This work identifies 10 putative Populus orthologs of Arabidopsis genes that encode structurally different 1,3-β-glucanases and shows that they localize at and around plasmodesmata. These enzymes are differently regulated by daylength, temperature, GA3, and GA4, providing a mechanistic explanation of how cell communication is modulated during the dormancy cycling in synchrony with the seasons.
Abstract
In trees, production of intercellular signals and accessibility of signal conduits jointly govern dormancy cycling at the shoot apex. We identified 10 putative cell wall 1,3-β-glucanase genes (glucan hydrolase family 17 [GH17]) in Populus that could turn over 1,3-β-glucan (callose) at pores and plasmodesmata (PD) and investigated their regulation in relation to FT and CENL1 expression. The 10 genes encode orthologs of Arabidopsis thaliana BG_ppap, a PD-associated glycosylphosphatidylinositol (GPI) lipid-anchored protein, the Arabidopsis PD callose binding protein PDCB, and a birch (Betula pendula) putative lipid body (LB) protein. We found that these genes were differentially regulated by photoperiod, by chilling (5°C), and by feeding of gibberellins GA3 and GA4. GA3 feeding upregulated all LB-associated GH17s, whereas GA4 upregulated most GH17s with a GPI anchor and/or callose binding motif, but only GA4 induced true bud burst. Chilling upregulated a number of GA biosynthesis and signaling genes as well as FT, but not CENL1, while the reverse was true for both GA3 and GA4. Collectively, the results suggest a model for dormancy release in which chilling induces FT and both GPI lipid-anchored and GA3-inducible GH17s to reopen signaling conduits in the embryonic shoot. When temperatures rise, the reopened conduits enable movement of FT and CENL1 to their targets, where they drive bud burst, shoot elongation, and morphogenesis.
INTRODUCTION
Developmental transitions in plants rely on endogenous mechanisms that are triggered by environmental cues. A well-studied transition is that of flowering, in which the shoot apical meristem (SAM) transforms into an inflorescence meristem that produces the inflorescence and floral buds. The perennial SAM has the additional and unique capacity to assume a dormant and freezing tolerant state in response to a declining photoperiod (Weiser, 1970; Welling et al., 1997; Rinne et al., 2001). The first signs preceding this response are the cessation of growth and the emergence of a bud, events that unfold rapidly, particularly in northern ecotypes (Böhlenius et al., 2006).
The sequence of events that culminates in photoperiod-induced transitions relies on remote communication, and research on flowering has identified the signaling peptide FLOWERING LOCUS T (FT) as a major long-distance signal. FT is produced in the vasculature of leaves by action of CONSTANS (CO) and sent out to molecular targets at the apex (An et al., 2004; Corbesier et al., 2007; Lin et al., 2007; Tamaki et al., 2007). In perennials, FT also functions in the seasonal arrest, but in a negative fashion as short photoperiod (short day [SD]) rapidly downregulates FT in the leaves (Böhlenius et al., 2006; Hsu et al., 2006; Ruonala et al., 2008). In Arabidopsis thaliana, the closely related but apex-expressed gene TERMINAL FLOWER1 (TFL1) is upregulated during bolting (Bradley et al., 1997), while its Populus ortholog CENTRORADIALIS-LIKE1 (CENL1) is downregulated during the cessation of stem elongation (Ruonala et al., 2008). Both TFL1 and CENL1 are expressed in a small domain just below the SAM. TFL1 encodes a putative signaling peptide that functions non-cell-autonomously during the transition to flowering in Arabidopsis (Bradley et al., 1997; Conti and Bradley, 2007), and the CENL1 peptide might function similarly during the transition to dormancy in Populus (Ruonala et al., 2008). As FT and TFL1/CENL1 both move through plasmodesmata (PD) in the postphloem track, these conduits may also modulate signal flow within the apex.
Crucially, PD in the apex become functionally and structurally modified during the gradual unfolding of the SD response. Early during SD exposure the size exclusion limit of PD in the SAM is lowered (Rinne et al., 2001; Ruonala et al., 2008), perhaps compromising the sink function of the SAM during the formation of a compressed embryonic shoot within the developing bud. After elongation has ceased and bud formation is completed, the SAM arrests itself in a dormant state, disconnecting all its cells by hermetically closing the PD with callosic sphincters (Rinne and van der Schoot, 1998; Rinne et al., 2001; Ruonala et al., 2008).
The early lowering of the size exclusion limit might reflect a gradual shift in callose turnover around the PD orifices, which is thought to be a factor in PD regulation (Lucas et al., 1993; Levy et al., 2007b; Maule, 2008; Epel, 2009). Net callose deposition is governed by the joint action of 1,3-β-glucansynthase (callose synthase) and 1,3-β-glucanase (glucan hydrolase family 17 [GH17]) (Rinne et al., 2005; Levy et al., 2007a; Simpson et al., 2009). Shifts in the balance of these enzymes are central to many developmental events and cellular defense responses (Rinne and van der Schoot, 2003). Dormancy is unique in the sense that the balance remains shifted toward net deposition as long as dormancy lasts, and callose is deposited not only extracellularly around the PD but also intracellularly inside the PD (Rinne et al., 2001).
Dormancy is a state that prevents growth and development under growth-promoting conditions, and release from this state requires nonfreezing chilling temperatures (Vegis, 1964). Such conditions prevail in autumn, and, as a consequence, dormant buds are often released in advance of winter. The effect of chilling is dual, however, and chilling of dormant buds also initiates freezing tolerance, permitting survival through winter (Rinne et al., 2001). Dependent on species and ecotype, temperatures from 0 to 8°C that last for 250 to 2000 h are required (Saure, 1985). Even within a single plant requirements vary, however,influencing crown architecture and productivity (Arora et al., 2003).
Despite the beneficial effects of chilling on dormant buds, it has remained largely elusive which cell biological processes are induced. The classic notion that chilling is nontransportable and that the bud or SAM itself must perceive chilling (Vegis, 1964) is in agreement with the observation of physical isolation of SAM cells during dormancy (Rinne and van der Schoot, 1998). To hydrolyze PD callose, chilling may thus recruit GH17 members locally in the dormant bud (Rinne et al., 2001). As de novo gene expression and protein synthesis might be challenging under conditions of low temperature and dehydration, part of the release mechanism might be installed in advance of winter freezing (i.e., during SD perception) only to be triggered by chilling (Rinne and van der Schoot, 2003).
Central in this putative mechanism are lipid bodies (LBs) that under SD are pinched off from the endoplasmic reticulum in high numbers, both in the SAM and rib meristem (RM) (Rinne et al., 2001). LBs store triacylglycerols and improve freezing tolerance (Siloto et al., 2006). Although the stored and readily available lipids may support membrane repair and bud burst in spring, LBs also carry proteins, among which are putative GH17 members. During chilling, they are collectively displaced to the cell walls, often in close proximity to callose deposits at PD (Rinne et al., 2001). Subsequent hydrolysis of PD callose and reestablishment of dye coupling between SAM cells may support the hypothesis that prior to bud burst LBs function in dormancy release (Rinne and van der Schoot, 2003). The identity and precise cellular function of the LB-associated GH17s and that of most other GH17 family members have remained elusive. However, a PD-associated Arabidopsis GH17 protein (BG_ppap) has been identified that may hydrolyze callose at PD (Levy et al., 2007a).
Hormones have been associated with dormancy based on concentration measurements and on expression analysis of hormone biosynthesis and hormone-responsive genes. Gibberellic acids (GAs) are particularly important, as they may function in the timing of dormancy establishment and chilling induced release (Hazebroek et al., 1993; Zanewich and Rood, 1995; Schrader et al., 2004). Interestingly, in both vernalization and dormancy release, GA application may substitute for chilling (Saure, 1985), although in the case of dormancy results have been equivocal. Thus, GAs could potentially recruit GH17 genes to release dormancy, reopen symplasmic paths, and kick start growth of the embryonic shoot.
We tested whether both GA3 and GA4 could replace chilling of dormant buds in hybrid aspen (Populus tremula × Populus tremuloides). Over 100 putative GH17 genes were identified in the Populus trichocarpa genome, 10 of which were selected for expression studies, including one lacking the GH17 family domain. The selection was based on homology to recently characterized genes encoding cell wall proteins in Arabidopsis (Bayer et al., 2006; Levy et al., 2007a) and a putative LB GH17 of birch (Betula pendula; Rinne et al., 2001, 2008). We analyzed the expression patterns of Populus FT and CENL1, which are sequentially downregulated during the dormancy cycle (Böhlenius et al., 2006; Ruonala et al., 2008). Similarly, we examined several Populus GA biosynthesis genes, two GA receptor genes, a DELLA-like growth repressor gene, and a GA-response gene, GIP-like1.
Our results show that SD, chilling, GA3, and GA4 differentially regulate the selected genes. Some GH17 genes were responsive to SD and/or chilling, while others, mostly growth-expressed, were not. Chilling affected GA biosynthesis by upregulating specific members of the GA3-oxidase and GA20-oxidase families, whereas GA feeding downregulated them. Moreover, chilling hyperinduced FT, but CENL1 was unaffected. CENL1 was upregulated only in sufficiently chilled buds after transfer to 18°C and prior to bud burst. By contrast, GA3 and GA4 feeding dramatically increased CENL1, bypassing natural chilling requirement, but only GA4 rapidly induced bud burst and elongation. Nevertheless, in the natural situation, GA3 (or its analog GA1) is likely to be the GA required for dormancy release as chilling upregulates the same LB-associated GH17s. We present a heuristic model that depicts components of the mechanism that drives dormancy cycling in response to environmental factors. The model highlights the importance of signal conduits and shows that effective signaling with FT and CENL1 is a function of both signal production and conduit regulation.
RESULTS
Chilling Impacts Differently on Embryonic Leaves, Bud Scales, and RM/SAM
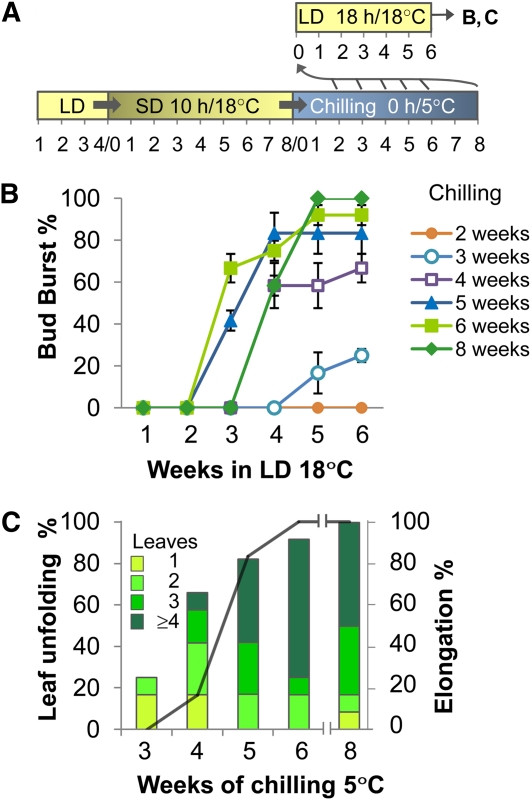
Although it is generally known that chilling at 0 to 8°C is sufficient to alleviate dormancy, chilling requirements are variable and species specific. To determine the chilling requirements for hybrid aspen, we placed dormant plants at 5°C and at weekly intervals tested their bud burst capacity under growth promoting conditions (Figure 1A). This revealed that chilling differentially affected the various parts of the dormant bud. While 3 weeks of chilling only occasionally promoted growth of the one to two oldest embryonic leaves (Figure 1B), a minimum of 4 weeks was required to induce a more complete opening and unfolding of more embryonic leaves. This stage was more readily achieved by buds from lower leaf axils. However, canonical or true bud burst, which results in internode elongation after leaves have unfolded, was only found after five or more weeks of chilling (Figure 1C). Six weeks was commonly optimal for dormancy removal as it always led to bud burst, internode elongation, and leaf initiation. When released buds were kept longer than 6 weeks at 5°C, it delayed bud burst (Figure 1B). In conclusion, bud scales and existing embryonic leaves were the first to respond to chilling, whereas the initiation of morphogenesis required in total about 6 weeks or ~1000 h.
Figure 1.
