This study shows that ClpT in chloroplasts is essential for plant viability and that ClpT specifically regulates the assembly of the Clp proteolytic core. Such a regulatory mechanism adds a new dimension to the functional importance of the chloroplast Clp protease, which has been considered exclusively as a constitutive housekeeping enzyme.
Abstract
The ATP-dependent caseinolytic protease (Clp) is an essential housekeeping enzyme in plant chloroplasts. It is by far the most complex of all known Clp proteases, with a proteolytic core consisting of multiple catalytic ClpP and noncatalytic ClpR subunits. It also includes a unique form of Clp protein of unknown function designated ClpT, two of which exist in the model species Arabidopsis thaliana. Inactivation of ClpT1 or ClpT2 significantly reduces the amount of Clp proteolytic core, whereas loss of both proves seedling lethal under autotrophic conditions. During assembly of the Clp proteolytic core, ClpT1 first binds to the P-ring (consisting of ClpP3-6 subunits) followed by ClpT2, and only then does the P-ring combine with the R-ring (ClpP1, ClpR1-4 subunits). Most of the ClpT proteins in chloroplasts exist in vivo as homodimers, which then apparently monomerize prior to association with the P-ring. Despite their relative abundance, however, the availability of both ClpT proteins is rate limiting for the core assembly, with the addition of recombinant ClpT1 and ClpT2 increasing core content up to fourfold. Overall, ClpT appears to regulate the assembly of the chloroplast Clp protease, revealing a new and sophisticated control mechanism on the activity of this vital protease in plants.
INTRODUCTION
Energy-dependent proteases perform a crucial role in protein quality control by removing short-lived regulatory proteins and misfolded or otherwise damaged polypeptides. Most of these proteases consist of two distinct components, one with chaperone characteristics that uses ATP for substrate selection, unfolding, and translocation and the other a proteolytic core for degradation (Baker and Sauer, 2006). Many of these also share a common architecture and are best exemplified by the 26S proteasome in eukaryotes and the Clp protease in eubacteria. The Ser-type Clp protease in Escherichia coli consists of a central proteolytic core comprising two apposing heptameric rings of ClpP. This cylindrical structure forms an internal chamber that houses the proteolytic active sites within (Wang et al., 1997). The proteolytic core is flanked on one or both ends by a single hexameric ring of an Hsp100 partner, either ClpA or -X (Grimaud et al., 1998). In an energy-dependent manner, the Hsp100 chaperone translocates the unfolded protein substrate through the narrow aperture of the core complex into the inner cavity for rapid degradation (Ishikawa et al., 2001; Kim et al., 2001; Ortega et al., 2002).
Consistent with their endosymbiotic origin, plant chloroplasts have a range of proteases of bacterial ancestry. These include the FtsH and Deg proteases associated with the thylakoid membranes and Clp protease localized within the stroma (Adam and Clarke, 2002). The chloroplast Clp protease in vascular plants is by far the most structurally diverse member of the Clp protease family. In the model species Arabidopsis thaliana, it consists of a single proteolytic core complex with 11 distinct subunits (ClpP1, ClpP3-6, ClpR1-4, and ClpT1-2), along with three potential chaperone partners (ClpC1-2 and ClpD) (Adam et al., 2001; Peltier et al., 2004; Clarke et al., 2005). All of these proteins with the exception of ClpP1 are nuclear encoded; thus, they are posttranslationally imported into chloroplasts (Adam et al., 2001; Adam and Clarke, 2002). The protease is also one of the most functionally important, being the principle constitutive housekeeping protease in the stroma whose activity is essential for plant viability (Shikanai et al., 2001; Kuroda and Maliga, 2003; Sjögren et al., 2006; Zheng et al., 2006; Koussevitzky et al., 2007; Kim et al., 2009). Up to 25 stromal proteins have now been identified as potential substrates for the Clp protease, ranging from different regulatory proteins to various metabolic enzymes (Sjögren et al., 2006; Stanne et al., 2009).
