of proteins extracted from constitutively active (MKK9DD) (Figures 1A and 1B) and inactive (MKK9KR) (Figures 1C and 1D) MKK9 mutant transgenic plants. Approximately 1462 and 1593 highly reproducible protein spots were visualized in the protein extracts from MKK9KR and MKK9DD plants, respectively. After analysis of the 2D gel profiles using PD-Quest Version 7.2, 33 protein spots were identified as differentially expressed in MKK9DD transgenic plants before and after MKK9DD protein induction. By contrast, induction of MKK9KR expression in MKK9KR transgenic plants failed to induce the changes. Among the 33 differentially induced protein spots, 10 protein spots were newly and specifically induced by MKK9DD because the corresponding protein positions in the 2D gels from either MKK9DD and MKK9KR plant extracts before dexamethasone (DEX) treatment or MKK9KR plant extracts after DEX treatment were empty.
Figure 1.
2D Electrophoresis Profiling of Proteins Extracted from MKK9DD and MKK9KR Transgenic Seedlings.
Gels (A) and (C) are separations of protein samples from MKK9DD and MKK9KR transgenic seedlings, respectively, before transgene induction (−DEX). Gels (B) and (D) are separations of protein samples from MKK9DD and MKK9KR transgenic seedlings, respectively, after transgene induction (+DEX). Protein spots that showed significant differences between (A), (B), (C), and (D) are circled and numbered.
After in-gel digestion, the proteins were identified by matrix-assisted laser-desorption ionization time of flight mass spectrometry (MALDI-TOF MS), and the data from MALDI-TOF MS were searched against the National Center for Biotechnology Information nonredundant database using the MASCOT search engine (http://www.matrixscience.com/). Twenty-seven out of the 33 proteins were positively identified with significance threshold P < 0.05. However, following the rule given by Mann et al. (2001), 25 proteins with sequence coverage higher than 15% and more than five peptide matches were considered as significant and listed in Table 1. These proteins were classified into three groups based on their biological functions, with 12 proteins in stimuli responses, four proteins in metabolic processes, and nine proteins in cellular processes. Multiple GSTFs, including GSTF2, GSTF6, and GSTF7, accumulated at high levels following MKK9DD protein induction. GSTs are a class of important enzymes that are involved in plant growth, development, and stress responses (Moons, 2005; Dalton et al., 2009; Dixon and Edwards, 2009; Dixon et al., 2009, 2010; Sappl et al., 2009; Townsend et al., 2009). Interestingly, each of the above-mentioned GSTFs was represented by two protein spots: spots 14 and 22 represented GSTF2, spots 18 and 21 represented GSTF6, and spots 9 and 11 represented GSTF7. This phenomenon, which has been observed previously (Sappl et al., 2004; Foley et al., 2006), may be the result of a posttranslational modification of these GST proteins. These results raised the question as to what are the GST functions upon MKK9 activation.
Table 1.
MKK9 Activation-Induced Proteins Identified by MS
| Spot No. | Protein Name | AGI Code | Score | Coverage Rate (%) | Matched Peptides |
| Response to stimulus | |||||
| 9 | GSTF7 | At1g02920 | 148 | 46 | 10 |
| 10 | RGP2 | At5g15650 | 94 | 20 | 7 |
| 11 | GSTF7 | At1g02920 | 202 | 69 | 15 |
| 13 | DSBA | At5g38900 | 98 | 48 | 10 |
| 14 | GSTF2 | At4g02520 | 321 | 93 | 24 |
| 18 | GSTF6 | At1g02930 | 216 | 69 | 15 |
| 19 | Acid phosphatase class B family protein | At5g44020 | 95 | 21 | 7 |
| 21 | GSTF6 | At1g02930 | 236 | 71 | 16 |
| 22 | GSTF2 | At4g02520 | 325 | 96 | 24 |
| 23 | Defense-related protein | At4g30530 | 97 | 36 | 7 |
| 24 | Defense-related protein | At4g30530 | 188 | 52 | 10 |
| 43 | YLS5 | At2g38860 | 72 | 16 | 6 |
| Metabolic process | |||||
| 3 | TPX2 | At1g65970 | 98 | 43 | 7 |
| 31 | PDIL2-1 | At2g47470 | 84 | 33 | 7 |
| 35 | NADP-ME2 | At5g11670 | 128 | 18 | 14 |
| 37 | ASA1 | At5g05730 | 128 | 19 | 14 |
| Cellular process | |||||
| 38 | EIF3F | At2g39990 | 116 | 49 | 10 |
| 39 | PXMT1 | At1g66700 | 71 | 24 | 7 |
| 40 | O-methyltransferase | At1g21110 | 133 | 32 | 12 |
| 41 | TSA1 | At3g54640 | 94 | 44 | 10 |
| 42 | O-methyltransferase | At1g21110 | 280 | 53 | 21 |
| 44 | ATP sulfurylase | At3g22890 | 121 | 32 | 15 |
| 47 | ASP5 | At4g31990 | 338 | 64 | 29 |
| 48 | Rubisco activase | At2g39730 | 353 | 58 | 37 |
| 51 | PSAT | At4g35630 | 156 | 44 | 16 |
AGI, Arabidopsis Genome Initiative; Rubisco, ribulose-1,5-bisphosphate carboxylase/oxygenase.
Both GST Activity and GSH Are Required for MKK9-Induced Camalexin Biosynthesis
To explore the function of MKK9-induced GSTs in camalexin biosynthesis, we monitored the changes in the activities of GSTs following MKK9 activation. Because GSTs display a large variation in substrate specificities as previously reported (Wagner et al., 2002; Dixon et al., 2009), the appropriate substrates for GSTF2, GSTF6, and GSTF7 must be used for the GST activity assays. The recombinant GSTF2, GSTF6, and GSTF7 proteins exhibited GST activity toward 1-chloro-2,4-dinitrobenzene (CDNB) (see Supplemental Figures 1A and 1B online) as previously reported (Dixon et al., 2009). Therefore, CDNB was used as the substrate for subsequent GST activity assays. The results showed that GST activity in extracts from MKK9DD plants was markedly increased following MKK9DD induction (Figure 2B). MKK9DD plants were pretreated with Cibacron Blue 3GA, a broadly used reversible inhibitor of GST activity (Axarli et al., 2004), and GST activities and camalexin content were measured. Treatment with 3GA effectively inhibited, but did not completely abolish, GST activity (Figure 2B). Interestingly, camalexin contents were reduced at the time points tested in the presence of 3GA. Camalexin content was reduced 38% 12 h after MKK9DD induction compared with that in the absence of 3GA (Figure 2A). The inhibitor did not affect the protein levels of the GSTFs (Figure 2C). Therefore, GST activities may be required for MKK9-induced camalexin biosynthesis.
Figure 2.
GST Activity Is Required for MKK9-Induced Camalexin Biosynthesis.
Twelve-day-old MKK9DD seedlings were pretreated with 10 μM 3GA (inhibitor of GST activity) for 72 h before DEX treatment. The camalexin concentration and GST activity in MKK9DD seedlings in the presence (open circles) or absence (closed circles) of 3GA were measured.
(A) MKK9-induced camalexin biosynthesis was effectively inhibited by 3GA treatment.
(B) MKK9-induced GST activity was effectively inhibited by 3GA treatment. In (A) and (B), data represent the means ± sd of three biological replicates. Asterisks indicate statistically significant differences between MKK9DD seedlings treated with DEX alone and those treated with DEX plus 3GA. **P < 0.01 (paired sample t test).
(C) Gel regions indicated that the MKK9DD seedlings accumulated comparable levels of GSTF2 (spots 14 and 22), GSTF6 (spots 18 and 21), and GSTF7 (spots 9 and 11) in the presence or absence of 3GA. Proteins were extracted from the seedlings 8 h after DEX treatment.
Changes in GSH levels were also detected. The results showed that GSH content was greatly decreased in response to MKK9DD protein induction (Figure 3B). Increases in GST activities and GSH consumption coincided with camalexin accumulation. Therefore, GST-catalyzed GSH consumption may be required for MKK9-induced camalexin biosynthesis.
Figure 3.
GSH Is Required for MKK9-Induced Camalexin Biosynthesis.
Twelve-day-old MKK9DD seedlings were pretreated with 0.1 mM BSO (inhibitor of γ-glutamylcysteine synthetase, the enzyme responsible for biosynthesis of GSH) for 4 h before DEX treatment. After DEX treatment, the camalexin concentration and GSH content in MKK9DD seedlings in the presence (open circles) or absence (closed circles) of BSO and MKK9DD/pad2 (open triangles) were measured.
(A) MKK9-induced camalexin biosynthesis was effectively inhibited by BSO treatment and by crossing with the pad2 mutant.
(B) GSH content was greatly reduced in MKK9DD seedlings during camalexin biosynthesis. BSO-treated MKK9DD seedlings and MKK9DD/pad2 seedlings produced lower levels of GSH. In (A) and (B), data represent the means ± sd of three biological replicates. Asterisks indicate statistically significant differences between MKK9DD seedlings treated with DEX alone and those treated with DEX plus BSO, between MKK9DD and MKK9DD/pad2. **P < 0.01 (paired sample t test).
(C) Gel regions indicated that the MKK9DD/pad2 and MKK9DD seedlings in the presence or absence of BSO accumulated comparable levels of GSTF2 (spots 14 and 22), GSTF6 (spots 18 and 21), and GSTF7 (spots 9 and 11). Proteins were extracted from the seedlings 8 h after DEX treatment.
To further confirm the requirement of GSH in camalexin biosynthesis, GSH biosynthesis in MKK9DD plants was blocked by either the GSH biosynthesis enzyme inhibitor BSO (dl-buthionine-[S,R]-sulfoximine) (inhibitor of γ-glutamylcysteine synthetase, the enzyme responsible for biosynthesis of GSH) (Griffith and Meister, 1980) or by crossing MKK9DD with pad2 (a mutant plant with mutated γ-glutamylcysteine synthetase gene that only produced 22% of the GSH found in wild-type plants) (Parisy et al., 2007). Camalexin levels were then measured. MKK9DD/pad2 plants and BSO-pretreated MKK9DD plants produced very low levels of GSH and camalexin (Figures 3A and 3B), while the levels of the GSTF proteins in these plants remained comparable (Figure 3C). However, feeding the MKK9DD/pad2 and pad2 plants with exogenous GSH greatly increased the camalexin production in a GSH concentration-dependent manner (Figures 4A and 4B). Twelve hours after MKK9DD induction, MKK9DD/pad2 plants fed with 1 mM exogenous GSH produced 24.8 μg/g fresh weight (FW) of camalexin, which was 40.8% of the amount of camalexin produced by MKK9DD plants. This level of camalexin was 31 times greater than that in unfed MKK9DD/pad2 plants. These results suggest that MKK9-induced camalexin formation is dependent on GSH and further supported a previous finding that deficiency in GSH biosynthesis caused a large reduction in pathogen-induced camalexin production in pad2 plants (Parisy et al., 2007).