Characteristics of Bud Burst after Chilling-Induced Release from Dormancy.
(A) Conditions for dormancy cycling (long bar) and bud burst testing (short bar). The numbers under each bar refer to weeks under indicated conditions in LDs, SDs, and chilling. After 2, 3, 4, 5, 6, and 8 weeks of chilling, bud burst capacity was tested at LD 18 h/18°C (arrows to short upper bar); results of the tests are shown in (B) and (C).
(B) Timing and degree of bud burst following 1 to 8 weeks of chilling (5°C) (except for week 7) and subsequent bud burst testing under LDs as indicated in (A). No bud burst was achieved until after 2 weeks of chilling (first week not shown). Values are means (±se) of three to five biological replicates, with 18 to 30 buds per time point.
(C) The quality of bud burst in relation to time in chilling conditions. Quality refers to leaf unfolding and stem elongation and was monitored after a 6-week growth period under LDs as indicated in (A). Bar colors in (C) indicate the number of leaves unfolding from bursting buds. The solid line indicates the percentage of buds that showed elongation growth.
Dormancy Release by Chilling Is Related to Effects of GA4 and GA3
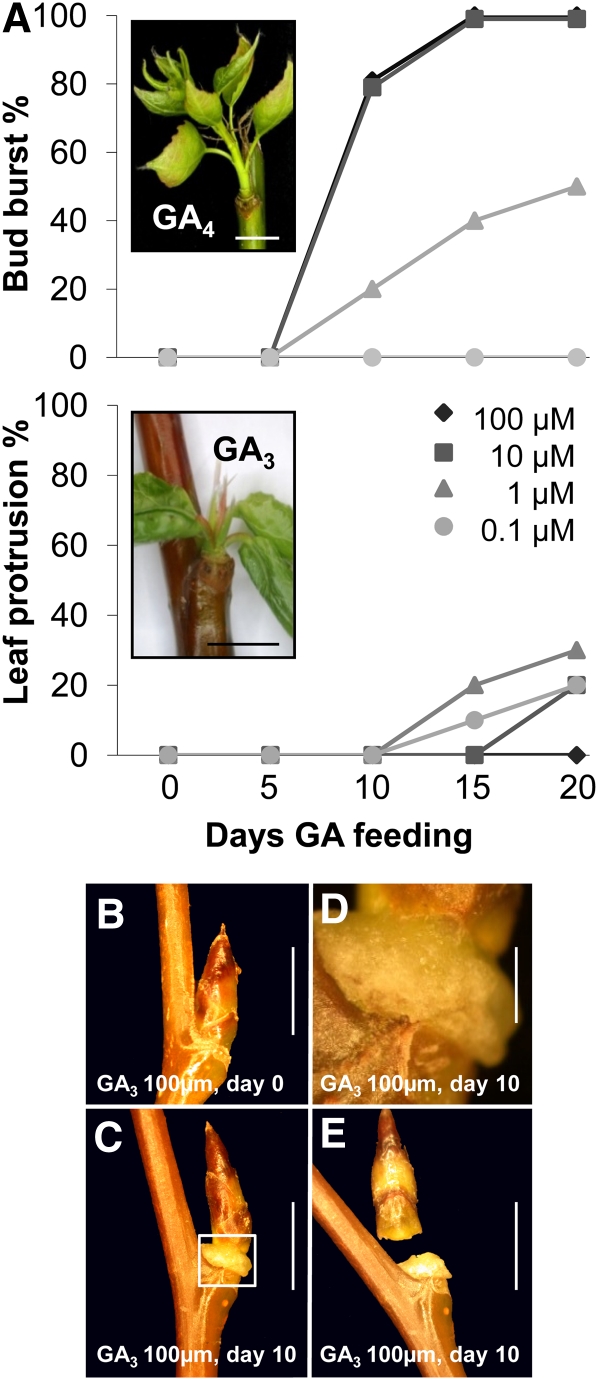
Reports that GA application can substitute for chilling in dormancy release suggest that chilling recruits GA in dormant buds. As the literature is ambiguous regarding this point, we investigated if the type of GA is important. GA4 and GA3, as well as its analog GA1, are the main biologically active GAs in shoot elongation, but the relative roles of the various forms are unclear (Eriksson et al., 2006; Yamaguchi, 2008). GA4 and GA1 are synthesized in parallel pathways from the respective precursors, GA12 and GA53, after a few oxidations by GA20-oxidase (GA20ox) and a final oxidation by GA3-oxidase (GA3ox) (Yamaguchi, 2008). Because in hybrid aspen GA4 is thought to be the main GA in shoot elongation (Israelsson et al., 2004), we selected GA4 and GA3 to compare their capacity to substitute for chilling. The results showed that GA4 induced canonical bud burst and development in a concentration-dependent way. Bud swelling started after 5 d, and buds were clearly elongating by day 10, while GA3 failed to induce the same response (Figure 2A). At high concentrations (100 μM), GA3 induced a disorganized cell mass at the base of the bud, resembling a callus (Figures 2B to 2E). The callus-like tissue formed at the junction of bud and stem, probably at leaf bases, and often resulted in bud abscission (Figure 2E), suggesting that the junction between bud scales and stem is very sensitive to GA3 feeding. Lower GA3 concentrations caused less abscission and an occasional protrusion of existing embryonic leaves. None of the GA3 concentrations used promoted canonical bud burst (Figure 2A). The GA4-induced shoots appeared morphologically normal, although their leaves were smaller and paler than those emerging from sufficiently chilled buds.
Figure 2.
Capacity of GA3 and GA4 to Induce Dormancy Release/Bud Burst and to Initiate Development.
(A) Bud burst after incubation in 0.1 to 100 μM GA4 and GA3. With higher concentrations of GA4, most buds show canonical or true bud burst (i.e., elongating internodes) and de novo formation of leaves (inset) at around 10 d. Low concentrations of GA3 (0.1 to 10 μM) could induce leaf protrusion and unfolding, which was not accompanied by formation of leaves and stem elongation (inset).
(B) to (E) Abscission events induced by 100 μM GA3. Within 1 to 2 weeks after the initial application of GA3 to dormant buds (B), cell proliferation was visible at the base of the bud (C), producing a callus-like cell mass (detail in [D]), along which buds often abscised (E).
Bars = 5 mm in (A) to (C) and (E), and 1 mm in (D).
Chilling and GA4 Recover the Impaired Transport Capacity of Dormant Buds
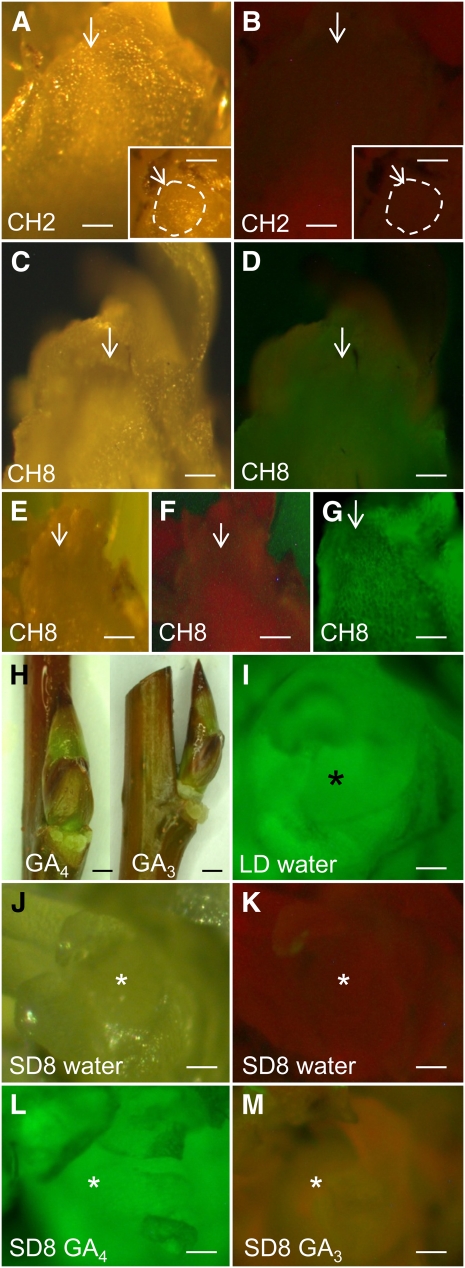
As dormancy is enforced by the physical obstruction of symplasmic paths in both the phloem (Aloni et al., 1991; Aloni and Peterson, 1997) and the apex (Rinne et al., 2001), we investigated if the dormancy releasing effect of chilling and GA4 is based on the reopening of these paths. We therefore monitored the accessibility of the transport paths between stem and apex at various dormancy phases by investigating the potential movement of the fluorescent tracer dye calcein into the apex (Figure 3). We compared bud-internode units taken from plants that were exposed to insufficient (2 weeks, dormant) and sufficient chilling (8 weeks, released from dormancy). In addition, we used bud-internode units that were preexposed to GA3/GA4 for 5 d, at which point bud burst was not yet visibly initiated. The results showed that calcein was not transported in insufficiently chilled plants (Figures 3A and 3B) resembling the situation in dormant plants (Figures 3J and 3K), while calcein did reach the apex in fully chilled plants (Figures 3C and 3D). Pretreatment of the internodes with 100 μmol GA4 (Figures 3H and 3L) improved entry of fluorescent tracers into the apex up to the level found in proliferating apices (Figure 3I) and, to a lesser degree, in fully chilled buds (Figure 3D). By contrast, pretreatment with 10 to 100 μmol GA3 did not promote any calcein transport to the apex (Figure 3M). Thus, transport paths in the symplasm, cell walls, or both were obstructed during dormancy, but chilling and GA4 were able to reverse this situation.
Figure 3.
Effect of Chilling and GA3 and GA4 Preincubation on Transport of the Fluorescent Dye Calcein (0.1%) to the Shoot Apex.
(A) and (B) Longitudinally cut shoot apex from 2 weeks chilled plants (CH2). Arrow points to the SAM under white light (WL) (A) and blue violet light (BVL) (B).
(B) No fluorescence is visible in the apex indicating absence of calcein import. Insets show a surface view of the SAM (encircled) before it was cut longitudinally.
(C) to (G) Longitudinally cut shoot apex from 8 weeks chilled plants (CH8) and viewed under WL ([C] and [E]) and BVL ([D], [F], and [G]).
(D) Calcein fluorescence is visible in the apex but has not entered all leaves.
(F) No calcein incubation. Note the greenish autofluorescence of the veins.
(G) Calcein was directly pipetted onto the cut apical surface. The hue is dimmer when the dye is transported via the stem (D).
(H) to (M) The bases of the bud-internode units (H) were preincubated in 10 μM GA4, GA3, or water (control) for 5 d before transfer to calcein and viewed under WL (J) and BLV ([I] and [K] to [M]).
(I) Nondormant LD apex preincubated in water. Intense green calcein fluorescence at the SAM.
(J) to (M) Shoot apex from dormant bud (SD8).
(J) and (K) Preincubation in water. No fluorescence visible in the SAM.
(L) Preincubation in GA4 showing intense green calcein fluorescence.
(M) Preincubation in GA3. Faint greenish fluorescence visible in overarching leaves but not in the SAM.
Arrows in (A) to (G) and asterisks in (I) to (M) indicate SAM. Bars = 50 μm in (A) to (G) and (I) to (M), and 1 mm (H).