One of the most intriguing features of the chloroplast Clp protease is the intricate subunit composition of the central proteolytic core complex. By analogy to bacterial orthologs, the core complex presumably consists of twin heptameric rings, one of which contains ClpP1 and ClpR1-4 and the other ClpP3-6 (Sjögren et al., 2006). Little redundancy exists within this configuration (Stanne et al., 2009), with loss of any one subunit typically disrupting the assembly of the entire protease. Even more perplexing is the association of the unusual ClpT accessory proteins that have so far been found only in plant chloroplasts and whose function remains a mystery. The relatively small ClpT proteins have high sequence similarity to the N-terminal region of ClpC but lack the AAA+ ATPase domains common to Hsp100 proteins (Kress et al., 2009). Two ClpT paralogs exist in Arabidopsis, with a single subunit of each thought to associate with the peripheral surface of the Clp proteolytic core complex (Peltier et al., 2004). Within the core structure, ClpT1 binds to the ClpP3-6 ring, whereas the location of ClpT2 has yet to be resolved (Sjögren et al., 2006). In this study, we demonstrate that ClpT is a crucial component of the chloroplast Clp protease in Arabidopsis. ClpT2 is shown to associate with the ClpP3-6 ring along with ClpT1, although most of both proteins exist as homodimers within the stroma. Overall, the availability of ClpT1 and ClpT2 appears to regulate the assembly of the Clp proteolytic core, revealing a previously undiscovered control mechanism on the formation of the chloroplast Clp protease in vascular plants.
RESULTS
Verification and Phenotypic Appearance of Arabidopsis clpT T-DNA Insertion Lines
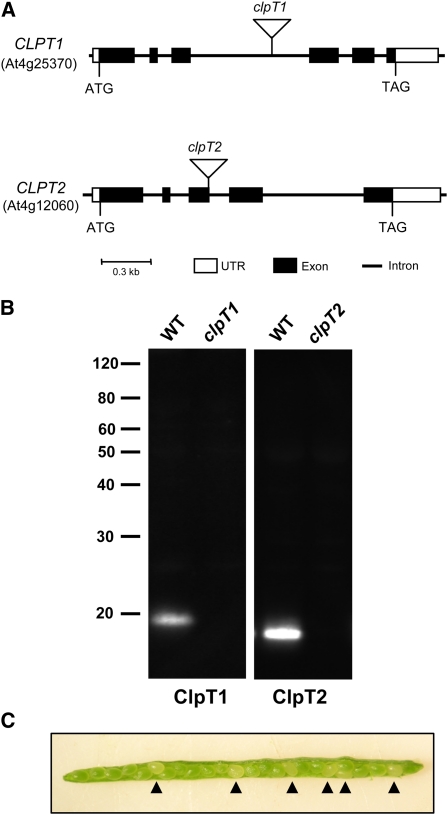
To investigate the function of ClpT in chloroplasts, we first screened the various Arabidopsis T-DNA insertion collections for possible clpT1 and clpT2 null mutants. Single putative null mutants were eventually identified for each of the CLPT genes, both of which were confirmed by segregation analysis as homozygous lines with a single T-DNA insertion (data not shown). For the clpT1 null line, the T-DNA insertion was located within the third intron of the CLPT1 gene, while for the clpT2 mutant the insertion was at the end of the third exon of the CLPT2 gene (Figure 1A). Loss of the corresponding ClpT protein in each of the two null mutants was later confirmed by immunoblotting using specific antibodies (Figure 1B). Under standard growth conditions, neither of the two null mutants showed any obvious phenotypic changes from the wild type throughout the life cycle.
Figure 1.
Null Mutations Affecting the Arabidopsis CLPT1 and CLPT2 Genes.
(A) Schematic representation of the two CLPT genes (with accession number underneath each) and the location of the T-DNA insertion (triangle) in the corresponding clpT1 and clpT2 mutants. Protein-coding exons and untranslated regions (UTRs) are represented by black and white boxes, respectively, with introns drawn as thin lines between the boxes.