Figure 4.
The Camalexin-Deficient Phenotype of MKK9DD/pad2 and pad2 Is Complemented by GSH Feeding in a Concentration-Dependent Manner.
Seedlings were fed with 0.2, 0.5, or 1 mM exogenous GSH 4 h before DEX and B. cinerea treatments.
(A) Camalexin production by MKK9DD/pad2 seedlings fed with various concentrations of GSH 12 h after DEX treatment.
(B) Camalexin production by pad2 seedlings fed with various concentrations of GSH 16 h after inoculation with B. cinerea spores. Data represent the means ± sd of three biological replicates. WT, wild type.
The requirement of both GST activity and GSH for MKK9-induced camalexin accumulation revealed a correlation between GST-catalyzed GSH consumption and camalexin biosynthesis.
GSTF6 Plays an Important Role in Botrytis cinerea– and MKK9-Induced Camalexin Production
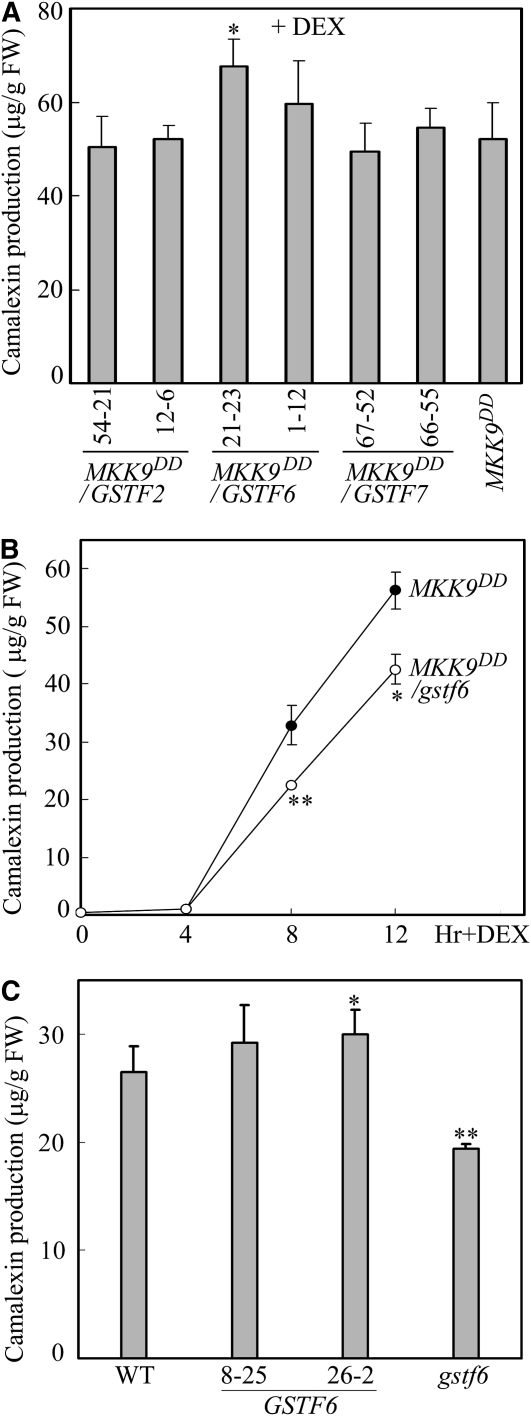
Because GST activity is required for camalexin production and since the Arabidopsis GSTF proteins GSTF2, GSTF6, and GSTF7 greatly accumulated during camalexin biosynthesis, we analyzed the roles of these GSTFs in camalexin biosynthesis. We generated transgenic plants by overexpressing individual Flag-tagged GSTFs in the MKK9DD transgenic background and detected camalexin in GSTF2/MKK9DD, GSTF6/MKK9DD, and GSTF7/MKK9DD plants after MKK9DD induction. The camalexin levels from two independent homozygous transgenic lines for each of the GSTF genes were monitored (Figure 5A). Increases in camalexin levels were observed in GSTF6/MKK9DD plants after MKK9DD induction. Line 21-23 and line 1-12 accumulated 29.7 and 14.7% more camalexin, respectively, compared with MKK9DD plants. Through 2D gel electrophoresis and MALDI-TOF MS analyses, Flag-GSTF6 protein spots were further identified from the samples extracted from line 21-23 and line 1-12 plants following MKK9DD induction (see Supplemental Figure 2C online). The levels of Flag-GSTF6 proteins in line 21-23 and line 1-12 plants were only 31.4 and 25.8% of the levels of MKK9-induced GSTF6 proteins, respectively, as calculated by scanning densitometry. The increased camalexin accumulation in line 21-23 and line 1-12 plants than in MKK9DD plants could possibly be a reflection of the Flag-GSTF6 protein accumulation caused by transgene expression.
Figure 5.
Regulation of Camalexin Biosynthesis by GSTF6.
(A) MKK9-induced camalexin production after overexpression of GSTF2, GSTF6, and GSTF7 on the MKK9DD background. Camalexin production was measured 12 h after DEX treatment. Two homozygous transgenic lines for each GST gene were examined.
(B) Knockout of GSTF6 on the MKK9DD background (MKK9DD/gstf6) significantly reduced MKK9-induced camalexin production. Closed circles and open circles represented MKK9DD and MKK9DD/gstf6, respectively.
(C) Overexpression of GSTF6 increased and knockout of GSTF6 reduced B. cinerea–induced camalexin production. Camalexin production was measured 16 h after inoculation with B. cinerea spores. Two homozygous transgenic lines for the GSTF6 gene were examined. Data represent the means ± sd of three biological replicates. Asterisks indicate statistically significant differences between MKK9DD and MKK9DD/GSTFs seedlings, between MKK9DD and MKK9DD/gstf6, and between Col wild-type WT and GSTF6 or gstf6 seedlings. *P < 0.05; **P < 0.01 (paired sample t test)
To further address the importance of GSTF6 in MKK9DD-induced camalexin biosynthesis, MKK9DD was crossed with gstf6, a GSTF6 T-DNA insertion mutant, to generate the MKK9DD/gstf6 double mutant. After MKK9DD induction, the proteins were extracted and separated on IEF/SDS-PAGE gels. The protein spot pair corresponding to GSTF6 was absent, while the spot pairs corresponding to GSTF2 and GSTF7 were comparable with those in MKK9DD plants (see Supplemental Figure 3 online). Therefore, the gstf6 mutant used in this study was a null mutant. MKK9DD/gstf6 plants produced less camalexin than did MKK9DD plants at all the time points examined. The camalexin produced by MKK9DD/gstf6 plants was reduced by 28% 12 h after MKK9DD induction (Figure 5B).
To analyze the effect of GSTF6 on pathogen-induced camalexin production, Flag-GSTF6–overexpressing transgenic plants were generated in the Columbia (Col) wild-type background. The wild type, the gstf6 mutant, and two lines of homozygous Flag-GSTF6 transgenic plants were treated with B. cinerea, and camalexin levels were measured. Sixteen hours after B. cinerea treatment, Flag-GSTF6–overexpressing plants produced 10.2 and 13.4% more camalexin, and gstf6 mutant plants produced 26.9% less camalexin than the wild-type plants (Figure 5C).
Overexpression and knockout of GSTF6 markedly affected MKK9- and B. cinerea–induced camalexin production, suggesting that GSTF6 plays an important role in the camalexin biosynthesis pathway. However, knockout of GSTF6 did not abolish camalexin biosynthesis, likely due to the functional redundancy within the Arabidopsis GST superfamily. Changes in the expression levels of 13 GSTFs and 28 GSTUs were detected through quantitative PCR (qPCR). Expression of GSTF9, GSTF10, GSTF11, and GSTF12 in the GSTF class and GSTU16, GSTU17, GSTU18, and GSTU27 in the GSTU class was upregulated in MKK9DD/gstf6 plants compared with MKK9DD plants after the MKK9DD induction (see Supplemental Figure 3B online). Upregulation of these genes suggests that loss of function of GSTF6 in MKK9DD/gstf6 plants might be, at least partially, complemented by some of these genes.
GSTF6 Catalyzes the Conjugation of GSH with IAN in Yeast
IAN and Cys(IAN) are two key intermediates in the camalexin biosynthesis pathway (Nafisi et al., 2007; Böttcher et al., 2009). However, it is unknown whether Cys itself or Cys derivative is conjugated with IAN, how the conjugate is catabolized to subsequently form Cys(IAN), and which enzyme(s) catalyzes the reaction(s). Based on the following data, we hypothesize that GSH is a possible Cys derivative and that GSTF6 is at least one of the enzymes that catalyzes the formation of GSH(IAN) during camalexin biosynthesis. (1) pad2, a mutant deficient in GSH biosynthesis, but containing 5 times more Cys than wild-type plants, produced low levels of camalexin upon pathogen infection (Parisy et al., 2007). (2) GSH(IAN) was identified in plants following treatment with AgNO3 to induce camalexin (Böttcher et al., 2009). (3) Inhibition of either GST activity or GSH biosynthesis significantly reduced MKK9- and B. cinerea–induced camalexin. (4) Overexpression of GSTF6 increased and knockout of GSTF6 reduced MKK9- and B.cinerea–induced camalexin production, respectively.
To confirm this hypothesis, we examined the ability of GSTF6 to catalyze the conjugation of GSH with IAN in yeast cells. A plasmid carrying the GSTF6 gene was transformed into yeast cells (pJ69a strain). The yeast cells transformed with green fluorescent protein (GFP) were used as a negative control. After expression of the transgenes, the cells were fed with IAN and GSH. Cells were collected 10 h after feeding and extracted with methanol/formic acid. Compounds in the extracts were separated and identified through liquid chromatography–mass spectrometry (LC-MS). The results showed that yeast cells with either the GSTF6 or GFP gene could produce GSH(IAN) compared with the synthesized GSH(IAN) standard. However, yeast cells expressing GSTF6 produced 40% more GSH(IAN) than did yeast cells expressing GFP (Figure 6A). The increased GSH(IAN) formation in GSTF6-expressing cells was likely due to the activity of GSTF6 and not the different cell densities, since GSTF6-expressing cells and GFP-expressing cells had comparable growth curves (see Supplemental Figure 4 online).