Selecting Candidate GH17 Genes Involved in Dormancy Cycling in Populus
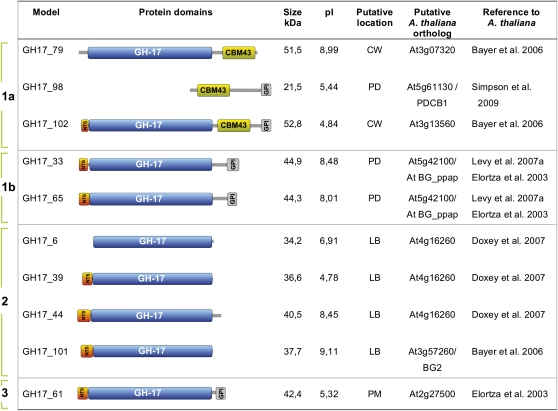
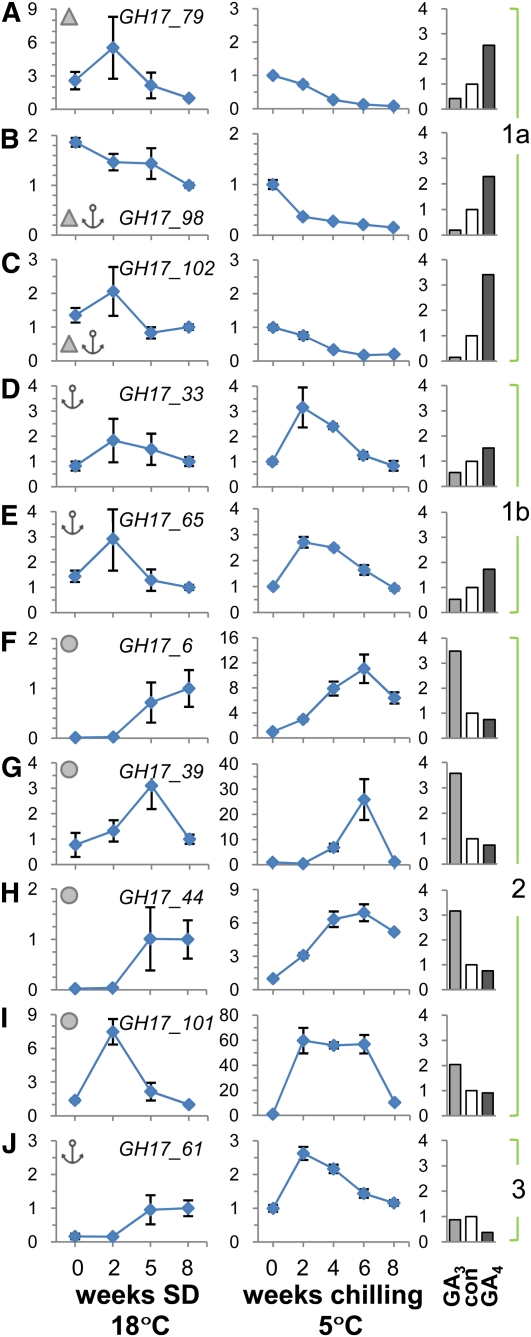
Our previous studies using immunolocalization showed that chilling induced the accumulation of 1,3-β-glucanases (GH17, glucan hydrolase family 17 proteins) in the SAM of birch (Rinne et al., 2001). To identify the PD-related GH17 members in the P. trichocarpa genome, we followed two approaches. First, we performed proteomic studies with isolated birch lipid body proteins (LBPs) (P.L.H. Rinne, unpublished). The sequence information acquired by tandem mass spectrometry sequencing of retained LBPs was compared with a birch expressed sequence tag database (birchEST, University of Helsinki, Finland). This information was used to search orthologs in the P. trichocarpa database. The search resulted in hits for nine putative GH17 genes, five of which had EST support, and four of which (GH17_6, GH17_39, GH17_44, and GH17_101) were chosen for further studies based on preliminary expression studies. In addition, we searched for P. trichocarpa orthologs of Arabidopsis GH17 proteins that have been reported to reside in the cell wall (Bayer et al., 2006) and in the plasma membrane (Elortza et al., 2003). Based on these analyses, we identified another six P. trichocarpa genes. An exception among the encoded proteins is GH17_98, which has a carbohydrate binding module (CBM43), previously known as the X8 domain, and a C-terminal sequence characteristic of many other GH17 proteins, but lacks the GH17 family domain. Nonetheless, GH17_98 is of interest, as Simpson et al. (2009) identified the Arabidopsis ortholog At5g61130 as a PD callose binding protein. The structural organization of the 10 Populus proteins is detailed in Figure 4. The genes encoding cell wall–related GH17 proteins constitute ~10% of all GH17 genes in both Arabidopsis (Doxey et al., 2007) and P. trichocarpa (Geisler-Lee et al., 2006). Phylogenetically, the Arabidopsis GH17 proteins are subdivided into three main clades (α, β, and γ) (Doxey et al., 2007), and our selected P. trichocarpa GH17 genes fall into two of them (see Supplemental Figure 1 and Supplemental Data Set 1 online). The putative Arabidopsis BG_ppap orthologs from P. trichocarpa, GH17_33 and GH17_65, and a related member, GH17_61, are similar to the α-clade of GH17 members like GH17_79 and GH17_102, although the two latter belong to more distant subgroups. Nevertheless, all the P. trichocarpa α-clade members, except GH17_79, have a glycosylphosphatidylinositol (GPI) anchor through which they may attach to the luminal or extracellular surfaces of the plasma membrane, putatively assisting PD association, as in the case of BG_ppap (Levy et al., 2007a). Like GH17_98, GH17_79 and GH17_102 have a CBM43/X8 motif, which may allow them to bind callose at PD (Simpson et al., 2009). Based on structural organization of the Populus GH17 proteins (Figure 4) and expression patterns during dormancy cycling (Figure 5), we refer to these α-clade Populus orthologs as group 1a (GH17_79, GH17_98, and GH17_102), group 1b (GH17_33 and GH17_65), and group 3 (GH17_61).
Figure 4.
Characteristics of Selected Populus GH17 Family and PD-Associated Proteins.
Nine GH17 family members and the GH17_98 protein that lacks the glycosyl hydrolase domain are shown. Proteins were selected based on their homology with Arabidopsis cell wall (CW), plasma membrane (PM), or birch LB-associated GH17 proteins. Protein domain architectures: N-terminal signal peptide (NTS), a carbohydrate binding module family 43 (CBM43), previously known as the X8 domain, the core glycosyl hydrolase family 17 domain (GH-17), and a hydrophobic C-terminal sequence that may encode a transient transmembrane domain involved in GPI anchor attachment (GPI). Size of the proteins is given in kilodaltons. pI is the isoelectric point. Based on structure and expression patterns, proteins were subdivided into three groups.
Figure 5.
qRT-PCR Analysis of Selected Genes Encoding Populus GH17 Family and PD-Associated Proteins.
The analysis was performed with apices and buds of hybrid aspen under conditions that mimic those during the phases of the annual dormancy cycle. The values are means (±se) of three biological replicates and represent fold changes (y axis) of indicated genes.
(A) to (J) Relative expression levels of indicated genes when plants were exposed to dormancy-inducing SD conditions for up to 8 weeks (line graphs on the left) and dormancy-releasing chilling conditions for up to 8 weeks (line graphs in the middle). Bar graphs on the right show fold changes after exposure to 10 μM GA3 and GA4, or water as a control (con), applied for 5 d to internodes with dormant buds. The genes were clustered into the functional groups 1 (subdivided into 1a and 1b), 2, and 3 based on their specific expression patterns and on the structural organization of their proteins.
(A) to (E) Group 1 GH17 members have a putative carbohydrate binding domain CBM43/X8 (triangles) ([A], [B], and [E]) and/or a GPI lipid anchor (anchors) ([B] to [E]).
(F) to (I) Group 2 GH17 members are putative lipid body proteins (circles).
(J) The group 3 member has a putative GPI lipid anchor. Line diagrams represent values relative to a control time point SD8. This time point was included separately in both SD and chilling experiments.
The P. trichocarpa genes with high homology to a birch gene encoding a putative LBP grouped into the γ-clade. They showed sequence similarity with Arabidopsis genes encoding proteins with endomembrane, vacuolar, and/or cell wall localization (e.g., BG1-3; see Bayer et al., 2006) and include the so-called pathogenesis-related group (PR-2) of GH17 proteins (Dong et al., 1991; Uknes et al., 1992). In addition, the LB-related sequences of P. trichocarpa showed similarity to ethylene- and salicylic acid–inducible GH17 enzymes in tobacco (Nicotiana tabacum; Felix and Meins, 1987; Leubner-Metzger et al., 1998). Based on LB association (Figure 4) and expression patterns during dormancy cycling (Figure 5), we refer to these γ-clade Populus orthologs as group 2 GH17 proteins.
GA3 and GA4 Induce Different Chilling-Responsive GH17 Subsets
Quantitative RT-PCR (qRT-PCR) analysis of GH17 genes in hybrid aspen showed that they were differentially affected by SD, chilling, and GA application, indicating that they function in distinct phases of the dormancy cycle. SD affected the expression of all GH17 genes. During the first 2 weeks of SD, most long-day (LD)-expressed GH17 genes with a GPI anchor and/or carbohydrate binding domain, CBM43/X8, belonging to groups 1a and 1b, as well as a member from LB-related group 2 (GH17_101) were transiently upregulated and thereafter downregulated (Figures 5A, 5C to 5E, and 5I). An exception in group 1a was GH17_98 (Figure 5B), which was gradually downregulated under SD. The remaining group 2 and group 3 genes were relatively little expressed under LD, and SD enhanced their expression, but only late in SD, at around the time when dormancy becomes established (Figures 5F to 5H and 5J).
Chilling of dormant seedlings modified the GH17 expression levels that were reached after 8 weeks of SD. Chilling upregulated seven of the GH17 genes, to various degrees and at various time points of the chilling period. Group 1a genes, downregulated around dormancy establishment, were further downregulated by chilling (Figures 5A to 5C), suggesting that these GPI-anchored and/or CBM43/X8-equipped proteins are not part of a chilling-induced dormancy release mechanism. By contrast, the transcripts belonging to GPI-anchored group 1b were sensitive to chilling in that their expression peaked early during chilling (Figures 5D and 5E). A similar early chilling-induced upregulation was observed for another GPI-anchored group 3 member (Figure 5J). Compared with groups 1b and 3, the LB-related group 2 members showed a more gradual and significant upregulation during chilling, with the highest detected expression levels at around 6 weeks (Figures 5F to 5I). In all cases of chilling-enhanced GH17 expression, the levels declined by 8 weeks (Figures 5D to 5J), perhaps due to a reduced availability of substrate at this point in time when dormancy is released (Figure 1; see Supplemental Figure 2 online). Upregulation of GH17 during chilling was highest in case of LB-associated GH17_101 and GH17_39 by 60- and 25-fold, respectively (Figures 5G and 5I). However, the highest normalized expression levels were observed for LB-associated GH17_44, a paralog to GH17_6 (Figure 5H; see Supplemental Figure 3 online).
The expression of the GH17 genes was further examined in dormant buds after GA feeding. The rationale was to establish if the capacity of GA to substitute for chilling is mediated by GH17 expression. While GA3 was not able to induce canonical bud burst (Figures 2 and 3), both GA3 and GA4 had specific effects on GH17 expression. The results showed that groups 1a and 1b responded uniquely to supplied GA3 and GA4. GA4 enhanced expression of group 1 genes, coding for GPI lipid-anchored and/or CBM43/X8-equipped GH17s, while the same transcripts were substantially reduced by GA3 compared with control levels (Figures 5A to 5E). On the other hand, GA3 was efficient in upregulation all LB-related group 2 GH17s (Figures 5F to 5I), while GA4 tended to downregulate them. By contrast, the group 3 GH17 was not upregulated by any GA but rather was downregulated by GA4 (Figure 5J).