(B) Immunoblot analysis of ClpT1 and ClpT2 in the wild type (WT) and clpT mutant lines. Total leaf protein was extracted from each plant and separated by denaturing-PAGE on the basis of equal chlorophyll content. ClpT1 (19.5 kD) and ClpT2 (19 kD) were detected by immunoblotting with specific antibodies as indicated below each panel. Molecular mass markers (kD) are shown on the left.
(C) A typical, mature silique from a heterozygous F1 clpT1 clpT2 double-knockout line containing both normal (green) and abnormal (white; arrowheads) seeds.
[See online article for color version of this figure.]
We next attempted to produce a double clpT1 clpT2 null line by crossing the confirmed clpT1 and clpT2 mutants. Examination of developing siliques from the resulting F1 heterozygous plants revealed several white seeds among the normal green ones (Figure 1C), suggesting that the double CLPT mutation might compromise seed viability. Later screening of 100 F2 progeny identified 22 clpT1 and 21 clpT2 single null mutants as expected but no double clpT1 clpT2 knockouts. Statistical analysis (χ2 test) indicated that with 99% certainty inactivation of both CLPT genes is seedling lethal under autotrophic conditions.
Relative Abundance of Chloroplast Clp Proteins in the Absence of ClpT1 or ClpT2
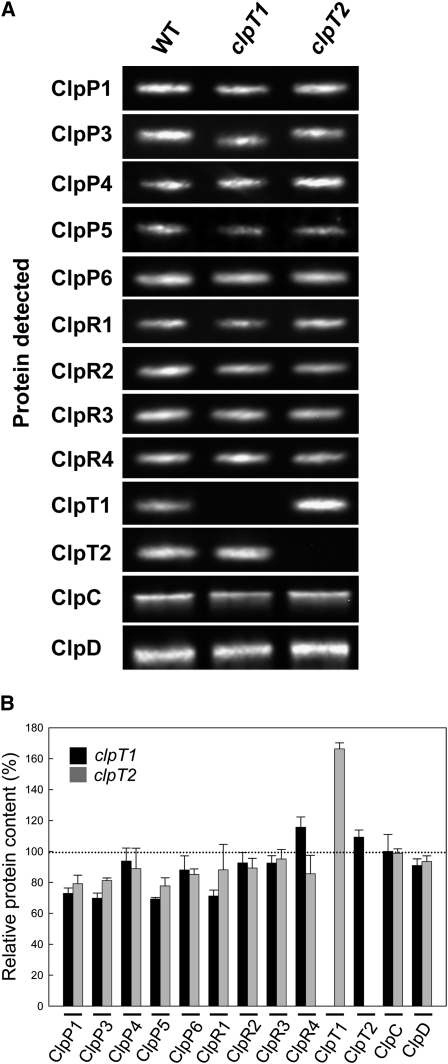
Given that ClpT1 and ClpT2 associate with the Clp proteolytic core, we next investigated whether the loss of either protein affected the levels of the other chloroplast Clp proteins. For this analysis, stromal proteins were extracted from wild-type and clpT mutant leaves, and the amount of each Clp protein was determined by immunoblotting using specific antibodies (Figure 2A). As shown in Figure 2B, loss of either ClpT1 or ClpT2 had little or no effect on the amount of the other subunits of the Clp proteolytic core (i.e., ClpP/R) or on the potential chaperone partners (ClpC/D). Interestingly, loss of ClpT2 did cause an increase in the level of ClpT1 (1.7-fold), suggesting a possible compensatory response, but no corresponding upregulation of ClpT2 was observed in the clpT1 mutant.
Figure 2.
Relative Amounts of Chloroplast Clp Proteins in Wild-Type Arabidopsis and clpT Null Mutants.