Figure 6.
GSTF6 Catalyzes GSH(IAN) Formation in Yeast Cells.
(A) Yeast cells transformed with either GSTF6, GSTF2, or GFP were fed with 0.2 mM GSH and 0.1 mM IAN. Ten hours after feeding, cells were extracted with methanol/formic acid. GSH(IAN) was identified by LC-MS and quantified (inset; top). The GSH(IAN) standard concentrations and LC-MS peak areas were plotted (inset; bottom).
(B) Yeast cells transformed with either GSTF6 or GFP were disrupted after transgene induction, and GSH and IAN were added. The boiled disrupted cells were used as a nonenzymatic control. After the reaction, GSH(IAN) was extracted and identified by LC-MS. Data represent the means ± sd of three biological replicates. Asterisks indicate statistically significant differences between GSTF2 and GFP transformed yeast cells and between GSTF6 and GFP transformed yeast cells. **P < 0.01 (paired sample t test). N.D., not detectable.
To determine if the background levels of GSH(IAN) produced in yeast cells were caused by nonenzymatic reactions or were catalyzed by yeast GSTs, the yeast cells were disrupted after transgene expression and used in later experiments. The complete disruption of the yeast cells was confirmed by microscopy observation. GSH and IAN were added to the reaction mixture with the disrupted cells, and GSH(IAN) was detected following the reaction. The results showed that GSH(IAN) was still produced by the disrupted cells expressing GFP or GSTF6. Expression of GSTF6 led to 61% more GSH(IAN) formation than did the expression of GFP (Figure 6B). As a nonenzymatic control, disrupted cells were boiled before the addition of GSH and IAN. There was no detectable GSH(IAN) produced by these controls following the reaction (Figure 6B), excluding the nonenzymatic formation of GSH(IAN) in whole-cell or disrupted-cell assays. These results suggest that GSTF6 expressed in yeast cells catalyzed GSH(IAN) formation and that GSH(IAN) produced in the yeast cells expressing GFP could be the result of endogenous yeast GST activity. Interestingly, GSTF6 in whole yeast cells catalyzed more effective conjugation of GSH with IAN than in disrupted cells (Figures 6A and 6B). This implied that the conjugation reaction might be coupled with another reaction that is likely catalyzed, for example, by a cytochrome P450 enzyme as previously speculated (Böttcher et al., 2009).
To test the ability of GSTF2 or GSTF7 to catalyze GSH(IAN) formation, two GSTFs that were also induced in MKK9DD plant after MKK9DD induction (Figure 1), GSTF2 and GSTF7, were expressed in yeast cells. In developing yeast cell cultures for the GSH and IAN feeding experiment, we found that yeast cells transformed with GSTF2 grew normally, as did the cells transformed with either GSTF6 or GFP; however, the cells transformed with GSTF7 grew markedly slower (growth curves are shown in Supplemental Figure 4 online). Therefore, the yeast cells transformed with GSTF7 were not used for further experiments. The yeast cells transformed with GSTF2 produced almost comparable levels of GSH(IAN), as did the cells transformed with GFP (Figure 6A), suggesting that GSTF2 might not be able to catalyze GSH(IAN) formation.
GSH(IAN) Is an Intermediate of Camalexin Biosynthesis
To confirm that GST-catalyzed GSH(IAN) formation participates in camalexin biosynthesis, we examined GSH(IAN) in plants undergoing camalexin biosynthesis and demonstrated that GSH(IAN) is an intermediate during camalexin biosynthesis in planta. Through LC-MS, we found that high levels of GSH(IAN) accumulated in MKK9DD plants after MKK9DD induction and in wild-type plants after B. cinerea infection (Figure 7). By contrast, GSH(IAN) was undetectable in MKK9DD prior to MKK9DD induction and in uninfected wild-type plants. As expected, MKK9DD/pad2 and pad2 plants produced only very low levels of GSH(IAN) after MKK9DD induction and B. cinerea infection. Therefore, MKK9DD/pad2 and pad2 plants were selected for the feeding experiments to minimize the effect of endogenous GSH formed GSH(IAN). Plants were fed with GSH(IAN) at a final concentration of 0.20 mM, and camalexin production was monitored. Feeding of GSH(IAN) markedly increased camalexin accumulation in MKK9DD/pad2 plants after MKK9DD induction and in pad2 plants after B. cinerea infection (Figure 8). The MKK9DD/pad2 and pad2 plants only produced 0.8 μg/g FW and 3.2 μg/g FW of camalexin, respectively, after treatments without GSH(IAN) feeding. Camalexin production increased to 20.1 μg/g FW in MKK9DD/pad2 plants and 10.9 μg/g FW in pad2 plants following feeding with GSH(IAN). The identification of GSH(IAN) in vivo and its function in restoring camalexin formation revealed that GSH(IAN) is an intermediate in the camalexin biosynthetic pathway in planta.
Figure 7.
Identification of GSH(IAN) in Seedlings Undergoing Camalexin Biosynthesis.
Both MKK9 activation and B. cinerea infection led to accumulation of GSH(IAN). Identification and quantification (inset) of GSH(IAN) in MKK9DD and MKK9DD/pad2 seedlings 12 h after DEX treatment (top) and in wild-type WT and pad2 seedlings 16 h after inoculation with B. cinerea spores (bottom). Data represent the means ± sd of three biological replicates. Asterisks indicate statistically significant differences between MKK9DD and MKK9DD/pad2 seedlings and between Col wild-type and pad2 seedlings. **P < 0.01 (paired sample t test).
Figure 8.
In Vivo Feeding of MKK9DD/pad2 and pad2 Seedlings with GSH(IAN), γGluCys(IAN), and (IAN)CysGly.
Seedlings were fed with the indicated components 4 h before DEX treatment or inoculation with B. cinerea spores.
(A) Camalexin production by MKK9DD/pad2 12 h after DEX treatment.
(B) Camalexin production by pad2 16 h after inoculation with B. cinerea spores. Data represent the means ± sd of three biological replicates. WT, wild type.
GGTs and PCS Are Involved in Camalexin Biosynthesis
The Glu and Gly residues of GSH(IAN) must be released to form Cys(IAN), which is subsequently converted to camalexin by CYP71B15 (Böttcher et al., 2009). Therefore, (IAN)CysGly and/or γGluCys(IAN) could be intermediates in the camalexin biosynthesis pathway. The MKK9DD/pad2 and pad2 plants were used in in vivo feeding experiments. Feeding MKK9DD/pad2 and pad2 plants with (IAN)CysGly or γGluCys(IAN) could greatly increase camalexin production in MKK9DD/pad2 plants after MKK9DD induction and in pad2 plants after B. cinerea infection (Figure 8). The plants used (IAN)CysGly and γGluCys(IAN) more efficiently than GSH(IAN), suggesting that these components could be downstream intermediates of GSH(IAN) during camalexin biosynthesis. Additionally, we preformed LC-MS to identify the components from plants undergoing camalexin biosynthesis. γGluCys(IAN) accumulated in MKK9DD plants after MKK9DD induction and in wild-type plants after B. cinerea infection (see Supplemental Figure 5 online). However, we could not detect (IAN)CysGly in the plants. Since the plants used (IAN)CysGly more efficiently than γGluCys(IAN) in the feeding experiments, (IAN)CysGly was potentially converted to derivative metabolites very rapidly to form camalexin in vivo.
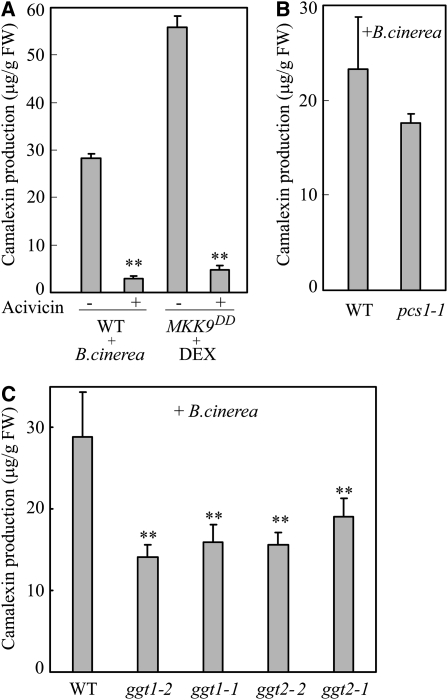
In plants, degradation of GSH conjugates could initiate from either the N terminus to remove Glu by GGT (Ohkama-Ohtsu et al., 2007a) or from the C terminus to remove Gly by phytochelatin synthase (PCS) (Blum et al., 2007). A GGTs inhibitor, acivicin, was used to test the effect of GGT activity on camalexin biosynthesis. Treatment with acivicin greatly reduced, but did not abolish, camalexin production in MKK9DD plants after MKK9DD induction and in wild-type plants infected with B. cinerea (Figure 9A). Therefore, GGT activity is likely more important than PCS activity for camalexin biosynthesis.
Figure 9.
GGT and PCS Are Involved in MKK9- and B. cinerea–Induced Camalexin Biosynthesis.
(A) Inhibition of GGT activity greatly reduced MKK9- and B. cinerea–induced camalexin production. Seedlings were pretreated with 55 μM acivicin 4 h before DEX treatment or inoculation with B. cinerea spores. Camalexin production in MKK9DD and wild-type seedlings (WT) was determined 12 h after DEX treatment and 16 h after B. cinerea inoculation.
(B) Camalexin production in wild-type and pcs1-1 seedlings 16 h after B. cinerea inoculation.
(C) Camalexin production in wild-type, GGT1, and GGT2 mutant seedlings was determined 16 h after inoculation with B. cinerea spores. Data represent the means ± sd of three biological replicates. Asterisks indicate statistically significant differences between acivicin-treated and untreated MKK9DD or Col wild-type seedlings and between Col wild-type seedlings and mutant seedlings after B. cinerea inoculation. **P < 0.01 (paired sample t test).