Genes for GA Biosynthesis, Deactivation, and Signaling Are Differentially Regulated by Chilling and GA
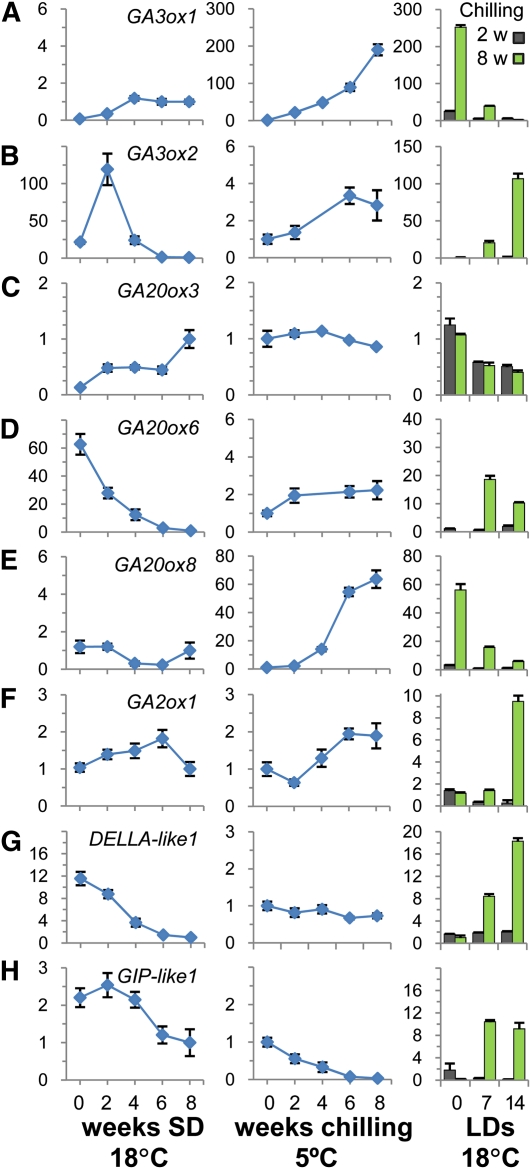
To assess if not only applied GAs but also endogenous GAs could be involved in chilling induced release from dormancy, we analyzed the expression of several GA biosynthesis genes. Transcripts of two genes encoding GA3ox and GA20ox family members (GA3ox1 and GA20ox8), which have very low abundance in the apex under LD and SD, appeared to be upregulated significantly during chilling (Figures 6A and 6E). Notably, other genes of these families were upregulated only during growth and bud burst (Figures 6B and 6D), showing that members of the GA biosynthesis gene families have nonredundant functions during the activity-dormancy cycle. Genes involved in GA deactivation, such as GA2ox1 (Thomas et al., 1999), were also upregulated toward the end of the chilling (Figure 6F) as in the case of GA feeding (>50-fold), which may reflect increased GA biosynthesis during chilling. This would be in line with the view that GA metabolism is under homeostatic control. Interestingly, expression of two Populus orthologs of the rice (Oryza sativa) GA receptor GIBBERELLIN INSENSITIVE DWARF1 (GID1), GID1.1 and GID1.3 (Mauriat and Moritz, 2009), were substantially increased under SD (see Supplemental Figure 4 online), suggesting that the dormant system is sensitized to GA. This was probably further enhanced by downregulation of the gene DELLA-like1 (Figure 6G). Also, a GIBBERELLIN INDUCIBLE PROTEIN-like1 (GIP-like1) (Israelsson et al., 2005) was downregulated late in SDs and further during chilling (Figure 6H). However, their expression levels immediately increased under warmer conditions (18°C), provided sufficient chilling was received (Figures 6G and 6H). After the chilling requirement was fulfilled, at around 6 weeks, prolonged chilling upregulated the GA repressor DELLA-like1, while bud burst was delayed (Figure 1B). Hypothetically, DELLA-like1 and related DELLA-like negative regulators of growth could thus be part of a safety mechanism that prevents precocious canonical burst of nondormant, quiescent buds in spring.
Figure 6.
qRT-PCR Analysis of Selected Populus GA Biosynthesis, Signaling, and Response Genes.
The analysis was performed with apices and buds of hybrid aspen under conditions that mimic those during the phases of the annual dormancy cycle. The values are means (±se) of three biological replicates and represent fold changes (y axis) of indicated genes. Line graphs on the left show relative expression levels in apices when plants are exposed to dormancy-inducing SD conditions for up to 8 weeks. Dormancy is reached after ~6 weeks of SDs. Line graphs in the middle show fold changes during the subsequent dormancy-releasing chilling conditions for up to 8 weeks. Bar graphs on the right show fold changes in top buds during the subsequent 2-week period in forcing conditions (18°C, LD), after insufficient (2 weeks) and sufficient chilling (8 weeks). Bud burst takes place only in the latter case, when dormancy has been fully removed.
(A) and (B) Members of GA3ox family (GA3ox1 and GA3ox2).
(C) to (E) Members of GA20ox family (GA20ox3, GA20ox6, and GA20ox8).
(F) GA catabolizing GA2ox (GA2ox1), selected because of its highest expression level under LDs (see Supplemental Figure 4 online).
(G) Growth repressor gene DELLA-like1.
(H) Gibberellin-responsive gene GIP-like1. Ct values are normalized with an actin gene. Diagrams represent values relative to a control treatment, SD8. This data point is assessed separately in the two experiments.
Expression of FT, CO, and CENL1 during Chilling and Bud Burst
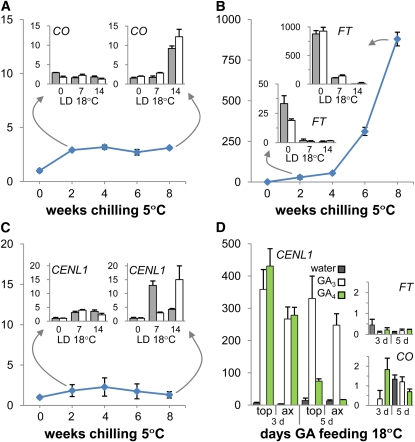
In hybrid aspen, FT is expressed in the vasculature of the leaves, and the protein moves to apex to support elongation growth (Böhlenius et al., 2006). Because under SDs, FT is completely downregulated and all the leaves are shed, this begs the question how and where the CO/FT module is recruited to drive elongation of the embryonic shoot after dormancy release. We investigated if the CO/FT module becomes operational in the leaves of the embryonic shoot itself and found that in the dormant bud, CO was upregulated ~3-fold after 2 weeks of chilling and that it remained at that level throughout the rest of chilling period (Figure 7A). After a sufficient chilling period (8 weeks at 5°C), transfer to LD at 18°C enhanced CO expression further (Figure 7A, inset). In contrast with CO, FT showed a gradual but dramatic chilling-induced upregulation of >800-fold. However, early during bud burst (i.e., within 1 to 2 weeks of LD), FT transcripts decreased to the very low level that is characteristic of growing apices (Figure 7B). This showed that in dormant buds, embryonic leaves are the source of FT and that chilling might activate FT independently from CO.
Figure 7.
qRT-PCR Analysis of CO, FT, and CENL1 during Dormancy Release.
Dormancy was released by 8 weeks of chilling ([A] to [C]) and by GA feeding (D). The values are means (±se) of three biological replicates and represent fold changes (y axis) of indicated genes. The bar graphs in (A) to (C) are snapshots of expression levels in axillary buds (gray bars) and apical buds (white bars) exposed to LDs and higher temperatures (18 h; 18°C) after insufficient (2 weeks) and sufficient (8 weeks) periods of chilling. Values in snapshots are means (±sd) of technical repeats of a pooled sample (three plants). (D) shows fold changes of CENL1, in apical (top) and axillary buds (ax), and fold changes of FT and CO in axillary buds after 3 and 5 d of GA feeding or water relative to nontreated dormant buds.
CENL1, a P. trichocarpa ortholog of Arabidopsis TFL1, encodes a signaling peptide that resembles FT, but its expression domain is in the RM subjacent to the SAM. Throughout the chilling period, CENL1 remained at a very low level (Figure 7C), comparable to that in nonchilled dormant buds (Ruonala et al., 2008). CENL1 was upregulated only after sufficient chilling and subsequent exposure to 18°C (Figure 7C), similarly to genes that function mostly under LDs such as CO, GA3ox2, GA20ox6, GA2ox1, DELLA-like1, and GIP-like1 (Figures 6B, 6D, and 6F to 6H). A 12- to 15-fold peak in CENL1 expression was detected preceding canonical bud burst. The peak occurred somewhat earlier in axillary buds, which also burst earlier than top buds (Figure 7C, inset). CENL1 thus appeared to be a marker of bud burst.
Both GA3 and GA4 strongly promoted upregulation of CENL1 in nonchilled dormant buds, whereas CO and FT expression was unaffected (Figure 7D, insets). Compared with the upregulation of CENL1 by 18°C in buds that have been released from dormancy by chilling (Figure 7C), the upregulation induced by GAs was dramatic. However, like in case of natural bud burst, the GA4-promoted dramatic peak in CENL1 expression was transient and preceded bud burst. In clear contrast, GA3-induced upregulation of CENL1 remained high (Figure 7D) probably due to lack of downstream effects, such as signal movement, that may be required for bud burst. GA3 also induced ectopic CENL1 expression at the bud-stem junction where callus-like tissue formed (Figures 2B to 2E), at levels similar to those in the bud.
GA4- and Chilling-Induced Structural Changes Partly Overlap
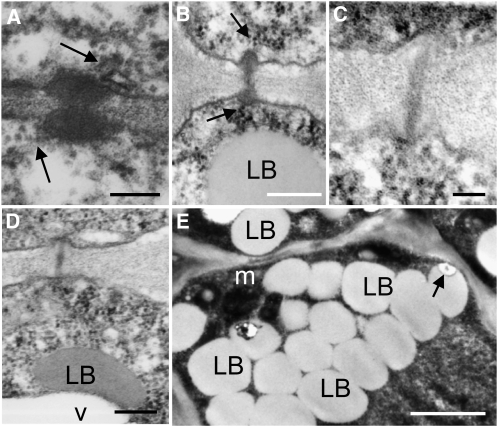
The question arises if GA4-induced release from dormancy, bud break, and elongation involves the LB-based mechanism that was previously proposed (Rinne et al., 2001). Central to this mechanism are preformed SD-induced GH17-decorated LBs that remain nonfunctional until chilling moves them to their targets at the cell wall. We therefore investigated if GA4 feeding affected displacement of GH17-decorated LBs to PD and cell walls. The results revealed that after 5 d of GA4 feeding, when the buds started to swell, LBs were not aligning with the plasma membrane and PD, while the dormancy sphincters that obstruct PD during dormancy had disappeared (Figures 8C and 8D). The large numbers of LBs that characterize the SAM and RM during dormancy (see Supplemental Figure 5 online) had all disappeared from GA4-treated buds. At the earlier 3-d time point, LBs were observed only rarely at PD (Figure 8E). In the rib zone, they were commonly observed to merge with vacuole-like structures. Collectively, and in connection with previous studies on birch (Rinne et al., 2001), the results suggest that GA4-induced PD reopening may not involve LBs and LB-associated group 2 GH17 proteins. Although GA4 feeding was very efficient in inducing bud burst and elongation, it may have impacted negatively on SAM organization. It reduced SAM size while the RM appeared normal or somewhat larger (see Supplemental Figure 5 online), perhaps reflecting the >400-fold increase in CENL1 expression (Figure 7D).
Figure 8.
Transmission Electron Micrographs of Dormant Meristems of Hybrid Aspen after Treatment with GA4.
(A) and (B) Examples of PD between L1 cells (A) and between corpus cells (B) in dormant meristems after 7 d treatment with water. The arrows point to a strand of endoplasmic reticulum at the PD.
(C) and (D) Callose-containing sphincters were no longer visible at PD after 5 d of GA feeding. Examples of PD between corpus cells (C) and rib meristem cells (D) resembling those in actively growing meristems. LBs were typically merging into vacuoles (D).
(E) At an earlier time point of 3 d, LBs are still present, but LB–cell wall connections were scarce.
Bars = 200 nm in (A) to (C), 100 nm in (D), and 1 μm in (E). For overviews, controls, and details, see Supplemental Figure 5 online.
Functional Studies with Populus Groups 1b, 2, and 3 GH17 Members
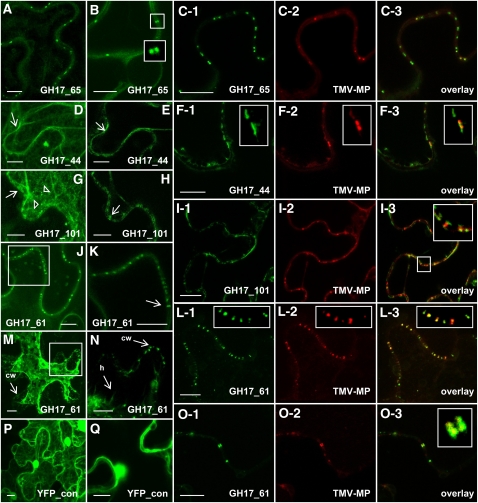
For localization studies, we selected two GPI-anchored GH17s of group 1b (GH17_65) and group 3 (GH17_61) on grounds of their phylogenetic distance and differential regulation by ambient conditions and GA. On the same grounds, we selected two LB-associated GH17 proteins of group 2 (GH17_44 and GH17_101). Constructs encoding GH17–enhanced green fluorescent protein (eGFP) fusions were agroinfiltrated into Nicotiana benthamiana leaves to investigate transient expression and localization. In additional experiments, constructs were coinfiltrated with a second construct encoding the movement protein of Tobacco mosaic virus (TMV MP-RFP) to identify PD sites.