Stromal proteins from 3-week-old plants were separated by denaturing-PAGE on the basis of equal protein content. The various chloroplast Clp proteins were detected by immunoblotting using specific polyclonal antibodies (A) and the relative amounts of each were then quantified (B). Values shown are averages ± se (n = 3) with the wild-type (WT) values set to 100%. The molecular masses in kilodaltons of the proteins detected are as follows: 20.5 (ClpP1), 28.5 (ClpP3), 27 (ClpP4), 22.5 (ClpP5), 21.5 (ClpP6), 28 (ClpR1), 25 (ClpR2), 28.5 (ClpR3), 24.5 (ClpR4), 19.5 (ClpT1), 19 (ClpT2), 93 (ClpC), and 96 (ClpD).
Loss of ClpT1 or ClpT2 Reduces the Amount of the Clp Proteolytic Core Complex
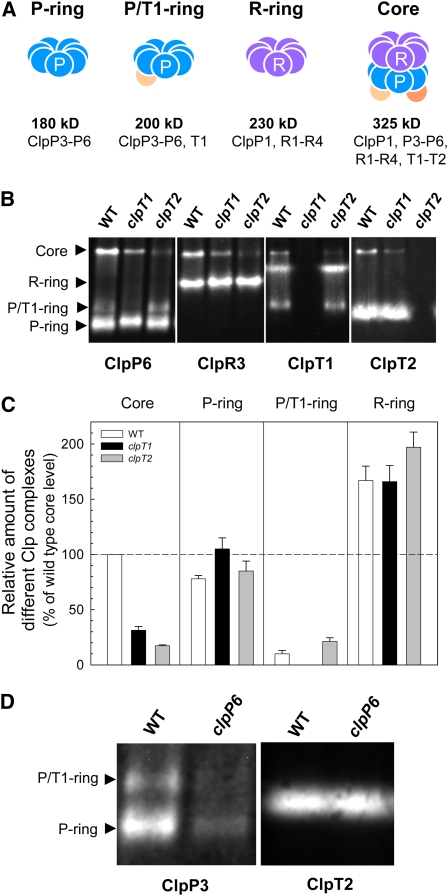
Despite the lack of changes in the amount of ClpP/R subunits in the clpT mutants, we checked for possible effects on the assembly of the Clp proteolytic core. Stromal proteins were isolated from young leaves of the wild type and clpT mutants and separated by native-PAGE. The Clp proteolytic core complex and the various assembly intermediates were detected by immunoblotting using antibodies for specific marker subunits along with those for ClpT1 and ClpT2 (i.e., ClpP6 for the core and ClpP3-6 [P-ring] and ClpP3-6, T1 [P/T1-ring] subcomplexes; ClpR3 for the core and ClpP1, R1-4 subcomplex [R-ring]) (Figure 3A). In the wild type, the pattern of all Clp subcomplexes and proteolytic core was as previously shown (Sjögren et al., 2006; Stanne et al., 2009). ClpP6 is equally distributed between the P-ring and the core complex, with a small amount in the P/T1-ring (~5% of the total ClpP6 content). Similarly, around 35% of the ClpR3 resides in the core, while the remaining is in the R-ring (Figure 3C). Of the total ClpT1 content, 30% resides in the P/T1-ring and only 10% in the core complex, while the remaining protein (60%) is in a seemingly large homogeneous complex of 260 kD, an oligomer known not to contain any other chloroplast Clp protein (Sjögren et al., 2006). Using the new ClpT2 antibody, we can now reveal that the proportion of ClpT2 residing in the core is even lower than that of ClpT1, with most of the ClpT2 protein (>95%) located within a 200-kD complex (Figure 3B). This ClpT2 complex did not correspond to either the P- or P/T1-rings since its level remained unchanged in clpP6 antisense lines despite ~90% reduction in both the P- and P/T1-rings (Figure 3D).
Figure 3.
Clp Protein Complexes in Wild-Type Arabidopsis and clpT Null Mutants.
(A) Schematic representation of the chloroplast Clp proteolytic core and its various subcomplexes as previously identified, indicating relative size and subunit composition.