The Arabidopsis genome contains three functional GGTs, GGT1, GGT2, and GGT3 (Ohkama-Ohtsu et al., 2007b), and two functional PCSs, PCS1 and PCS2 (Blum et al., 2007). To identify which GGTs and PCSs are involved in catalyzing the release of Glu/Gly residues from GSH(IAN) during camalexin biosynthesis, we examined B. cinerea–induced camalexin production in GGT or PCS knockout mutants. We screened T-DNA insert mutants of various GGTs and PCSs on the Col background. For both GGT1 and GGT2, two alleles of T-DNA insert mutants (ggt1-1 and ggt1-2, ggt2-1 and ggt2-2) and one allele for PCS1 (pcs1-1) were obtained. The homozygous null mutants were screened using genomic PCR and confirmed by RT-PCR (see Supplemental Figure 6 online). The mutant and Col wild-type plants were challenged with B. cinerea, and camalexin content was measured. Compared with wild-type plants, camalexin accumulation in ggt1-1, ggt1-2, ggt2-1, ggt2-2, and pcs1-1 plants was greatly reduced (Figures 9B and 9C). These results suggest that both GGT and PCS activities are involved in camalexin biosynthesis.
Regulation of GSTF6, GGT1, GGT2, and PCS1 Transcript Levels
MPK3 and MPK6 are downstream MAPKs of MKK9 in planta (Xu et al., 2008; Yoo et al., 2008; Zhou et al., 2009). Our previous study indicated transcription of genes involved in the formation of IAOx (CYP79B2/CYP79B3), IAN (CYP71A13), DHCA, and camalexin (CYP71B15 or PAD3) are coordinately upregulated in MKK9-induced camalexin biosynthesis. Additionally, loss of MPK3 or MPK6 activity compromised MKK9-induced camalexin biosynthesis and transcript levels of these genes transcription (Xu et al., 2008). In this study, the mRNA transcript levels of GSTF6, GGT1, GGT2, and PCS1 were detected using qPCR. GSTF6, GGT1, GGT2, and PCS1 transcript levels were coordinately and rapidly upregulated in MKK9DD plants, while as control, the transcript levels of the genes did not show significant changes in MKK9KR plants (Figure 10). The greatest increases in transcript levels were seen for GSTF6 (213-fold), GGT1 (25-fold), GGT2 (19-fold), and PCS1 (15-fold) in MKK9DD plants 4 h after MKK9DD induction. The induction of GSTF6, GGT1, GGT2, and PCS1 was partially compromised in MKK9DD/mpk3 and MKK9DD/mpk6 plants. These results suggest that all genes that function in the pathway are fully activated to trigger camalexin biosynthesis.
Figure 10.
qPCR Detection of Genes Responsible for Camalexin Biosynthesis in MKK9 Mutant Transgenic Seedlings and Crossed Seedlings.
GSTF6, GGT1, GGT2, PCS1, GSH1, and GSH2 transcript levels in seedlings were monitored by qPCR. Data represent the means ± sd of three biological replicates.
DISCUSSION
MAPK cascades play pivotal roles in mediating signals during plant responses to pathogens and pathogen-associated molecular patterns (Pitzschke et al., 2009). Phytoalexins are produced in many plant species upon infection by a variety of pathogens or treatment with pathogen-associated molecular patterns (Böttcher et al., 2009; Rauhut et al., 2009; Kishi-Kaboshi et al., 2010; Kurusu et al., 2010). Recent reports have demonstrated that activation of MAPK cascades is sufficient to activate expression of specific genes that results in the biosynthesis of phytoalexins, including camalexin in Arabidopsis and diterpenoid in rice (Oryza sativa; Qiu et al., 2008; Ren et al., 2008; Xu et al., 2008; Kishi-Kaboshi et al., 2010). In the camalexin biosynthesis pathway, the genes that are currently known to catalyze the enzymatic reactions converting Trp to form camalexin include CYP79B2/CYP79B3, CYP71A13, and CYP71B15. These genes are transcriptionally activated by MPK3, MPK6, and MPK4 through phosphorylation by their upstream MKKs (Qiu et al., 2008; Ren et al., 2008; Xu et al., 2008).
In this study, proteomic analyses revealed that multiple members of the Arabidopsis GSTF class of proteins, including GSTF2, GSTF6, and GSTF7, accumulated at high levels in transgenic plants following activation of MKK9, an upstream MKK of MPK3 and MPK6 (Figure 1B). Enzyme activity assays using CDNB as the substrate indicated that activation of MKK9 led to increased GST activity (Figure 2). The activation of MKK9 was previously reported to induce leaf senescence (Zhou et al., 2009), ethylene biosynthesis and signaling (Xu et al., 2008; Yoo et al., 2008), and camalexin biosynthesis (Xu et al., 2008). In plants, GSTs are multifunctional proteins that can function as GSH-dependent peroxidases, GSH-dependent isomerases, and GSH-dependent oxidoreductases (Edwards and Dixon, 2005) or as noncatalytic binding proteins (Bilang and Sturm, 1995; Mueller et al., 2000; Gonneau et al., 2001; Smith et al., 2003). However, GSH-dependent catalytic activities are the most important characteristics of GSTs. When GST activity was inhibited by a GST inhibitor, camalexin production was reduced significantly (Figure 2). GSH levels were rapidly reduced in plants undergoing camalexin biosynthesis (Figure 3). The inhibition of GSH production by either a GSH1 inhibitor or a GSH1 gene mutation (MKK9DD/pad2) nearly abolished MKK9-induced camalexin accumulation (Figure 3). Our results support the observation that a lack of GSH biosynthesis in the pad2 mutant causes camalexin deficiency (Parisy et al., 2007). The detoxification of xenobiotics in animal cells is typically accompanied by an upregulation of GSH formation that leads to a net increase in GSH content (Woods and Ellis, 1995; Obrador et al., 1997). However, we did not observe a net increase in GSH content in our experiments, even though expression of GSH1 and GSH2 genes was upregulated (Figure 10). A possible explanation is that the rapid camalexin biosynthesis following MKK9DD induction uses more GSH, and the newly synthesized GSH is not enough to complement the loss of GSH.
IAN is an intermediate for camalexin biosynthesis and is only converted to indole-3-acetic acid when excess IAN accumulates in Arabidopsis (Vorwerk et al., 2001; Nafisi et al., 2007). GSH(IAN), the product of IAN glutathionylation, was identified in MKK9DD plants following MKK9DD induction and in wild-type plants following pathogen infection but was undetectable in control plants. MKK9DD/pad2 and pad2 plants produced low levels of GSH(IAN), and feeding with GSH(IAN) or GSH could partially restore the camalexin deficiency phenotype. Together, these findings suggest that GSH is the Cys derivative and that GST(s) is the enzyme that catalyzes the conjugation of the Cys derivative with the indolic camalexin precursor IAN.
Proteins of the plant GST superfamily have been divided into seven classes, namely, phi (F), tau (U), zeta (Z), theta (T), lambda (L), glutathione-dependent dehydroascorbate reductase, and tetrachloro-p-hydroquinone dehydrogenase-related (Dixon et al., 2010). A variety of plant GST genes are found to be transcriptionally regulated by biotic and abiotic stresses, hormones, and developmental changes (Seppänen et al., 2000; Bianchi et al., 2002; Dixon et al., 2002a; Moons, 2003, 2005; Frova, 2006; Sappl et al., 2009). However, though the roles of GSTs in detoxification of xenobiotics have been well studied (Lamoureux and Rusness, 1993; Coleman et al., 1997), we still know very little about the endogenous functions of individual plant GSTs (Dixon et al., 2010). Arabidopsis contains 54 GSTs that are classified as 13 GSTFs, 28 GSTUs, three GSTTs, two GSTZs, three GSTLs, four glutathione-dependent dehydroascorbate reductases, and one tetrachloro-p-hydroquinone dehydrogenase-related (Dixon et al., 2010). Enzymatic activity assays using recombinant GST proteins with the common substrates have revealed that 41 Arabidopsis GSTs are functional as GSH-dependent enzymes in vitro (Dixon et al., 2002b, 2009; Wagner et al., 2002; Smith et al., 2003; Dixon and Edwards, 2006). In the phi class GSTs (GSTF) of Arabidopsis, GSTF2 was induced by ethylene, auxin, salicylic acid (SA), herbicides, and pathogens (Maleck et al., 2000; DeRidder et al., 2002; Wagner et al., 2002; Lieberherr et al., 2003; Sappl et al., 2009) and involved in auxin transport by binding flavonoids (Smith et al., 2003). GSTF6 was induced by jasmonate, SA, H2O2, and pathogens, and weakly induced by ethylene (Wagner et al., 2002). Knockout of GSTF6 did not show any visible phenotype (Wangwattana et al., 2008), while coreducing the expression of GSTF6/GSTF7/GSTF9/GSTF10 resulted in altered metabolism of carbon and nitrogen compounds (Sappl et al., 2009). GSTF8 (formerly as GST6) was induced by multiple stresses (Chen et al., 1996; Wagner et al., 2002; Sappl et al., 2004, 2009). We showed that both GST activity and GSH are required for MKK9- and B. cinerea–induced camalexin biosynthesis and that GSTF2, GSTF6, and GSTF7 accumulate during camalexin biosynthesis. Additionally, overexpression of GSTF6 significantly increased and knockout of GSTF6 significantly reduced the MKK9- and B. cinerea–induced camalexin production. By feeding the yeast cells expressing Arabidopsis GSTF6 with IAN and GSH, we demonstrated that GSTF6 could catalyze the conjugation of IAN and GSH to form GSH(IAN). These findings suggest that one important function of GSTF6 in planta is to catalyze GSH IAN) formation, thereby participating in camalexin biosynthesis.
GSTF2 and GSTF7 were also highly induced by MKK9 activation. GSTF2 did not catalyze GSH(IAN) formation in yeast cells, and overexpression of GSTF2 or GSTF7 did not elevate camalexin production in transgenic plants. Therefore, GSTF2 may not be involved in MKK9-induced camalexin biosynthesis. MKK9 activation was previously shown to induce ethylene biosynthesis and ethylene responses (Xu et al., 2008; Yoo et al., 2008), and GSTF2 was reported to be induced by multiple stimuli, including ethylene. Thus, GSTF2 induction may be caused, at least partially, by ethylene signaling. GSTF7 was previously reported to be significantly induced by SA and H2O2 (Sappl et al., 2004, 2009). However, further studies are needed to determine whether MKK9 induces GSTF7 accumulation through SA or H2O2 signaling.