Transiently expressed GH17_65, a putative GPI lipid-anchored protein representative of group 1 GH17s, localized in a punctate pattern to the cell walls (Figures 9A and 9B). Fluorescence was confined to twin spots at opposite sides of the cell walls, indicative of the ends of single PD (Figure 9B, insets). The twin spots colocalized with the red fluorescent punctae of TMV MP-RFP (Figures 9C1 to 9C3). Transiently expressed GH17_44, a putative LB-associated protein of group 2, localized at the plasma membrane (Figures 9D and 9E), often in patches that colocalized with TMV MP at PD (Figures 9F1 to 9F3). Another putative group 2 protein, GH17_101, localized to LB-like cytoplasmic structures as well as to the plasma membrane (Figure 9G), where it sandwiched on the opposite sides of the cell TMV MP (Figures 9I1 to 9I3). The putative GPI lipid-anchored protein GH17_61 localized to the cell wall in a punctate pattern (Figures 9J and 9K) and colocalized with TMV MP (Figures 9L1 to 9L3). This could indicate that the GH17_61, just like the MP of TMV, preferentially accumulates inside the PD channel, whereas the LB-associated GH17_101 tends to cover the channel in small patches (detail in Figure 9I3). That the colocalization with TMV MP was inside the PD was demonstrated further by plasmolyzing cells, which left GH17_61 inside the cell wall after withdrawal of the plasma membrane (Figures 9M and 9N). Further experiments showed that both GH17_61 and the TMV MP could reside simultaneously inside the PD orifices (Figures 9O1 to 9O3). The 35S-expressed free yellow fluorescent protein, used as a control, localized throughout the cell (Figures 9P and 9Q).
Figure 9.
Localization of Hybrid Aspen GH17 Proteins and Colocalization with TMV MP in Epidermal Cells of N. benthamiana Infiltrated with Agrobacterium Carrying GH17_eGFP or/and TMV MP-RFP.
(A) and (B) GH17_65, a GPI lipid-anchored protein of group 1b, localizes at the cell wall in a punctate pattern (A). Twin spots suggest localization at ends of single PD ([B], insets).
(C1) to (C3) Colocalization of GH17_65 with TMV MP at PD.
(D) and (E) GH17_44, a LB-associated protein of group 2, localizes at the plasma membrane (arrow) in a combined Z-stack (D) and in patches (arrow) in an optical section (E).
(F1) to (F3) GH17_44 partly colocalizes with TMV MP at PD; boxed areas are details.
(G) and (H) GH17_101, a LB-associated protein of group 2, localizes in LB-like structures (triangles) and at the plasma membrane (arrow) in a combined Z-stack (G) and in optical section (H).
(I1) to (I3) GH17_101 colocalizes with TMV MP at PD.
(J) and (K) GH17_61, a GPI lipid-anchored protein of group 3 localizes at the cell wall in a punctate pattern (J).
(K) Boxed area in (J) showing GH17_61 in structures that cross the cell wall (arrow).
(L1) to (L3) GH17_61 colocalizes with TMV MP at PD; boxed areas are details.
(M) and (N) GH17_61 remains in the PD after plasmolysis and plasma membrane withdrawal (M); boxed area is detailed in (N). cw, cell wall with fluorescent punctate; h, Hechtian strand.
(O1) to (O3) GH17_61 colocalizes at cell walls in twin spots with TMV MP at PD (inset in [O3]).
(P) and (Q) Control. YFP is distributed unspecifically throughout the cell in a combined Z-stack (P) and in optical sections (Q).
Bars = 10 μm.
DISCUSSION
The cycling of perennial plants through phases of growth and dormancy is based on a series of developmental events that are adjusted to the seasons. Although global expression analysis has identified a number of genes, which are differentially expressed in apices and cambium during SD (Schrader et al., 2004; Ruttink et al., 2007) and dormancy release (Druart et al., 2007; Mathiason et al., 2009), the dormancy cycle has remained poorly characterized, particularly at the level of cellular mechanisms. As illustrated in Figure 10A, the initial response to a critically changing photoperiod is the downregulation of FT in the phloem of the leaves, which deprives the apex of FT and its downstream effects (Böhlenius et al., 2006). At the apex, this is followed by the initiation of scale leaves and an embryonic shoot (Rohde et al., 2002), the downregulation of CENL1 at the RM, and the narrowing of SAM PD (Ruonala et al., 2008). PD narrowing may constrain the capacity of the SAM to function as a sink and contribute to the miniaturization of the shoot system emerging within the bud, as depicted in Figure 10B.
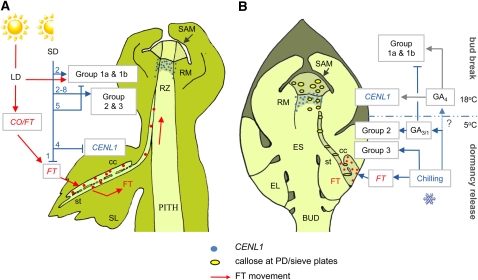
Figure 10.
Heuristic Model of Signal Production and Conduit Regulation during the Dormancy Cycle in Populus.
(A) Dormancy induction trajectory. In LDs (red arrows) the clock-regulated module CO/FT produces FT in the companion cells (cc) of source leaves (SL). FT is loaded into the sieve tubes and transported to the shoot apex, unloaded into the rib zone (RZ), and transported to the RM. Sieve plate pores and plasmodesmata are maintained in a conductive state mainly by group 1a and 1b GH17 proteins (see below). Under SDs (blue arrows), FT is completely downregulated within a single week (1), which eventually results in the establishment of dormancy. At about the second week of SDs (2), members of groups 1 and 2 GH17s are transiently upregulated to maintain symplasmic paths to facilitate bud formation, but downregulated before week 5. Some other GH17 members of groups 2 and 3 become upregulated at this point. CENL1 is downregulated from week 4 onwards, and dormancy is established at week 6.
(B) Dormant bud and the enclosed embryonic shoot (ES). Chilling (blue arrows and bar) upregulates FT in the companion cells of embryonic leaves (EL) and group 3 GH17s. Meanwhile, chilling-enhanced production of GA increases GA3-inducible group 2 GH17s, which remove dormancy callose from sieve plate pores and PD in the RM and SAM, thereby providing functional conduits for FT transport to the RM. After sufficient chilling, elevated temperatures (18°C), mediated by GA4 (gray arrows), upregulate growth-related groups 1a and 1b GH17s. Arrows indicate upregulation, and T-bars indicate downregulation of genes.
During dormancy establishment at the SAM of the embryonic shoot, net deposition of callose around and inside the PD channel obstructs further symplasmic transport and signaling (Rinne and van der Schoot, 1998; Ruonala et al., 2008). In addition, sieve tubes become occluded with dormancy callose (Levy and Epel, 2009). Together, this may preclude precocious return to a proliferative state. Sufficient chilling reverses this situation, and callose hydrolysis reopens symplasmic paths in the apex and the phloem (Aloni et al., 1991; Aloni and Peterson, 1997; Rinne et al., 2001). The GH17s in apex and phloem may be highly similar; those recently isolated from the phloem of Populus (Dafoe et al., 2009) and rice (Hao et al., 2008) closely resemble the group 1 and group 2 proteins described here. Based on homology with Arabidopsis cell wall/plasma membrane proteins (Elortza et al., 2003; Bayer et al., 2006) and birch LBP proteins (Rinne et al., 2008), we identified 10 GH17 candidates for PD callose turnover. The selected GH17 genes, clustered in three ad hoc groups based on their organization (Figure 4), expression patterns (Figure 5), and localization (Figure 9), appeared to function in distinct phases of the activity-dormancy cycle, as illustrated in Figure 10.
Early SD Expression of GH17 Genes and Homeostasis
Genes of group 1 encode enzymes that putatively target the plasma membrane and/or PD via their GPI lipid anchor and/or CBM34 domain (Levy et al., 2007a; Simpson et al., 2009). These genes, predominantly expressed in LDs (Figures 5A to 5E), could function in PD formation and maintenance as well as cell plate formation (Levy and Epel, 2009). The distinctive peak at 2 weeks of SD, typical for group 1 genes as well as for the LD-expressed LB-related group 2 gene GH17_101 (Figure 5I), indicates a temporary increase in the demand for such GH17s. Given the fact that GH17_65 and GH17_101 colocalize with TMV MP at PD (Figures 9A to 9C and 9G to 9I), their peak expression at this early SD time point might serve to temporarily prolong PD function.
This early SD-induced increase in GH17 transcript levels occurs at around the same time as the narrowing of the SAM PD, which at a later stage become occluded with callose during dormancy establishment (Ruonala et al., 2008). An earlier microarray study with Populus apices indicates that transcripts for a callose synthase are upregulated already after 1 week of SD (Ruttink et al., 2007). As callose deposition depends on the relative activities of GH17 enzymes and callose synthases (Rinne and van der Schoot, 2003; Levy et al., 2007a), it seems feasible that the early SD peak in GH17 expression offsets an increase in callose production. If so, it is the callose turnover that is increased in early SD, whereas net callose deposition is postponed until dormancy establishment. The fact that GA4 feeding upregulated the same group 1 GH17 genes suggests that GA4 plays a role in callose breakdown during early SD. This is supported by the upregulation of the GA biosynthesis gene GA3ox2 and the GA-response gene GIP-like1 at this time (Figures 6B and 6H).
The downregulation of several GH17 transcripts later in SD, at around week 5, may tip the balance toward net callose deposition at all PD. Although some GH17 genes of group 2 (Figures 5F to 5H) also become expressed at this time point, the encoded proteins are putatively associated with LB. Since their displacement to the cell wall and PD requires a sufficiently long period of chilling (Rinne et al., 2001), it renders them ineffective under SD alone. The modest SD-induced increase of the group 3 gene, encoding the GPI lipid-anchored protein GH17_61 (Figure 5I) (Geisler-Lee et al., 2006), is puzzling, but it remains to be seen if its movement is compromised by the dehydrated cytoplasmic matrices (Welling et al., 1997) in dormant buds.
Differential Regulation of GH17 Predicts Localization
The question how supplied GA can substitute for chilling in dormancy release has remained unanswered. Possibly, GA production is a downstream effect of chilling, as proposed for dormant seeds (Hazebroek et al., 1993; Yamauchi et al., 2004). Our finding that chilling of dormant buds increased the expression of GA biosynthesis genes (Figures 6A and 6E) is in support of this. The remarkable difference we found in the capacity of GA3 and GA4 to induce canonical bud burst (Figure 2A) might be due, at least in part, to their selective effects on the expression of specific sets of GH17 genes. That different GA species have such distinct effects on gene expression could explain some of the inconsistencies in the reported results of GA treatments in compensating vernalization or chilling in various plant species (Mutasa-Göttgens and Hedden, 2009).
In dormant buds, GA4 feeding upregulated all group 1 genes and completely abolished dormancy without any need for chilling. Of these only a subset, group 1b genes (GH17_33 and GH17_65), were also upregulated by early chilling (Figures 5D and 5E), while GA biosynthesis genes were not yet highly upregulated (Figure 6). This suggests that group 1b genes are regulatable by both GA4 and chilling, while group 1a members (GH17_79, GH17_102, and GH17_98) might require both GA4 and higher temperatures for their expression. GA4-inducible members in group 1 probably substitute for chilling by bypassing the LB-mediated reopening of PD, as the encoded proteins may directly target the plasma membrane aided by a GPI lipid anchor or the CBM43/X8 carbohydrate/callose binding domain (Levy et al., 2007a; Simpson et al., 2009), present in group 1 and group 3 proteins. The GPI lipid anchor may tether these proteins to lipid rafts, which could serve as sorting devices for delivery to the PD channel (Tilsner et al., 2010). It remains to be established if proteins in general reach PD on lipid rafts like the recently characterized PD-located proteins (Amari et al., 2010).