(B) Clp proteolytic core complexes in stromal fractions from 3-week-old wild type (WT) and clpT null mutants were separated by native-PAGE on the basis of equal protein content. The different complexes of the Clp proteolytic core (indicated on the left) were visualized by immunoblotting with antibodies specific for selected marker subunits of each as indicated below the panels (ClpP6 for the core, P-ring, and P/T1-ring; ClpR3 for the core and R-ring; ClpT1 for the core, P/T1-ring, and unknown T1 oligomer; ClpT2 for the core and unknown T2 oligomer).
(C) Quantification of the relative amount of the Clp proteolytic core and other Clp subcomplexes in the clpT null mutants based on the immunoblots with the ClpP6 and ClpR3 antibodies. Values shown are averages ± se (n = 6) and plotted relative to the wild-type value for the core complex, which was set to 100%.
(D) Detection of the 200-kD ClpT2 oligomer in stromal fractions from 3-week-old wild-type Arabidopsis and a clpP6 antisense line in which the Clp core, P-ring, and ClpP/T1-ring are reduced to ~10% wild-type levels (as confirmed using the ClpP3 antibody).
[See online article for color version of this figure.]
In the clpT1 mutant, loss of ClpT1 had no marked effect on the assembly of the 180-kD P-ring, but the 200-kD P/T1-ring was absent as expected. As for the P-ring, the level of the R-ring also remained unchanged, as did the ClpT2 complex (Figure 3B). Despite wild-type levels of both P- and R-rings, however, Clp core content decreased to 30% of that in the wild type (Figure 3C). An even more severe drop in core content was observed in the clpT2 mutant (15% of wild-type level), again despite the lack of changes in the amount of the various Clp subcomplexes (P-ring, P/T1-ring, and R-ring). There was an accumulation of the 260-kD ClpT1 complex in the clpT2 mutant, however, suggesting that most if not all of the additional ClpT1 protein observed in Figure 2 resides in this homogeneous oligomer.
Purification of Recombinant ClpT1 and ClpT2 Proteins
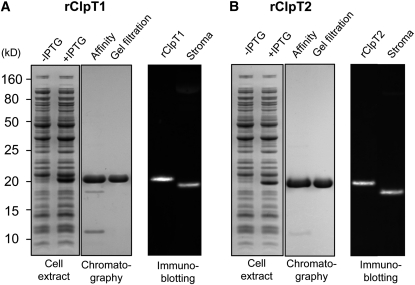
To test the characteristics of the ClpT proteins more closely, we purified recombinant forms of both Arabidopsis ClpT1 and -T2 by overexpression in E. coli. Each recombinant ClpT protein (rClpT) was expressed from the predicted mature N terminus and contained a His6-tag at the C terminus. Both rClpT proteins were isolated to >95% purity, and their molecular masses corresponded to those of the native proteins plus the His-tag (Figure 4).
Figure 4.
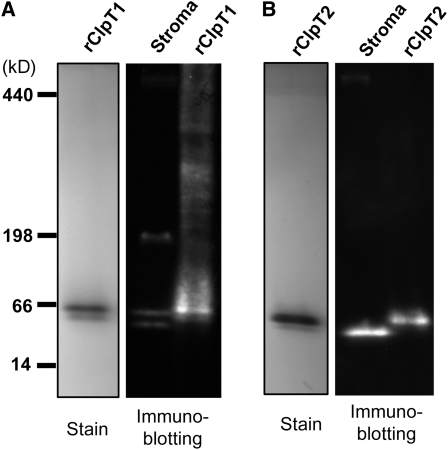
Purification of Recombinant ClpT1 and ClpT2.
Soluble C-terminally His-tagged Arabidopsis ClpT1 (A) and ClpT2 (B) were overexpressed in E. coli upon induction with IPTG. Each ClpT protein was purified from cell lysates by sequential Ni2+ affinity chromatography and gel filtration. Fractions before (−IPTG) and after (+IPTG) induction in E. coli and after each chromatography step were analyzed by denaturing-PAGE and Coomassie blue staining. The molecular masses of the recombinant ClpT1 (rClpT1) and ClpT2 (rClpT2) were then compared with those of the native stromal ClpT proteins (stroma) isolated from wild-type Arabidopsis by immunoblotting with specific ClpT antibodies.