Cys(IAN) is an intermediate in the camalexin biosynthetic pathway and serves as a substrate of the multifunctional enzyme CYP71B15 to form camalexin through a two-step reaction (Böttcher et al., 2009). Glu and Gly are subsequently released from GSH(IAN) by certain enzyme(s) to form Cys(IAN). Feeding the MKK9DD/pad2 and pad2 plants with either γGluCys(IAN) or (IAN)CysGly could restore camalexin production, suggesting that both γGluCys(IAN) and (IAN)CysGly are intermediates downstream of GSH(IAN) in the camalexin biosynthetic pathway. We identified γGluCys(IAN) in plants undergoing camalexin biosynthesis. In mammalian cells, catabolism of glutathione-S-conjugates is initiated by the removal of the N-terminal Glu residue catalyzed by γ-glutamyl transpeptidase, which is the rate-limiting step (Meister, 1995). Two modes of glutathione-S-conjugates catabolism have been proposed in plants (Wünschmann et al., 2010). One mode is that the initial step is the removal of the Glu residue from the N terminus catalyzed by γ-glutamyl transpeptidases, for example, GGTs in Arabidopsis (Ohkama-Ohtsu et al., 2007a, 2007b). The other mode is that the initial step is the removal of the Gly residue from the C terminus catalyzed by PCS, for example, PCSs in Arabidopsis (Beck et al., 2003). Both MKK9- and B. cinerea–induced camalexin production are nearly abolished using a GGT activity inhibitor, suggesting that much of GSH(IAN) catabolism is controlled by GGTs during camalexin biosynthesis. However, we failed to identify (IAN)CysGly from plants undergoing camalexin biosynthesis, possibly because (IAN)CysGly may be unstable during the extraction processes or it might be rapidly converted to another intermediate used for camalexin biosynthesis. Arabidopsis contains three functional GGT-encoding genes, GGT1 to GGT3 (Ohkama-Ohtsu et al., 2007a, 2007b), and two PCS genes, PCS1 and PCS2 (Blum et al., 2007). Analysis of the single gene knockout mutants inoculated with B. cinerea indicated that in comparison with wild-type plants, ggt1, ggt2, and pcs1 mutants produced less camalexin. These findings demonstrated the involvement of GGT1, GGT2, and PCS1 in GSH(IAN) catabolism in the camalexin biosynthetic pathway.
Currently, CYP79B2/CYP79B3, CYP71A13, and CYP71B15 are known to function in the camalexin biosynthesis pathway (Glawischnig et al., 2004; Schuhegger et al., 2006; Nafisi et al., 2007; Böttcher et al., 2009). However, overexpression of these individual genes did not lead to camalexin formation (Glawischnig et al., 2004; Schuhegger et al., 2006). This suggests that all of the genes within camalexin biosynthetic pathway need to be activated to keep the pathway on. This conclusion is supported by several lines of evidence. During camalexin biosynthesis in Arabidopsis induced by pathogen infection, AgNO3 treatment, or activation of MAPK cascades, transcription of all of the above-mentioned genes are coordinately activated, and loss of function of any one gene causes camalexin deficiency (Glawischnig et al., 2004; Schuhegger et al., 2006; Nafisi et al., 2007; Qiu et al., 2008; Ren et al., 2008; Xu et al., 2008; Böttcher et al., 2009). Recently, activation of MAPK cascades was shown to induce diterpenoid phytoalexin formation through activation of diterpenoid pathway genes in rice (Kishi-Kaboshi et al., 2010). In this study, we demonstrated that GSTs, GGTs, and AtPCS are involved in the camalexin biosynthetic pathway through catalysis of GSH conjugation to an indolic camalexin intermediate and further catabolism of the GSH conjugate. As revealed by qPCR, GSTF6, GGT1, GGT2, and PCS1, which function in catalyzing camalexin biosynthesis as indicated in this study, were also coordinately upregulated in MKK9-induced camalexin biosynthesis. These findings suggest that activation of the complete pathway is required to produce camalexin.
Based on previous studies and our data, we proposed a modified model for the camalexin biosynthetic pathway (Figure 11; Böttcher et al., 2009). After subsequent conversion of Trp to IAN by CYP79B2/CYP79B3 and CYP71A13, GSH is conjugated to IAN by GSTs, including GSTF6, to form GSH(IAN). Oxidation of the methylene group adjacent to the nitrile by certain enzymes, for example, cytochrome P450 as speculated previously (Böttcher et al., 2009), is likely still needed before the conjugation reaction. Either the N-terminal Glu residue is released from GSH(IAN) to form (IAN)CysGly by GGT1 and GGT2 or the C-terminal Gly is released from GSH(IAN) to form γGluCys(IAN) by PCS1. The remaining amino acid residues, except for Cys, are released from γGluCys(IAN) and (IAN)CysGly by currently unidentified enzymes, such as dipeptidase, to form Cys(IAN). Cys(IAN) is then converted to DHCA and finally to camalexin by CYP71B15. However, a single gene knockout of GSTF6, GGT1, GGT2, or PCS1 could not completely abolish camalexin biosynthesis, possibly due to functional redundancy of the genes. Identification of the functionally redundant genes, generation of double or triple mutants, and identification of the genes required for the conversion of γGluCys(IAN) and (IAN)CysGly to Cys(IAN) will be the focus of future studies.
Figure 11.
Proposed Model of the GSH- and IAN-Dependent Camalexin Biosynthetic Pathway.
METHODS
Plant Materials
Arabidopsis thaliana (ecotype Col-0) wild-type, mutants, and transgenic seeds were surface sterilized. After cold treatment at 4°C for 3 d, the seeds were germinated on plates with 0.5× Murashige and Skoog (MS) with 1% agar and 1% sucrose. For soil growth, seedlings were transferred from plates to soil and grown at 22°C in a growth room with a 12-h photoperiod at a photon flux density of 100 μE/m−2 s−1. For camalexin assays, seedlings were transferred to liquid culture medium with 0.5× MS, 0.025% MES, and 0.25% sucrose and were treated as previously described (Xu et al., 2008). For inhibitor treatments, 12-d-old seedlings were pretreated and 5 × 105 Botrytis cinerea spores per vial, and 2 μM DEX was added.
Agrobacterium tumefaciens-Mediated Transformation
Agrobacterium-mediated Arabidopsis transformation was performed using the flower-dipping method (Clough and Bent, 1998). Transgenic plants were selected on 0.5× MS-agar plates with 15 mg/L hygromycin or 50 mg/L kanamycin. To test the expression of transgenes, the fully expanded leaves from 4-week-old soil-grown plants were taken and quick frozen in liquid nitrogen and stored at −80°C until use.
T-DNA Insertional Mutants and Genetic Crosses
MKK9DD, MKK9DD/mpk3, and MKK9DD/mpk6 mutant plants were generated as previously described (Xu et al., 2008). The T-DNA insert mutants used in this study were obtained from the ABRC. Homozygous null mutants were screened using genomic PCR, and gene knockouts were confirmed by RT-PCR using gene-specific primers. Mutants of pad2 and gstf6 were crossed onto the MKK9DD background, and the transgenic plants were selected using genomic PCR and MKK9DD expression. The GSTF2-, GSTF6-, and GSTF7-overexpression transgenic plants were generated on either the wild-type or MKK9DD backgrounds by Agrobacterium-mediated transformation. Homozygous plants were screened by hygromycin and kanamycin resistance and transgene expression. The sequences of primers are listed in Supplemental Table 1 online.
Vector Constructs
Total RNA was isolated from samples with Trizol reagent (Invitrogen). Reverse transcription was performed using oligo dT(16) as a primer and total RNA as the template. M-MLV reverse transcriptase (Promega) was used to reverse transcribe the poly(A+) mRNAs.
Coding regions of the indicated GST genes were obtained by PCR and cloned into pGEM-T easy vectors. The primer pairs used are as follows: GSTF2-OF and GSTF2-OB for GSTF2, GSTF6-OF and GSTF6-OB for GSTF6, and GSTF7-OF and GSTF7-OB for GSTF7. GST genes with an NdeI site added before the first ATG were cloned into a modified pBluescript vector with a Flag-epitope tag coding sequence at the 5′-end. The GST genes with Flag-epitope tag coding sequences were inserted into a modified pBI121 binary vector with SpeI/XhoI cloning sites. The Ω sequence from tobacco mosaic virus was placed before the Flag-epitope tag coding sequence. All resultant constructs were electroporated into Agrobacterium.
For the yeast expression vector construct, coding regions of the GSTF2, GSTF6, and GFP genes were obtained by PCR. The primer pairs used were GSTF6-YF and GSTF6-YB for GSTF6, GSTF2-YF and GSTF2-YB for GSTF2, and GFP-YF and GFP-YB for GFP. PCR fragments were further digested with BamHI/XhoI and inserted into pYES2 vector. The resultant constructs were transformed into yeast (pJ69a strain) by heat shock.
For the Escherichia coli expression vector construct, coding regions of the GST genes in the pGEM-T easy vector were digested with NdeI/SalI and inserted into pET28a(+). The resultant constructs were transformed into E. coli (BL21 strain) by heat shock.
The sequences of primers used for vector constructs are listed in Supplemental Table 1 online.
Real-Time Quantitative RT-PCR
Twelve-day-old seedlings were used for real-time quantitative RT-PCR (qPCR). Total RNA was isolated from MKK9KR, MKK9DD, MKK9DD/mpk3, MKK9DD/mpk6, and MKK9DD/gstf6 plants with Trizol reagent (Invitrogen). Reverse transcription was performed as mentioned above. qPCR was performed in the presence of SYBR Green Mix (Takara). Amplification was monitored in real time with a 7500 real-time PCR system (Applied Biosystems). The expression level was normalized to the UBIQUITIN5 (UBQ5) control. Primers used for the experiments are listed in Supplemental Table 1 online.