GA3 reduced the expression of all group 1 genes, but in contrast upregulated all group 2 genes, which are putative orthologs of the birch LB-related GH17 (Figures 5A to 5I). Group 2 genes were also upregulated by chilling, but mostly during the later part of the chilling period, concomitant with the upregulation of GA biosynthesis genes (Figures 6A, 6B, and 6E). This indicates that it is GA3, rather than GA4, which is involved in the upregulation of LB-related transcripts during chilling. Despite the fact that GA3 closely mimicked chilling-regulated GH17 transcription, it lacked the capacity, in contrast with GA4, to induce canonical bud burst when supplied to a dormant bud. As in the case of SDs, enhanced expression of GH17 by GA3 alone is insufficient for callose degradation. The group 2 GH17 proteins can only be effective in releasing dormancy (i.e., removing PD callose) in combination with chilling, which displaces LBs to the cell wall (Rinne et al., 2001). Dormancy release thus not only requires the transcription of relevant genes but also the transport of the encoded proteins to their intracellular destinations.
The fact that LB-related group 2 members (GH17_44 and GH17_101) localized at the cell wall in distinct sandwich-like patches (Figures 9D, 9E, 9G, and 9H), not in the punctate pattern typical of GPI lipid-anchored proteins (Figures 9A, 9B, 9J, and 9K), might be characteristic for delivery by LBs which expectedly unload their cargo into a wider area of the plasma membrane. The narrowness of the PD channel might preclude precise docking and fusion of LBs, and group 2 GH17 proteins potentially move to PD through lateral diffusion. GPI lipid-anchored proteins might target lipid rafts and also move via lateral diffusion to PD, as suggested for other PD proteins (Tilsner et al., 2010). LB-associated GH17s were often present over the PD orifices, sandwiching TMV-MP in the PD channel (Figures 9F and 9I, insets). We hypothesize that GPI lipid-anchored GH17s might function extracellularly because the GPI group anchors a protein to the extracellular site of the PM, while the LB-associated GH17s function intracellularly after diffusional entry into the PD channel.
FT and CENL1 Are under Complex Regulation by SD, Temperature, and GA
Previous studies have found that SD reduces GA levels in the young stem, thereby contributing to cessation of stem elongation (Junttila and Jensen, 1988). In support of this, higher GA levels achieved by genetic modifications delay the process (Eriksson et al., 2000). Our results demonstrate that apices respond in more complex ways. For example, GA feeding can upregulate CENL1 (Figure 7D), as does SD exposure (Ruonala et al., 2008). This might be a more general response, as the same factors upregulate CENL1/TFL1 orthologs in pea (Pisum sativum; Wang et al., 2009). Therefore, GA signaling in the apex might actually be increased during the first weeks under SD. This is further supported by the fact that transcript abundance of the GA receptors GID1.1 and GID1.3 in apices increased under SD (see Supplemental Figure 4 online). In case GA levels decrease under SD, this would be counteracted by an increase in GA sensitivity that could account for the transient upregulation of CENL1 (Ruonala et al., 2008) and the GA4-responsive group 1 GH17 genes in early SD (Figures 5A to 5E).
The distinctness of the expression patterns of FT and CENL1, with FT being strongly upregulated during chilling and CENL1 requiring elevated temperatures (Figures 7B and 7C) is mutatis mutandis, also found during early SD exposure (Böhlenius et al., 2006; Ruonala et al., 2008). At first sight, our observation that chilling upregulated GA biosynthesis genes concomitantly with FT seemed to suggest that GA could promote FT transcription. However, GA3 and GA4 failed to upregulate FT in dormant buds, implying that either GA-mediated upregulation of FT requires chilling or that chilling independently promotes FT. Also in Arabidopsis, GA application can directly promote FT transcription in varieties that do not require vernalization (Mutasa-Göttgens and Hedden, 2009), whereas vernalization-induced demethylation of FT repressors is needed to promote flowering in winter annuals of Arabidopsis (Jiang et al., 2008).
CENL1, in contrast with FT, was promptly upregulated in dormant buds by both GA3 and GA4 application without requirement for chilling (Figure 7D). As judged from the elevated expression of GA biosynthesis genes during chilling (Figures 6A, 6B, and 6E), one would expect CENL1 transcripts to accumulate during this treatment, while they do so only at subsequently elevated temperatures. However, the expression domain of CENL1 in Populus (Ruonala et al., 2008) as well as that of TFL1 in Arabidopsis (Bradley et al., 1997) is a small area subjacent to the SAM. This domain overlaps with the expression domain of the GA deactivating enzyme GA2ox (Jasinski et al., 2005; Sakamoto et al., 2001, 2006), which prevents entry of leaf-produced GAs into the SAM (Bolduc and Hake, 2009). The GA2ox1 gene was somewhat upregulated by chilling (Figure 6F), and in principal, high activity of these genes could prevent GA effects on CENL1 at this restricted expression domain, even if GA levels elsewhere rise.
When GA was applied to the base of the bud-internode unit, GA2ox1 was upregulated 50-fold in the dormant bud, indicating that GA2ox functions locally to regulate GA homeostasis. Continuous GA feeding might saturate the capacity of the GA2ox system and disrupt GA homeostasis, thereby enhancing CENL1 expression, like observed here during the first 5 d of GA feeding (Figure 7D). Saturation of the GA2ox system might also be the reason for the observed size reduction of the SAM, as some GA might possibly pass the GA2ox barrier (see Supplemental Figure 5 online). GA3 is inefficient in binding to its receptors compared with GA4 (Ueguchi-Tanaka et al., 2007), and it is not subject to stringent degradation by GA2ox (Sakamoto et al., 2006). Therefore, GA3 might more easily pass through the GA2ox domain on its way to the SAM. The peak of CENL1 expression, occurring prior to bud burst (Figure 7C; Mohamed et al., 2010), may thus be promoted by elevated GA levels in spring, after the chill has subsided.
Modifications of Signaling Conduits Drive Dormancy Cycling
This work shows that applied GA4 is a potent experimental inducer of bud burst of fully dormant buds and that this involves the induction of LD-expressed GH17 genes. However, this situation is unnatural in the sense that GA4 application might induce simultaneous callose removal via PD-localized GPI lipid-anchored GH17 proteins and cell elongation, which are normally separated in time. By contrast, GA3, the slow growth hormone, mimics the natural situation of chilling-induced release from dormancy, in that it promotes the production of the same LB-related GH17 transcripts as chilling does during release from dormancy. In contrast with GA4, GA3-induced GH17 may use LBs as a transport vehicle, which makes their function dependent on chilling.
A heuristic scenario for dormancy release (Figure 10B) is that chilling may trimethylate histones in FT chromatin and simultaneously induce GA biosynthesis. As GA3/1 are less effected by the chilling-upregulated GA2ox, they might be most important at this point. However, FT protein, produced in phloem of embryonic leaves, can initially not move due to the presence of phloem winter callose and callosic PD sphincters in the sink areas of the apex. As chilling upregulates directly GPI lipid-anchored group 1b and group 3 genes, and together with GA3 LB-associated group 2 genes, callose becomes hydrolyzed at the PD and sieve plate pores enabling chilling-produced FT protein to move to the apex. At growth-permissive temperatures, GA4 promotes CENL1 expression at the RM required for elongation of the embryonic shoot. GA4 seems to be the dominant force for the initiation of bud burst due to its capacity to induce a specific subset of GH17 enzymes (group 1) that are expressed under LD. Within the bud, FT and CENL1, genes crucial in vegetative and generative growth, respond differentially to chilling and GA application, probably due to their chromatin status and disjoined expression domains, but both require concerted action of GH17 proteins at sieve plate pores and PD to reach their targets.
METHODS
Plant Material and Growing Conditions
Hybrid aspen (Populus tremula × Populus tremuloides) clone T89 used in the experiments has a critical daylength of ~16 h (Böhlenius et al., 2006). It was micropropagated in vitro, planted in a mixture of sterilized peat and perlite (4:1 [v/v]), and fertilized with 4 g L−1 osmocote (Substral; Scotts). Plants were grown in a greenhouse under LDs (18 h light) at 18°C and 80% relative humidity (RH) and watered twice a day. Natural light was supplemented to a level of 200 μmol m−2 s−1 at 400 to 750 nm (Osram). After 4 weeks of LD, the 40- to 50-cm-tall plants were exposed to a short photoperiod (SD) of 10 h to induce growth cessation and dormancy. In natural conditions, temperatures may gradually fall off during the shortening of photoperiod. As SD and chilling have opposite effects on dormancy, thereby confounding their individual effects, we selected experimental conditions that allowed us to study both phenomena in separation. We exposed plants to 8 weeks of SD at constant 18°C, after which the dormant plants were exposed for 8 weeks to 5°C in darkness. The experimental design is illustrated in Figure 1A.
Dormancy Induction and Testing
Dormancy establishment under SD and dormancy release through chilling were assessed in bud burst experiments using a stringent culture system of bud-internode units under forcing conditions (18 h of LD with a PPFD of 200 μmol m−2 s−1, 18°C, and 85% relative humidity). At 1-week intervals, stems of SD-exposed plants were cut into segments, with the terminal or axillary bud at the higher end of a 1- to 2-cm-long internodal segment. The bases of the bud-internode units were supported by Styrofoam, allowing them to float with only the lower ends in solutions. Bud burst was scored and categorized based on the number of unfolding leaves and the capacity of the embryonic shoot to elongate during the 6-week observation period (Figure 1A). Dormancy status is expressed in bud burst percentage and elongation capacity. Three to five plants (18 to 30 buds) were used per time point.
GA Feeding
The potential capacity of GA to substitute for chilling in dormancy release was tested using the same stringent bud-internode unit system as used for the dormancy test (described above). The water medium was supplemented with GA3 (Sigma-Aldrich) or GA4 (Lew Mander). GA was supplied to the bud via the stem vasculature and not directly on the bud. GA1, its analog GA3, and GA4 are the main biologically active GAs in shoot elongation (Yamaguchi, 2008). We used GA3 and GA4 at concentrations ranging from 100 nM to 100 μM. As in case of dormancy, bud burst percentage and bud elongation were monitored. Three to five plants (18 to 30 buds) were used per test solution. Applications were repeated at least three times with similar results.
Probing the Conductivity of PD and Vascular Tissues with a Fluorescent Dye
As long-distance transport through both phloem and xylem cease during dormancy, we examined the capacity of the vasculature system in bud-internode units to conduct solutes to the apex. We used the fluorescent dye calcein added at a final concentration of 0.1%. In preliminary tests, it was established that calcein in this concentration was moving visibly and quickly to the apex of internode cuttings isolated from LD plants. For dormant systems, a 24-h incubation period was used to maintain a wide margin. The bud-internode units were placed in solutions so that only the base of the internode was in contact with the solution to make sure fluorescent probe only moved to the apex via the stem vasculature. Three to five plants (18 to 30 buds) were used per test solution. Applications were repeated two times with similar results.
RNA Extraction and Real-Time qRT-PCR Analysis
RNA was extracted from apices, and top and axillary buds of plants that were exposed to various environmental conditions from bud-internode cuttings treated with GAs. Sampling was performed during the last hour of the light period under SD and at the same time during chilling in darkness. Material for each time point was collected from 9 to 15 individual plants. Axillary buds were pooled from the four uppermost locations for further processing. RNA was extracted from 0.2 g frozen tissue and grinded in a mortar with 750 μL extraction buffer (Qiagen RTL buffer containing 1% polyvinylpyrrolidone-40). After addition of a 0.4 volume KoAC at pH 6.5 and further grinding, the solution was transferred to a 2-mL tube, incubated on ice for 15 min, and centrifuged at 13,400 rcf at 4°C for 15 min. The supernatant was transferred to a new 1.5-mL tube and a 0.5 volume of 100% ethanol was added. The mix was transferred to two RNeasy spin columns and further processed in accordance with instructions of the Qiagen Plant RNA isolation kit. RNA was DNase (Ambion) treated, cleaned using the total RNA purification system Purelink RNA mini kit (Invitrogen), and reverse transcribed using SuperScriptIII reverse transcriptase (Invitrogen). Real-time qRT-PCR analyses were performed with the ABI Prism 7500 Fast sequence detection system using SYBR Green PCR master mix (Applied Biosystems). RNA was in most cases extracted twice from each sample, both of which were analyzed in triplicate. Transcript levels were normalized using Populus actin. Gene-specific primer sequences for the qRT-PCR analysis were designed using Primer3 (http://frodo.wi.mit.edu/primer3/input.htm) (see Supplemental Table 1 online). qRT-PCR data in Figures 5 to 7 are based on three biological replicates, analyzed in three technical repeats, except for analyses of GA-treated buds and bud burst at 18°C. In these cases, one pooled sample was analyzed, which was divided into two before extraction and analysis.