Native Homogeneous ClpT Complexes
Since the native ClpT proteins appear to form large homogeneous complexes in addition to their association with the various other Clp complexes, we tested if the recombinant ClpT proteins would form similar homo-oligomers. To size the ClpT complexes accurately, we first used the same native-PAGE system previously optimized for the other Clp complexes (Sjögren et al., 2006). In principle, proteins are electrophoresed until they reach their pore limitation within the gel matrix, thereby negating the effect of net charge on the mobility of each protein complex. Preliminary tests, however, revealed that both the native ClpT protein complexes migrated much more slowly than the other Clp complexes did, so the electrophoresis conditions had to be modified. Later optimization of these conditions (see Supplemental Figure 1 online) revealed that the native ClpT complexes were considerably smaller than previously observed (Figure 5). Apart from the 200-kD P1/T-ring, the native ClpT1 protein was found in two similarly sized complexes between 45 and 60 kD. Native ClpT2 also formed a single complex in the same size range (45 kD), suggesting that both ClpT proteins form homogeneous dimers in vivo. In comparison, the rClpT proteins separated as a single complex, both slightly larger than their native counterparts. Taking into account the inclusion of the His-tags, the recombinant proteins also appear to form the same homogeneous dimers as the native ClpT1 and -T2. Interestingly, the protein with highest sequence similarity to ClpT, ClpC, is also known to form dimers in vivo in the absence of ATP (Clarke et al., 2005).
Figure 5.
Size Determination of the ClpT Oligomers.
Native and recombinant ClpT1 (A) and ClpT2 (B) were separated by a modified form of native-PAGE designed specifically to size slow migrating protein complexes accurately. The rClpT oligomers were visualized by Coomassie blue staining (Stain), the sizes of which were then compared with those of native ClpT1 and ClpT2 in stromal fractions (Stroma) from wild-type Arabidopsis by immunoblotting with ClpT-specific antibodies. Molecular mass standards in kilodaltons are shown on the left.
Recombinant ClpT Facilitates Assembly of the Clp Proteolytic Core Complex
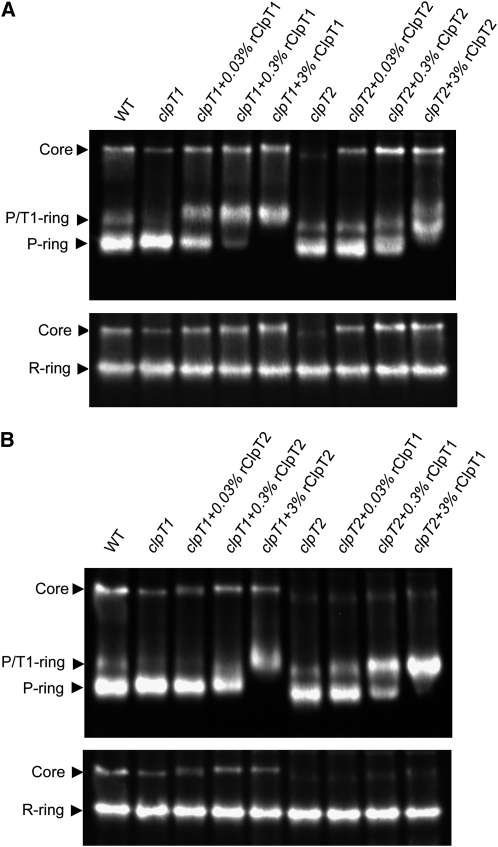
Next, we tested if the rClpT proteins could restore wild-type levels of the Clp proteolytic core within the clpT mutants. First, we determined the relative amount of ClpT1 in the wild type. By comparing known amounts of rClpT1 with the native protein, we determined that ClpT1 accounts for ~0.3% of the total protein in the stroma (see Supplemental Figure 2 online). Knowing the relative level of native ClpT1 in wild-type chloroplasts, different amounts of rClpT1 were added to stroma extracts from the clpT1 mutant (Figure 6A). Adding only 10% wild-type levels of rClpT1 restored most of the core complex (90%) that was lost in the clpT1 mutant. It also restored the formation of the 200-kD P/T1-ring, actually converting more of the 180-kD P-ring than observed in the wild type. Addition of rClpT1 to wild-type levels converted almost all the P-ring to the P/T1-ring but only further increased the amount of the core complex to the wild-type level. An excess amount of rClpT1 (3% of total stromal protein) converted all the P-ring to the P/T1-ring and again increased the core content to 20% above wild-type levels.