2D Gel and Image Analysis
Samples were ground to a fine powder in liquid nitrogen. The powder was added to a tube with precooled 12.5% TCA/acetone (1% β-mercaptoethanol), vortexed briefly, and incubated overnight at −20°C. After centrifugation at 5000g at 4°C for 10 min, the supernatants were discarded and the pellets were washed twice with precooled acetone (with 1% β-mercaptoethanol). After incubation at −20°C for 30 min, samples were centrifuged at 5000g at 4°C for 10 min. Final pellets were vacuum freeze-dried and resuspended in IEF rehydration solution (8.75 M urea, 2.5 M thiourea, 2.5% CHAPS, 25 mM DTT, trace of bromophenol blue, and 0.5% ampholine [Pharmalite 4-7; GE Healthcare Life Sciences]) to extract the proteins. The solution was centrifuged at 12,000g at 25°C for 10 min, and the supernatants were used for 2D gel electrophoresis. Total protein concentrations were estimated using 2D Quant kits (GE Healthcare Life Sciences). Protein (1 mg) in rehydration buffer was loaded onto 18-cm IEF strips (pH 4 to 7; GE Healthcare Life Sciences). Strips were rehydrated for 18 h at room temperature. IEF was performed on a Bio-Rad IEF Protean cell at 20°C. When IEF was complete, strips were equilibrated as previously described (Giribaldi et al., 2007). Proteins in the strips were separated on 10% polyacrylamide gels. Gels were stained with a blue silver staining method as previously described (Candiano et al., 2004), scanned with a UMAX2000 scanner, and analyzed with PD-Quest 7.2 software (Bio-Rad).
Protein Identification by MALDI-TOF MS
In-gel protein digestion was performed using trypsin (modified sequencing grade; Roche) as previously described with minor modifications (Giribaldi et al., 2007). Excess trypsin solution was removed, and gel pieces were submerged in working solution (50 mM NH4HCO3 and 1 mM CaCl2) and incubated at 37°C for 11 h. After brief centrifugation, the supernatants were collected. Gel pieces were extracted with working solution for 15 min with sonication. After brief centrifugation, the supernatant was collected. The extraction step was repeated twice. Supernatants were pooled and concentrated by freeze-drying. Samples were loaded onto an AnchorChip target plate (Bruker Daltonics) and dried. Matrix solution (0.5 μL), containing 1 mg/mL of α-cyano-4-hydroxycinnamic acid dissolved in 70% acetonitrile and 0.1% trifluoroacetic acid, was loaded onto the surface of the dried peptides. After fast evaporation, the peptide microcrystalline was washed twice with 0.1% trifluoroacetic acid and analyzed using an AutoFlexII TOF/TOF mass apectrometer (Bruker Daltonics). Measurements were performed in a positive ion reflection mode with a 20-kV accelerating voltage and 150-ns delayed extraction time. Spectrum masses of 800 to 4000 D were acquired with laser shots at 60/spectrum. All mass spectra were externally calibrated using a peptide calibration standard.
Monoisotopic peaks were collected and used for peptide fingerprinting identification with FlexAnalysis 3.0 software (Bruker Daltonics). Proteins were identified by searching the National Center for Biotechnology Information nonredundant database with the MASCOT search engine (http://www.matrixscience.com/). The following search parameters were applied: trypsin as cleaving enzyme, peptide mass tolerance 30 ppm, and one missed cleavage allowed. Carbamidomethylation of Cys and oxidation of Met were set as fixed modifications.
Immunoblot Assays
Protein extraction, separation, and immunoblot assays were performed as previously described (Xu et al., 2008). An anti-Flag M2 monoclonal antibody (1:10,000) was used as the primary antibody, and a horseradish peroxidase–conjugated goat anti-mouse antibody (1:10,000) was used as the secondary antibody. The protein membranes were visualized using an enhanced chemiluminescence kit (Roche) and exposed to x-ray films.
GST Activity Assays
GSTs were extracted and their activities were assayed using a GST Colorimetric Activity Assay kit (BioVision) following the manufacturer’s instructions. CDNB was used as the substrate. Protein concentrations of the GST extracts were determined using a protein assay kit (Bio-Rad) with BSA as the standard.
E. coli cells transformed with pET28a(+) constructs were inoculated in Luria-Bertani broth containing 50 mg/L kanamycin with 200 rpm shaking until the OD600 reached 0.4. Cultures were incubated at 28°C for an additional 8 h after the addition of isopropyl d-thiogalactopyranoside to a final concentration of 0.25 mM. The recombinant GST proteins were affinity purified using a Ni2+-chelating Sepharose Fast Flow column (Amersham Bio-Sciences). The activities of the purified recombinant GST proteins toward CDNB were measured as described above.
GSH Concentration Determination
GSH contents were determined using a Glutathione Colorimetric Detection kit (BioVision) following the manufacturer’s instructions with minor modifications. Briefly, 50 mg of plant materials were ground to a fine powder in liquid nitrogen, and 400 μL GSH buffer was added. One hundred microliters of 5% sulfosalicylic acid solution was then added and mixed gently. Samples were incubated on ice for 5 min and centrifuged at 10,000g at 4°C for 15 min. The supernatants were collected and serially diluted five times prior to GSH detection. Diluted samples or GSH standards (20 μL) were mixed gently with 160 μL of the reaction solution in microtiter plates. Substrate [20 μL of 5,5′-dithiobis(2-nitrobenzonic acid)] was added to each sample, and absorbencies at 420 nm were recorded per minute. GSH content was calculated according to the manufacturer’s instructions.
Identification of GSH(IAN), γGluCys(IAN), and (IAN)CysGly
Plant materials (100 mg) were ground into fine powder in liquid nitrogen. After the addition of 400 μL precooled extraction solution (methanol/formic acid, 99.875/0.125, v/v), the samples were sonicated for 15 min at room temperature and centrifuged at 20,000g at 4°C for 10 min. Chromatographic separations of the supernatants were performed on an HP1100 liquid chromatograph (Agilent Technologies). A Luna C18 (2) precolumn (4 × 2.0 mm; Phenimenex) and a Luna 3u C18 (2) 100A analytic column (150 × 2.0 mm; Phenimenex) were used at a flow rate of 200 μL/min. The mobile phase used 0.1% formic acid (v/v) in ultrapure water as solvent A and 0.1% formic acid in acetonitrile (v/v) as solvent B. The solvent gradient program used was 0 min, 95% A; 0 to 20 min, linear gradient 95 to 25% A; 20 to 25 min, 25% A; 25 to 26 min, linear gradient 25 to 95% A; and 26 to 30 min, 95% A. The eluted compounds were detected by a Bruker Esquire 6000 ion trap mass spectrometer with an electrospray ionization source. Vaporization was achieved with a nitrogen sheath gas (25 p.s.i.) and drying gas (10 L/min) at 300°C. Ions in the mass-to-charge (m/z) ratio range of 100 to 1000 were detected in positive mode, and the intermediates were identified according to their m/z [GSH(IAN) at 462.17, γGluCys(IAN) at 405.13, and (IAN)CysGly at 333.21]. GSH(IAN), γGluCys(IAN), and (IAN)CysGly were synthesized and used as standards (see Supplemental Methods online).
In Vivo Feeding with GSH, GSH(IAN), γGluCys(IAN), and (IAN)CysGly
Wild-type, pad2, and MKK9DD/pad2 seedlings growing in liquid media were used for these experiments. The seedlings were incubated with indicated compounds for 4 h before treatments. The wild-type and pad2 seedlings were then treated with B. cinerea spores (5 × 105 spores per vial), and MKK9DD/pad2 seedlings were treated with 2 μM DEX. Camalexin content in culture media was measured as previously described (Xu et al., 2008).
GST-Catalyzed GSH(IAN) Formation in Yeast
Yeast cells were grown to log phase at an OD600 of 0.8 in Ura-minus medium (F&Q) containing 1% glucose. Cells were collected by centrifugation at 6000g for 10 min. Cell pellets were resuspended and cultured overnight in Ura-minus medium containing 2% galactose and 1% raffinose to induce transgene expression. Cells were diluted to an OD600 of 0.2, and 100 μM IAN and 200 μM GSH were added to the medium. Cultures were incubated at 30°C for an additional 10 h with shaking at 200 rpm. Cells were collected by centrifugation, washed twice with milli-Q water, and extracted with extraction solution (methanol/formic acid, 99.875/ 0.125, v/v). The compounds in the solutions were analyzed on a HP1100 liquid chromatograph (Agilent Technologies) coupled to a Bruker Esquire 6000 ion trap mass spectrometer (Bruker Daltonics) with an ESI source as above. Synthetic GSH(IAN) was used as a standard.
The overnight-cultured yeast cells were diluted to an OD600 of 0.2 in Ura-minus medium containing 2% galactose and 1% raffinose and incubated at 30°C for an additional 10 h with shaking at 200 rpm. Cells were collected by centrifugation and washed twice with milli-Q water. Harvested yeast cells were quickly freeze/thawed five times followed by the addition of buffer (0.1 M Tris, pH 7.5, 0.2 M NaCl, 5 mM EDTA, 1 mM PMSF, and 1× protease inhibitors cocktail) and glass beads. Mixtures were vortexed vigorously for 20 s and placed on ice for 40 s. The above steps were repeated until the cells were completely disrupted, and fractions were used for further experiments. IAN and GSH were added to the disrupted cells or the boiled disrupted cells and incubated for 1 h at 30°C on a rotating mixer. The reaction mixtures were then extracted with precooled extraction solution, and the compounds in the solutions were analyzed as mentioned above.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: MKK9, At1g73500; GGT1, At4g39640; GGT2, At4g39650; GSTF6, At1g02930; GSTF7, At1g02920; PCS1, At5g44070; GSH1, At4g23100; GSH2, At5g27380; GSTF9, At2g30860; GSTF10, At2g30870; GSTF11, At3g03190; GSTF12, At5g17220; GSTU16, At1g59700; GSTU17, At1g10370; GSTU18, At1g10360; GSTU27, At3g43800; GSTF2, At4g02520; and UBQ5, At3g62250. T-DNA insertion lines used here are as follows: mpk3 (SALK_100651), mpk6 (SALK_127507), ggt1-1 (SAIL_1178_C01), ggt1-2 (SALK_133807C), ggt2-1 (SAIL_6_G02), ggt2-2 (SALK_139430), gstf6-1 (SALK_026398), and pcs1-1 (SAIL_650_C12).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Measurement of Purified Recombinant GSTF2, GSTF6, and GSTF7 Activities.
Supplemental Figure 2. Immunoblot Analysis of GSTs in Transgenic Plants and Two-Dimensional Electrophoresis Identification of Flag-GSTF6 in MKK9DD/GSTF6 Plants.
Supplemental Figure 3. Two-Dimensional Electrophoresis Identification of the GSTF6-Knockout Mutant and qPCR Detection of Upregulated GST Genes in the Mutant.