Light Microscopy and Electron Microscopy
Dormant buds were incubated for 5 d with and without GA3 and GA4 as described above (GA applications) and thereafter fixed overnight at 4°C in 2% (v/v) glutaraldehyde and 3% (v/v) paraformaldehyde in 100 mM phosphate citrate buffer, pH 7.2, as described earlier (Rinne et al., 2005). The samples were infiltrated gradually into LR White Resin and kept 4 d in 100% before polymerization at 55°C. For light microscopy 1- to 3-μm median longitudinal sections were stained with 1% aqueous Toluidine Blue. Ultrathin sections were taken from median longitudinal positions and examined at 60 kV with a transmission electron microscope (Jeol 1200-EXII).
Transient Expression Studies
To create fusion protein constructs, the coding sequences of hybrid aspen GH17_44 and GH17_101, 1116 and 1029 bp, respectively, were PCR amplified in two steps by gene-specific primers with partial attB1 and attB2 sites and subsequently with attB1/B2adapters for Gateway (Invitrogen) recombination reaction. The cDNA template for amplification was obtained from an apex tissue previously described (Ruonala et al., 2008). The obtained attB-products for GH17_44 and GH17_101 were cloned into the pDONR221 donor vector to create entry clones. Binary vector pK7FWG2,0 (Karimi et al., 2002) with eGFP was used as a destination vector to create the final GH17_44-eGFP and GH17_101-eGFP fusion protein constructs (C-terminal fusion to protein). Since GH17_61 and GH17_65 are GPI-anchored proteins, we used overlap PCR and Gateway technology (Tian et al., 2004) to tag eGFP internally to the coding sequences of GH17_61 and GH17_65. To amplify eGFP, the pK7FWG2,0 binary vector was used as a PCR template. The coding sequences of GH17_61 and GH17_65 were amplified with two sets of gene-specific primers, P1/P2 and P3/P4. P1 and P2 primers amplified the coding sequences from the start codon to the site preceding the predicted GPI anchor, 1041 and 1134 bp for GH17_61 and GH17_65, respectively. For both GH17s, P3 and P4 primers amplified first 30 bp of assumed low complexity region and the GPI anchor site to the stop codon, 108 and 105 bp for GH17_61 and GH17_65, respectively. Additionally, P1 and P4 primers extended the amplification products with the sequences that partially overlapped the Gateway primers (TT-GW-for and TT-GW-rev), while P2 and P3 primers extended the products with the sequences that overlapped eGFP. The three templates (eGFP, P1/P2, and P3/P4 PCR products) were joined by triple-template PCR with the Gateway primers as described (Tian et al., 2004), cloned into the pDONR221 donor vector, and linearized with EcoRV to eliminate kanamycin resistance and to confirm right insert sizes in entry plasmids. Binary vector pMDC32 (Curtis and Grossniklaus, 2003) was used as a destination vector to create the final fluorescent-tagged GH17_61-eGFP and GH17_65-eGFP constructs. All the primers used for vector constructions are listed in Supplemental Table 2 online. The sequence integrities of all constructs were confirmed by sequencing.
Agrobacterium tumefaciens strain CV3101 with pSoup was grown overnight at 5 mL of 28°C in L broth supplemented with 100 mg L−1 kanamycin (GH17_61, GH17_65, and TMV MP) or spectinomycin (GH17_101 and GH17_44), 50 mg L−1 gentamycin, and 20 mg L−1 rifampicin to a stationary phase. For infiltration, bacteria were sedimented by centrifugation at 3000g for 10 min at room temperature and resuspended in 10 mM MgCl2 and 200 μM acetosyringone to an approximate cell density of OD600 0.7. Cells were left in this medium for 2 to 3 h and then infiltrated into the abaxial air spaces of 2- to 4-week-old Nicotiana benthamiana plants. For colocalization, bacteria carrying TMV and GH17 plasmids were mixed in equal volumes after adjusting the OD600 to the same value. Images were acquired using a Leica TCS SP5 confocal microscope (Leica Microsystems) 48 to 96 h after incubation. The HXC PL APO 20x0.7 and 63x1.20 objectives were used for localization and colocalization images, respectively. A 488-nm laser was used for GFP (emission 490 to 550 nm) and a 561-nm laser for RFP (emission 584 to 627 nm) imaging. Plasmolysis was induced by incubating leaf discs in 1 M NaCl for 20 min.
Bioinformatics
Protein domains were predicted using ExPaSy InterProScan program (http://www.ebi.ac.uk/Tools/InterProScan/). The possible hydrophobic C-terminal sequence encoding the transient transmembrane domain for GPI anchor attachment was predicted using PredGPI (http://gpcr.biocomp.unibo.it/predgpi/pred.htm) (Pierleoni et al., 2008) and BIG_PI Plant Predictor (http://mendel.imp.ac.at/gpi/plant_server.html) (Eisenhaber et al., 2003) programs, and the low complexity region was identified with the SMART (http://smart.embl-heidelberg.de) program (Schultz et al., 1998). Molecular weight and isoelectric point were calculated using the ProtParam (http://au.expasy.org/tools/protparam.html) program in ExPaSy server (Gasteiger et al., 2005).
Phylogenetic Analysis
Phylogenetic analysis of GH17 proteins was performed with the full-length protein sequences. Arabidopsis thaliana sequences were originally reported by Doxey et al. (2007). GH17 proteins were initially aligned with ClustalW, and the MEGA Version 4.1 software (http://www.megasoftware.net/) was used to conduct a phylogenetic analysis based on the neighbor-joining method on 1000 bootstrap replications. Bootstrap percentages are shown on the dendrogram branch points.
Accession Numbers
The Populus gene model identifiers and/or sequence accessions used for qRT-PCR analysis are listed in Supplemental Table 1 online. Sequence data for the hybrid aspen GH17 genes that were used in localization studies can be found in the GenBank/EMBL data libraries under the following accession numbers: GH17_44 (HQ443266), GH17_61 (HQ443267), GH17_65 (HQ443268), and GH17_101 (HQ443269).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phylogenetic Analysis of GH17 Proteins.
Supplemental Figure 2. Presence of Callose at PD during Dormancy Cycling in Populus.
Supplemental Figure 3. qRT-PCR Analysis of Populus GH17 Genes.
Supplemental Figure 4. qRT-PCR Analysis of Populus GA Signaling Genes.
Supplemental Figure 5. TEM Micrographs of GA4-Treated Dormant Populus Buds.
Supplemental Table 1. Populus Genes, Model Identifiers, and Primer Pairs for qRT-PCR.
Supplemental Table 2. Primers for GH17 Vector Construction.
Supplemental Data Set 1. Text File of the Sequences and Alignment Used for the Phylogenetic Analysis Shown in Supplemental Figure 1.
Supplementary Material
Acknowledgments
We thank Mervi Lindman and Tuomas Puukko for technical help, Pekka Lönnqvist and Marit Siira for taking care of the plants, Karl Oparka and Jens Tilsner for the gift of the TMV MP vector construct, and two anonymous referees for valuable suggestions. We acknowledge the financial support of the Norwegian Research Council (C.v.d.S. and P.L.H.R.; ClimaDorm 171970 and BioDorm 192013) and the Academy of Finland (The Finnish Centre of Excellence Program [J.K.] and postdoctoral Project 1115280 [A.W.]).
References
- Aloni R., Peterson C.A. (1997). Auxin promotes dormancy callose removal from the phloem of Magnolia kobus and callose accumulation and early wood vessel differentiation in Quercus robur. J. Plant Res. 110: 37–44 [DOI] [PubMed] [Google Scholar]
- Aloni R., Raviv A., Peterson C.A. (1991). The role of auxin in the removal of dormancy callose and resumption of phloem activity in Vitis vinifera. Can. J. Bot. 69: 1825–1832 [Google Scholar]
- Amari K., et al. (2010). A family of plasmodesmal proteins with receptor-like properties for plant viral movement proteins. PLoS Pathog. 6: e1001119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An H.L., Roussot C., Suárez-López P., Corbesier L., Vincent C., Piñeiro M., Hepworth S., Mouradov A., Justin S., Turnbull C., Coupland G. (2004). CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131: 3615–3626 [DOI] [PubMed] [Google Scholar]
- Arora R., Rowland L.J., Tanino K. (2003). Induction and release of bud dormancy in woody perennials: A science comes of age. HortScience 38: 911–921 [Google Scholar]
- Bayer E.M., Bottrill A.R., Walshaw J., Vigouroux M., Naldrett M.J., Thomas C.L., Maule A.J. (2006). Arabidopsis cell wall proteome defined using multidimensional protein identification technology. Proteomics 6: 301–311 [DOI] [PubMed] [Google Scholar]
- Böhlenius H., Huang T., Charbonnel-Campaa L., Brunner A.M., Jansson S., Strauss S.H., Nilsson O. (2006). CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312: 1040–1043 [DOI] [PubMed] [Google Scholar]
- Bolduc N., Hake S. (2009). The maize transcription factor KNOTTED1 directly regulates the gibberellin catabolism gene ga2ox1. Plant Cell 21: 1647–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley D., Ratcliffe O., Vincent C., Carpenter R., Coen E. (1997). Inflorescence commitment and architecture in Arabidopsis. Science 275: 80–83 [DOI] [PubMed] [Google Scholar]
- Conti L., Bradley D. (2007). TERMINAL FLOWER1 is a mobile signal controlling Arabidopsis architecture. Plant Cell 19: 767–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbesier L., Vincent C., Jang S.H., Fornara F., Fan Q.Z., Searle I., Giakountis A., Farrona S., Gissot L., Turnbull C., Coupland G. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dafoe N.J., Zamani A., Ekramoddoullah A.K.M., Lippert D., Bohlmann J., Constabel C.P. (2009). Analysis of the poplar phloem proteome and its response to leaf wounding. J. Proteome Res. 8: 2341–2350 [DOI] [PubMed] [Google Scholar]
- Dong X.N., Mindrinos M., Davis K.R., Ausubel F.M. (1991). Induction of Arabidopsis defense genes by virulent and avirulent Pseudomonas syringae strains and by a cloned avirulence gene. Plant Cell 3: 61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxey A.C., Yaish M.W.F., Moffatt B.A., Griffith M., McConkey B.J. (2007). Functional divergence in the Arabidopsis β-1,3-glucanase gene family inferred by phylogenetic reconstruction of expression states. Mol. Biol. Evol. 24: 1045–1055 [DOI] [PubMed] [Google Scholar]
- Druart N., Johansson A., Baba K., Schrader J., Sjödin A., Bhalerao R.R., Resman L., Trygg J., Moritz T., Bhalerao R.P. (2007). Environmental and hormonal regulation of the activity-dormancy cycle in the cambial meristem involves stage-specific modulation of transcriptional and metabolic networks. Plant J. 50: 557–573 [DOI] [PubMed] [Google Scholar]
- Eisenhaber B., Wildpaner M., Schultz C.J., Borner G.H.H., Dupree P., Eisenhaber F. (2003). Glycosylphosphatidylinositol lipid anchoring of plant proteins. Sensitive prediction from sequence- and genome-wide studies for Arabidopsis and rice. Plant Physiol. 133: 1691–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elortza F., Nühse T.S., Foster L.J., Stensballe A., Peck S.C., Jensen O.N. (2003). Proteomic analysis of glycosylphosphatidylinositol-anchored membrane proteins. Mol. Cell. Proteomics 2: 1261–1270 [DOI] [PubMed] [Google Scholar]
- Epel B.L. (2009). Plant viruses spread by diffusion on ER-associated movement-protein-rafts through plasmodesmata gated by viral induced host β-1,3-glucanases. Semin. Cell Dev. Biol. 20: 1074–1081 [DOI] [PubMed] [Google Scholar]
- Eriksson M.E., Israelsson M., Olsson O., Moritz T. (2000). Increased gibberellin biosynthesis in transgenic trees promotes growth, biomass production and xylem fiber length. Nat. Biotechnol. 18: 784–788 [DOI] [PubMed] [Google Scholar]
- Eriksson S., Böhlenius H., Moritz T., Nilsson O. (2006). GA4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation. Plant Cell 18: 2172–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix G., Meins F. (1987). Ethylene regulation of β-1,3-glucanase in tobacco. Planta 172: 386–392 [DOI] [PubMed] [Google Scholar]
- Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M.R., Appel R.D., Bairoch A. (2005). Protein identification and analysis tools on the ExPASy server. The Proteomics Protocols Handbook, Walker J.M., (Totowa, NJ: Humana Press; ), pp. 571–607 [Google Scholar]
- Geisler-Lee J., et al. (2006). Poplar carbohydrate-active enzymes. Gene identification and expression analyses. Plant Physiol. 140: 946–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao P., Liu C., Wang Y., Chen R., Tang M., Du B., Zhu L., He G. (2008). Herbivore-induced callose deposition on the sieve plates of rice: An important mechanism for host resistance. Plant Physiol. 146: 1810–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazebroek J.P., Metzger J.D., Mansager E.R. (1993). Thermoinductive regulation of gibberellin metabolism in Thlaspi arvense L. II. Cold induction of enzymes in gibberellin biosynthesis. Plant Physiol. 102: 547–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C.Y., Liu Y.X., Luthe D.S., Yuceer C. (2006). Poplar FT2 shortens the juvenile phase and promotes seasonal flowering. Plant Cell 18: 1846–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelsson M., Mellerowicz E., Chono M., Gullberg J., Moritz T. (2004). Cloning and overproduction of gibberellin 3-oxidase in hybrid aspen trees. Effects on gibberellin homeostasis and development. Plant Physiol. 135: 221–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelsson M., Sundberg B., Moritz T. (2005). Tissue-specific localization of gibberellins and expression of gibberellin-biosynthetic and signaling genes in wood-forming tissues in aspen. Plant J. 44: 494–504 [DOI] [PubMed] [Google Scholar]
- Jasinski S., Piazza P., Craft J., Hay A., Woolley L., Rieu I., Phillips A., Hedden P., Tsiantis M. (2005). KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 15: 1560–1565 [DOI] [PubMed] [Google Scholar]
- Jiang D.H., Wang Y.Q., Wang Y.Z., He Y.H. (2008). Repression of FLOWERING LOCUS C and FLOWERING LOCUS T by the Arabidopsis Polycomb repressive complex 2 components. PLoS ONE 3: e3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junttila O., Jensen E. (1988). Gibberellins and photoperiodic control of shoot elongation in Salix. Physiol. Plant. 74: 371–376 [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Leubner-Metzger G., Petruzzelli L., Waldvogel R., Vögeli-Lange R., Meins F., Jr (1998). Ethylene-responsive element binding protein (EREBP) expression and the transcriptional regulation of class I β-1,3-glucanase during tobacco seed germination. Plant Mol. Biol. 38: 785–795 [DOI] [PubMed] [Google Scholar]
- Levy A., Epel B.L. (2009). Cytology of the (1,3)-β-glucan (callose) in plasmodesmata and sieve plate pores. Chemistry, Biochemistry, and Biology of (1,3)-β-Glucans and Related Polysaccharides, Bacic A., Fincher G.B., Stone B.A., (Burlington, MA: Academic Press, Elsevier; ), pp. 439–463 [Google Scholar]
- Levy A., Erlanger M., Rosenthal M., Epel B.L. (2007a). A plasmodesmata-associated β-1,3-glucanase in Arabidopsis. Plant J. 49: 669–682 [DOI] [PubMed] [Google Scholar]
- Levy A., Guenoune-Gelbart D., Epel B.L. (2007b). β-1,3-Glucanases. Plasmodesmal gate keepers for intercellular communication. Plant Signal. Behav. 2: 404–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M.K., Belanger H., Lee Y.J., Varkonyi-Gasic E., Taoka K.I., Miura E., Xoconostle-Cázares B., Gendler K., Jorgensen R.A., Phinney B., Lough T.J., Lucas W.J. (2007). FLOWERING LOCUS T protein may act as the long-distance florigenic signal in the cucurbits. Plant Cell 19: 1488–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas W.J., Ding B., van der Schoot C. (1993). Tansley Review No. 58 Plasmodesmata and the supracellular nature of plants. New Phytol. 125: 435–476 [DOI] [PubMed] [Google Scholar]
- Mathiason K., He D., Grimplet J., Venkateswari J., Galbraith D.W., Or E., Fennell A. (2009). Transcript profiling in Vitis riparia during chilling requirement fulfillment reveals coordination of gene expression patterns with optimized bud break. Funct. Integr. Genomics 9: 81–96 [DOI] [PubMed] [Google Scholar]
- Maule A.J. (2008). Plasmodesmata: Structure, function and biogenesis. Curr. Opin. Plant Biol. 11: 680–686 [DOI] [PubMed] [Google Scholar]
- Mauriat M., Moritz T. (2009). Analyses of GA20ox- and GID1-over-expressing aspen suggest that gibberellins play two distinct roles in wood formation. Plant J. 58: 989–1003 [DOI] [PubMed] [Google Scholar]
- Mohamed R., Wang C.T., Ma C., Shevchenko O., Dye S.J., Puzey J.R., Etherington E., Sheng X.Y., Meilan R., Strauss S.H., Brunner A.M. (2010). Populus CEN/TFL1 regulates first onset of flowering, axillary meristem identity and dormancy release in Populus. Plant J. 62: 674–688 [DOI] [PubMed] [Google Scholar]
- Mutasa-Göttgens E., Hedden P. (2009). Gibberellin as a factor in floral regulatory networks. J. Exp. Bot. 60: 1979–1989 [DOI] [PubMed] [Google Scholar]
- Pierleoni A., Martelli P.L., Casadio R. (2008). PredGPI: A GPI-anchor predictor. BMC Bioinformatics 9: 392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinne P.L.H., Kaikuranta P.M., van der Schoot C. (2001). The shoot apical meristem restores its symplasmic organization during chilling-induced release from dormancy. Plant J. 26: 249–264 [DOI] [PubMed] [Google Scholar]
- Rinne P.L.H., Ruonala R., Ripel L., van der Schoot C. (2008). Dormancy release-proteins in the shoot apical meristem of Populus. Physiol. Plant. 133 [Google Scholar]
- Rinne P.L.H., van den Boogaard R., Mensink M.G.J., Kopperud C., Kormelink R., Goldbach R., van der Schoot C. (2005). Tobacco plants respond to the constitutive expression of the tospovirus movement protein NS(M) with a heat-reversible sealing of plasmodesmata that impairs development. Plant J. 43: 688–707 [DOI] [PubMed] [Google Scholar]
- Rinne P.L.H., van der Schoot C. (1998). Symplasmic fields in the tunica of the shoot apical meristem coordinate morphogenetic events. Development 125: 1477–1485 [DOI] [PubMed] [Google Scholar]
- Rinne P.L.H., van der Schoot C. (2003). Plasmodesmata at the crossroads between development, dormancy, and defense. Can. J. Bot. 81: 1182–1197 [Google Scholar]
- Rohde A., Prinsen E., De Rycke R., Engler G., Van Montagu M., Boerjan W. (2002). PtABI3 impinges on the growth and differentiation of embryonic leaves during bud set in poplar. Plant Cell 14: 1885–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruonala R., Rinne P.L.H., Kangasjärvi J., van der Schoot C. (2008). CENL1 expression in the rib meristem affects stem elongation and the transition to dormancy in Populus. Plant Cell 20: 59–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruttink T., Arend M., Morreel K., Storme V., Rombauts S., Fromm J., Bhalerao R.P., Boerjan W., Rohde A. (2007). A molecular timetable for apical bud formation and dormancy induction in poplar. Plant Cell 19: 2370–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T., Kobayashi M., Itoh H., Tagiri A., Kayano T., Tanaka H., Iwahori S., Matsuoka M. (2001). Expression of a gibberellin 2-oxidase gene around the shoot apex is related to phase transition in rice. Plant Physiol. 125: 1508–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T., Sakakibara H., Kojima M., Yamamoto Y., Nagasaki H., Inukai Y., Sato Y., Matsuoka M. (2006). Ectopic expression of KNOTTED1-like homeobox protein induces expression of cytokinin biosynthesis genes in rice. Plant Physiol. 142: 54–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saure M.C. (1985). Dormancy release in deciduous fruit trees. Hortic. Rev. (Am. Soc. Hortic. Sci.) 7: 239–300 [Google Scholar]
- Schrader J., Moyle R., Bhalerao R., Hertzberg M., Lundeberg J., Nilsson P., Bhalerao R.P. (2004). Cambial meristem dormancy in trees involves extensive remodelling of the transcriptome. Plant J. 40: 173–187 [DOI] [PubMed] [Google Scholar]
- Schultz J., Milpetz F., Bork P., Ponting C.P. (1998). SMART, a simple modular architecture research tool: Identification of signaling domains. Proc. Natl. Acad. Sci. USA 95: 5857–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siloto R.M.P., Findlay K., Lopez-Villalobos A., Yeung E.C., Nykiforuk C.L., Moloney M.M. (2006). The accumulation of oleosins determines the size of seed oilbodies in Arabidopsis. Plant Cell 18: 1961–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson C., Thomas C., Findlay K., Bayer E., Maule A.J. (2009). An Arabidopsis GPI-anchor plasmodesmal neck protein with callose binding activity and potential to regulate cell-to-cell trafficking. Plant Cell 21: 581–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki S., Matsuo S., Wong H.L., Yokoi S., Shimamoto K. (2007). Hd3a protein is a mobile flowering signal in rice. Science 316: 1033–1036 [DOI] [PubMed] [Google Scholar]
- Thomas S.G., Phillips A.L., Hedden P. (1999). Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc. Natl. Acad. Sci. USA 96: 4698–4703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G.W., et al. (2004). High-throughput fluorescent tagging of full-length Arabidopsis gene products in planta. Plant Physiol. 135: 25–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilsner J., Amari K., Torrance L. (2011). Plasmodesmata viewed as specialised membrane adhesion sites. Protoplasma 248: 39–60 [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M., Nakajima M., Motoyuki A., Matsuoka M. (2007). Gibberellin receptor and its role in gibberellin signaling in plants. Annu. Rev. Plant Biol. 58: 183–198 [DOI] [PubMed] [Google Scholar]
- Uknes S., Mauch-Mani B., Moyer M., Potter S., Williams S., Dincher S., Chandler D., Slusarenko A., Ward E., Ryals J. (1992). Acquired resistance in Arabidopsis. Plant Cell 4: 645–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vegis A. (1964). Dormancy in higher plants. Annu. Rev. Plant Physiol. 15: 185–224 [Google Scholar]
- Wang D.Y., Li Q., Cui K.M., Zhu Y.X. (2009). Occurrence of the transition of apical architecture and expression patterns of related genes during conversion of apical meristem identity in G2 pea. J. Integr. Plant Biol. 51: 13–20 [DOI] [PubMed] [Google Scholar]
- Weiser C.J. (1970). Cold resistance and injury in woody plants: Knowledge of hardy plant adaptations to freezing stress may help us to reduce winter damage. Science 169: 1269–1278 [DOI] [PubMed] [Google Scholar]
- Welling A., Kaikuranta P., Rinne P. (1997). Photoperiodic induction of dormancy and freezing tolerance in Betula pubescens. Involvement of ABA and dehydrins. Physiol. Plant. 100: 119–125 [Google Scholar]
- Yamaguchi S. (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59: 225–251 [DOI] [PubMed] [Google Scholar]
- Yamauchi Y., Ogawa M., Kuwahara A., Hanada A., Kamiya Y., Yamaguchi S. (2004). Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. Plant Cell 16: 367–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanewich K.P., Rood S.B. (1995). Vernalization and gibberellin physiology of winter canola - Endogenous gibberellin (GA) content and metabolism of [3H] GA1 and [3H] GA20. Plant Physiol. 108: 615–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.