Supplemental Figure 4. Growth Curves of Yeast Transformed with Different GSTFs and GFP Genes before or after GSH and IAN Addition.
Supplemental Figure 5. LC-MS Identification of γGluCys(IAN).
Supplemental Figure 6. T-DNA Insertion of the GGT1, GGT2, and PCS1 Mutants.
Supplemental Table 1. Oligonucleotides Used in This Study.
Supplemental Methods. Synthesis of γGluCys(IAN), (IAN)CysGly, and GSH(IAN).
Supplementary Material
Acknowledgments
We thank Chengbin Xiang (University of Science and Technology of China, Anhui, China) and David J. Oliver (Iowa State University, Ames, IA) for providing the ggt2-1 mutant. This work was supported by grants from the National Natural Science Foundation of China (30721062, 31030010, and 30870220) to D.R. and the National Natural Science Foundation of China (30771124) to H.Y.
References
- Axarli I.A., Rigden D.J., Labrou N.E. (2004). Characterization of the ligandin site of maize glutathione S-transferase I. Biochem. J. 382: 885–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck A., Lendzian K., Oven M., Christmann A., Grill E. (2003). Phytochelatin synthase catalyzes key step in turnover of glutathione conjugates. Phytochemistry 62: 423–431 [DOI] [PubMed] [Google Scholar]
- Bianchi M.W., Roux C., Vartanian N. (2002). Drought regulation of GST8, encoding the Arabidopsis homologue of ParC/Nt107 glutathione transferase/peroxidase. Physiol. Plant. 116: 96–105 [DOI] [PubMed] [Google Scholar]
- Bilang J., Sturm A. (1995). Cloning and characterization of a glutathione S-transferase that can be photolabeled with 5-azido-indole-3-acetic acid. Plant Physiol. 109: 253–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum R., Beck A., Korte A., Stengel A., Letzel T., Lendzian K., Grill E. (2007). Function of phytochelatin synthase in catabolism of glutathione-conjugates. Plant J. 49: 740–749 [DOI] [PubMed] [Google Scholar]
- Böttcher C., Westphal L., Schmotz C., Prade E., Scheel D., Glawischnig E. (2009). The multifunctional enzyme CYP71B15 (PHYTOALEXIN DEFICIENT3) converts cysteine-indole-3-acetonitrile to camalexin in the indole-3-acetonitrile metabolic network of Arabidopsis thaliana. Plant Cell 21: 1830–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouizgarne B., et al. (2006). Early physiological responses of Arabidopsis thaliana cells to fusaric acid: Toxic and signalling effects. New Phytol. 169: 209–218 [DOI] [PubMed] [Google Scholar]
- Candiano G., Bruschi M., Musante L., Santucci L., Ghiggeri G.M., Carnemolla B., Orecchia P., Zardi L., Righetti P.G. (2004). Blue silver: A very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis 25: 1327–1333 [DOI] [PubMed] [Google Scholar]
- Chen W., Chao G., Singh K.B. (1996). The promoter of a H2O2-inducible, Arabidopsis glutathione S-transferase gene contains closely linked OBF- and OBP1-binding sites. Plant J. 10: 955–966 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colcombet J., Hirt H. (2008). Arabidopsis MAPKs: A complex signalling network involved in multiple biological processes. Biochem. J. 413: 217–226 [DOI] [PubMed] [Google Scholar]
- Coleman J., Blake-Kalff M., Davies E. (1997). Detoxification of xenobiotics by plants: Chemical modification and vacuolar compartmentation. Trends Plant Sci. 2: 144–151 [Google Scholar]
- Dalton D.A., Boniface C., Turner Z., Lindahl A., Kim H.J., Jelinek L., Govindarajulu M., Finger R.E., Taylor C.G. (2009). Physiological roles of glutathione S-transferases in soybean root nodules. Plant Physiol. 150: 521–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRidder B.P., Dixon D.P., Beussman D.J., Edwards R., Goldsbrough P.B. (2002). Induction of glutathione S-transferases in Arabidopsis by herbicide safeners. Plant Physiol. 130: 1497–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devys M., Barbier M., Kollmann A., Rouxel T., Bousquet J.-F. (1990). Cyclobrassinin sulphoxide, a sulphur-containing phytoalexin from Brassica juncea. Phytochemistry 29: 1087–1088 [Google Scholar]
- Dixon D., Lapthorn A., Edwards R. (2002a). Plant glutathione transferases. Genome Biol. 3: REVIEWS3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon D.P., Davis B.G., Edwards R. (2002b). Functional divergence in the glutathione transferase superfamily in plants. Identification of two classes with putative functions in redox homeostasis in Arabidopsis thaliana. J. Biol. Chem. 277: 30859–30869 [DOI] [PubMed] [Google Scholar]
- Dixon D.P., Edwards R. (2006). Enzymes of tyrosine catabolism in Arabidopsis thaliana. Plant Sci. 171: 360–366 [DOI] [PubMed] [Google Scholar]
- Dixon D.P., Edwards R. (2009). Selective binding of glutathione conjugates of fatty acid derivatives by plant glutathione transferases. J. Biol. Chem. 284: 21249–21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon D.P., Hawkins T., Hussey P.J., Edwards R. (2009). Enzyme activities and subcellular localization of members of the Arabidopsis glutathione transferase superfamily. J. Exp. Bot. 60: 1207–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon D.P., Skipsey M., Edwards R. (2010). Roles for glutathione transferases in plant secondary metabolism. Phytochemistry 71: 338–350 [DOI] [PubMed] [Google Scholar]
- Edwards R., Dixon D.P. (2005). Plant glutathione S-transferases. Glutathione Transferases and gama-Glutamyl Traspeptidases, Sies H., Packer L., (San Diego, CA: Elsevier Academic Press; ), pp. 169–186 [Google Scholar]
- Edwards R., Dixon D.P., Walbot V. (2000). Plant glutathione S-transferases: Enzymes with multiple functions in sickness and in health. Trends Plant Sci. 5: 193–198 [DOI] [PubMed] [Google Scholar]
- Foley R.C., Sappl P.G., Perl-Treves R., Millar A.H., Singh K.B. (2006). Desensitization of GSTF8 induction by a prior chemical treatment is long lasting and operates in a tissue-dependent manner. Plant Physiol. 142: 245–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frova C. (2003). The plant glutathione transferase gene family: Genomic structure, functions, expression and evolution. Physiol. Plant. 119: 469–479 [Google Scholar]
- Frova C. (2006). Glutathione transferases in the genomics era: New insights and perspectives. Biomol. Eng. 23: 149–169 [DOI] [PubMed] [Google Scholar]
- Giribaldi M., Perugini I., Sauvage F.-X., Schubert A. (2007). Analysis of protein changes during grape berry ripening by 2-DE and MALDI-TOF. Proteomics 7: 3154–3170 [DOI] [PubMed] [Google Scholar]
- Glawischnig E. (2007). Camalexin. Phytochemistry 68: 401–406 [DOI] [PubMed] [Google Scholar]
- Glawischnig E., Hansen B.G., Olsen C.E., Halkier B.A. (2004). Camalexin is synthesized from indole-3-acetaldoxime, a key branching point between primary and secondary metabolism in Arabidopsis. Proc. Natl. Acad. Sci. USA 101: 8245–8250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J., Ausubel F.M. (1994). Isolation of phytoalexin-deficient mutants of Arabidopsis thaliana and characterization of their interactions with bacterial pathogens. Proc. Natl. Acad. Sci. USA 91: 8955–8959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonneau M., Pagant S., Brun F., Laloue M. (2001). Photoaffinity labelling with the cytokinin agonist azido-CPPU of a 34 kDa peptide of the intracellular pathogenesis-related protein family in the moss Physcomitrella patens. Plant Mol. Biol. 46: 539–548 [DOI] [PubMed] [Google Scholar]
- Griffith O.W., Meister A. (1980). Excretion of cysteine and γ-glutamylcysteine moieties in human and experimental animal γ-glutamyl transpeptidase deficiency. Proc. Natl. Acad. Sci. USA 77: 3384–3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerschmidt R. (1999). Phytoalexins: What have we learned after 60 years? Annu. Rev. Phytopathol. 37: 285–306 [DOI] [PubMed] [Google Scholar]
- Hull A.K., Vij R., Celenza J.L. (2000). Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc. Natl. Acad. Sci. USA 97: 2379–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi-Kaboshi M., Okada K., Kurimoto L., Murakami S., Umezawa T., Shibuya N., Yamane H., Miyao A., Takatsuji H., Takahashi A., Hirochika H. (2010). A rice fungal MAMP-responsive MAPK cascade regulates metabolic flow to antimicrobial metabolite synthesis. Plant J. 63: 599–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto K., Matsui K., Ozawa R., Takabayashi J. (2006). Analysis of defensive responses activated by volatile allo-ocimene treatment in Arabidopsis thaliana. Phytochemistry 67: 1520–1529 [DOI] [PubMed] [Google Scholar]
- Kurusu T., et al. (2010). Regulation of microbe-associated molecular pattern-induced hypersensitive cell death, phytoalexin production, and defense gene expression by calcineurin B-like protein-interacting protein kinases, OsCIPK14/15, in rice cultured cells. Plant Physiol. 153: 678–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamoureux G.L., Rusness D.G. (1993). Glutathione in the metabolism and detoxification of xenobiotics in plants. Sulfur Nutrition and Assimilation in Higher Plants, De Kok L.J., Stulen I., Rennenberg H., Brunold C., Rauser W.E., (The Hague, The Netherlands: SPB Academic Publishing; ), pp. 221–237 [Google Scholar]
- Lieberherr D., Wagner U., Dubuis P.-H., Métraux J.-P., Mauch F. (2003). The rapid induction of glutathione S-transferases AtGSTF2 and AtGSTF6 by avirulent Pseudomonas syringae is the result of combined salicylic acid and ethylene signaling. Plant Cell Physiol. 44: 750–757 [DOI] [PubMed] [Google Scholar]
- Maleck K., Levine A., Eulgem T., Morgan A., Schmid J., Lawton K.A., Dangl J.L., Dietrich R.A. (2000). The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat. Genet. 26: 403–410 [DOI] [PubMed] [Google Scholar]
- Mann M., Hendrickson R.C., Pandey A. (2001). Analysis of proteins and proteomes by mass spectrometry. Annu. Rev. Biochem. 70: 437–473 [DOI] [PubMed] [Google Scholar]
- MAPK Group (2002). Mitogen-activated protein kinase cascades in plants: A new nomenclature. Trends Plant Sci. 7: 301–308 [DOI] [PubMed] [Google Scholar]
- Meister A. (1995). Glutathione metabolism. Methods Enzymol. 251: 3–7 [DOI] [PubMed] [Google Scholar]
- Mert-Turk F., Bennett M.H., Mansfield J.W., Holub E.B. (2003). Camalexin accumulation in Arabidopsis thaliana following abiotic elicitation or inoculation with virulent or avirulent Hyaloperonospora parasitica. Physiol. Mol. Plant Pathol. 62: 137–145 [Google Scholar]
- Mikkelsen M.D., Hansen C.H., Wittstock U., Halkier B.A. (2000). Cytochrome P450 CYP79B2 from Arabidopsis catalyzes the conversion of tryptophan to indole-3-acetaldoxime, a precursor of indole glucosinolates and indole-3-acetic acid. J. Biol. Chem. 275: 33712–33717 [DOI] [PubMed] [Google Scholar]
- Mishra N.S., Tuteja R., Tuteja N. (2006). Signaling through MAP kinase networks in plants. Arch. Biochem. Biophys. 452: 55–68 [DOI] [PubMed] [Google Scholar]
- Monde K., Sasaki K., Shirata A., Takasugi M. (1990). 4-Methoxybrassinin, a sulphur-containing phytoalexin from Brassica oleracea. Phytochemistry 29: 1499–1500 [Google Scholar]
- Moons A. (2003). Osgstu3 and osgtu4, encoding tau class glutathione S-transferases, are heavy metal- and hypoxic stress-induced and differentially salt stress-responsive in rice roots. FEBS Lett. 553: 427–432 [DOI] [PubMed] [Google Scholar]
- Moons A. (2005). Regulatory and functional interactions of plant growth regulators and plant glutathione S-tansferases (GSTs). Plant Hormones 72: 155–202 [DOI] [PubMed] [Google Scholar]
- Mueller L.A., Goodman C.D., Silady R.A., Walbot V. (2000). AN9, a petunia glutathione S-transferase required for anthocyanin sequestration, is a flavonoid-binding protein. Plant Physiol. 123: 1561–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nafisi M., Goregaoker S., Botanga C.J., Glawischnig E., Olsen C.E., Halkier B.A., Glazebrook J. (2007). Arabidopsis cytochrome P450 monooxygenase 71A13 catalyzes the conversion of indole-3-acetaldoxime in camalexin synthesis. Plant Cell 19: 2039–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H., Pitzschke A., Hirt H. (2005). Emerging MAP kinase pathways in plant stress signalling. Trends Plant Sci. 10: 339–346 [DOI] [PubMed] [Google Scholar]
- Obrador E., Navarro J., Mompo J., Asensi M., Pellicer J.A., Estrela J.M. (1997). Glutathione and the rate of cellular proliferation determine tumour cell sensitivity to tumour necrosis factor in vivo. Biochem. J. 325: 183–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkama-Ohtsu N., Radwan S., Peterson A., Zhao P., Badr A.F., Xiang C., Oliver D.J. (2007b). Characterization of the extracellular γ-glutamyl transpeptidases, GGT1 and GGT2, in Arabidopsis. Plant J. 49: 865–877 [DOI] [PubMed] [Google Scholar]
- Ohkama-Ohtsu N., Zhao P., Xiang C., Oliver D.J. (2007a). Glutathione conjugates in the vacuole are degraded by γ-glutamyl transpeptidase GGT3 in Arabidopsis. Plant J. 49: 878–888 [DOI] [PubMed] [Google Scholar]
- Parisy V., Poinssot B., Owsianowski L., Buchala A., Glazebrook J., Mauch F. (2007). Identification of PAD2 as a γ-glutamylcysteine synthetase highlights the importance of glutathione in disease resistance of Arabidopsis. Plant J. 49: 159–172 [DOI] [PubMed] [Google Scholar]
- Pitzschke A., Schikora A., Hirt H. (2009). MAPK cascade signalling networks in plant defence. Curr. Opin. Plant Biol. 12: 421–426 [DOI] [PubMed] [Google Scholar]
- Qiu J.-L., et al. (2008). Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J. 27: 2214–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauhut T., Luberacki B., Seitz H.U., Glawischnig E. (2009). Inducible expression of a Nep1-like protein serves as a model trigger system of camalexin biosynthesis. Phytochemistry 70: 185–189 [DOI] [PubMed] [Google Scholar]
- Ren D., Liu Y., Yang K.-Y., Han L., Mao G., Glazebrook J., Zhang S. (2008). A fungal-responsive MAPK cascade regulates phytoalexin biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 105: 5638–5643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M.C.S., Petersen M., Mundy J. (2010). Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 61: 621–649 [DOI] [PubMed] [Google Scholar]
- Sappl P.G., Carroll A.J., Clifton R., Lister R., Whelan J., Harvey Millar A., Singh K.B. (2009). The Arabidopsis glutathione transferase gene family displays complex stress regulation and co-silencing multiple genes results in altered metabolic sensitivity to oxidative stress. Plant J. 58: 53–68 [DOI] [PubMed] [Google Scholar]
- Sappl P.G., Oñate-Sánchez L., Singh K.B., Millar A.H. (2004). Proteomic analysis of glutathione S -transferases of Arabidopsis thaliana reveals differential salicylic acid-induced expression of the plant-specific phi and tau classes. Plant Mol. Biol. 54: 205–219 [DOI] [PubMed] [Google Scholar]
- Schuhegger R., Nafisi M., Mansourova M., Petersen B.L., Olsen C.E., Svatos A., Halkier B.A., Glawischnig E. (2006). CYP71B15 (PAD3) catalyzes the final step in camalexin biosynthesis. Plant Physiol. 141: 1248–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppänen M.M., Cardi T., Borg Hyökki M., Pehu E. (2000). Characterization and expression of cold-induced glutathione S-transferase in freezing tolerant Solanum commersonii, sensitive S. tuberosum and their interspecific somatic hybrids. Plant Sci. 153: 125–133 [DOI] [PubMed] [Google Scholar]
- Sheehan D., Meade G., Foley V.M., Dowd C.A. (2001). Structure, function and evolution of glutathione transferases: implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem. J. 360: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.P., Nourizadeh S.D., Peer W.A., Xu J., Bandyopadhyay A., Murphy A.S., Goldsbrough P.B. (2003). Arabidopsis AtGSTF2 is regulated by ethylene and auxin, and encodes a glutathione S-transferase that interacts with flavonoids. Plant J. 36: 433–442 [DOI] [PubMed] [Google Scholar]
- Thomma B.P.H.J., Nelissen I., Eggermont K., Broekaert W.F. (1999). Deficiency in phytoalexin production causes enhanced susceptibility of Arabidopsis thaliana to the fungus Alternaria brassicicola. Plant J. 19: 163–171 [DOI] [PubMed] [Google Scholar]
- Townsend D.M., Manevich Y., He L., Hutchens S., Pazoles C.J., Tew K.D. (2009). Novel role for glutathione S-transferase pi. Regulator of protein S-glutathionylation following oxidative and nitrosative stress. J. Biol. Chem. 284: 436–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji J., Jackson E.P., Gage D.A., Hammerschmidt R., Somerville S.C. (1992). Phytoalexin accumulation in Arabidopsis thaliana during the hypersensitive reaction to Pseudomonas syringae pv syringae. Plant Physiol. 98: 1304–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji J., Zook M., Somerville S.C., Last R.L., Hammerschmidt R. (1993). Evidence that tryptophan is not a direct biosynthetic intermediate of camalexin in Arabidopsis thaliana. Physiol. Mol. Plant Pathol. 43: 221–229 [Google Scholar]
- Vorwerk S., Biernacki S., Hillebrand H., Janzik I., Müller A., Weiler E.W., Piotrowski M. (2001). Enzymatic characterization of the recombinant Arabidopsis thaliana nitrilase subfamily encoded by the NIT2/NIT1/NIT3-gene cluster. Planta 212: 508–516 [DOI] [PubMed] [Google Scholar]
- Wagner U., Edwards R., Dixon D.P., Mauch F. (2002). Probing the diversity of the Arabidopsis glutathione S-transferase gene family. Plant Mol. Biol. 49: 515–532 [DOI] [PubMed] [Google Scholar]
- Wangwattana B., Koyama Y., Nishiyama Y., Kitayama M., Yamazaki M., Saito K. (2008). Characterization of PAP1-upregulated glutathione S-transferase genes in Arabidopsis thaliana. Plant Biotechnol. 25: 191–196 [Google Scholar]
- Woods J.S., Ellis M.E. (1995). Up-regulation of glutathione synthesis in rat kidney by methyl mercury. Relationship to mercury-induced oxidative stress. Biochem. Pharmacol. 50: 1719–1724 [DOI] [PubMed] [Google Scholar]
- Wünschmann J., Krajewski M., Letzel T., Huber E.M., Ehrmann A., Grill E., Lendzian K.J. (2010). Dissection of glutathione conjugate turnover in yeast. Phytochemistry 71: 54–61 [DOI] [PubMed] [Google Scholar]
- Xu J., Li Y., Wang Y., Liu H., Lei L., Yang H., Liu G., Ren D. (2008). Activation of MAPK kinase 9 induces ethylene and camalexin biosynthesis and enhances sensitivity to salt stress in Arabidopsis. J. Biol. Chem. 283: 26996–27006 [DOI] [PubMed] [Google Scholar]
- Yoo S.-D., Cho Y.-H., Tena G., Xiong Y., Sheen J. (2008). Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature 451: 789–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Williams C.C., Last R.L. (1998). Induction of Arabidopsis tryptophan pathway enzymes and camalexin by amino acid starvation, oxidative stress, and an abiotic elicitor. Plant Cell 10: 359–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Cai Z., Guo Y., Gan S. (2009). An Arabidopsis mitogen-activated protein kinase cascade, MKK9-MPK6, plays a role in leaf senescence. Plant Physiol. 150: 167–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zook M. (1998). Biosynthesis of camalexin from tryptophan pathway intermediates in cell-suspension cultures of Arabidopsis. Plant Physiol. 118: 1389–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zook M., Hammerschmidt R. (1997). Origin of the thiazole ring of camalexin, a phytoalexin from Arabidopsis thaliana. Plant Physiol. 113: 463–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.