Figure 6.
Restored Assembly of the Clp Proteolytic Core by Addition of Recombinant ClpT Proteins.
Stromal fractions from wild-type (WT) Arabidopsis and the two clpT mutants were isolated from 3-week-old plants. Different amounts of rClpT (0.03 to 3% of total stromal protein content) were then added to the stroma from the clpT mutants and left for 1 min at 22°C. Stromal fractions were then separated by native-PAGE and the various complexes of the Clp proteolytic core identified by immunoblotting with specific marker antibodies (ClpP6 and ClpR3).
(A) rClpT1 was added to stroma from the clpT1 mutant and rClpT2 to stroma from clpT2 mutant.
(B) rClpT1 was added to stroma from clpT2 and rClpT2 added to stroma from clpT1.
The corresponding experiments were performed for ClpT2, adding similar amounts of rClpT2 to stroma extracts from the clpT2 mutant (Figure 6A). As for rClpT1, low levels of rClpT2 (0.03%) restored almost wild-type levels (90%) of the core complex in the corresponding mutant. Interestingly, additional rClpT2 (0.3%) formed even more core complex; almost double that in the wild type. Excess rClpT2 (3%), however, had less effect on the core complex but appeared to convert all the P-ring to the P/T1-ring, suggesting it can either substitute for ClpT1 in this 200-kD complex or promote the binding of preexisting ClpT1. A novel 220-kD complex was also observed, almost certainly corresponding to a P/T1/T2-ring complex. It should be noted that all the changes described above occurred within 1 min of adding either rClpT1 or rClpT2 and that longer incubations (up to 20 min) had no observable effect (data not shown). Furthermore, the fact that the in vitro reconstitution experiments also complement the reduced core phenotypes of the clpT1 and clpT2 null mutants confirms the causal link between phenotype and gene mutation.
We next examined more closely the ability of the two ClpT proteins to compensate for each other in the assembly of the various Clp complexes (Figure 6B). First, increasing amounts of rClpT2 were added to stromal extracts from the clpT1 mutant. As the amount of added rClpT2 rose from 0.03 to 0.3% of the total stromal protein, more of the P-ring was converted to the 200-kD form. More of the core complex was also formed but still markedly less (65%) than the wild-type level. Excess rClpT2 fully converted the P-ring in the clpT1 mutant to the 220-kD form but failed to restore any additional core complex. This confirmed that high concentrations of ClpT2 can indeed substitute for ClpT1 in the P/T1-ring, but it is less capable of forming stable core complex. The corresponding experiment of adding rClpT1 to stromal extracts from the clpT2 mutant revealed that ClpT1 has little or no ability to substitute for ClpT2. As shown in Figure 6B, increasing amounts of rClpT1 converted the P-ring until all formed the P/T1-ring, but only a marginal amount of the core complex was restored. It should be noted that the slightly larger sizes of the core and P-ring complexes when rClpT proteins are bound is due to reduced mobility of the oligomers on the standard native-PAGE conditions, which is used for optimal resolution of the various Clp complexes. Extended electrophoresis conditions similar to those used in Figure 5 confirmed that these complexes with rClpT match the sizes of their native equivalents.
ClpT1 and ClpT2 Regulate the Assembly of the Clp Proteolytic Core
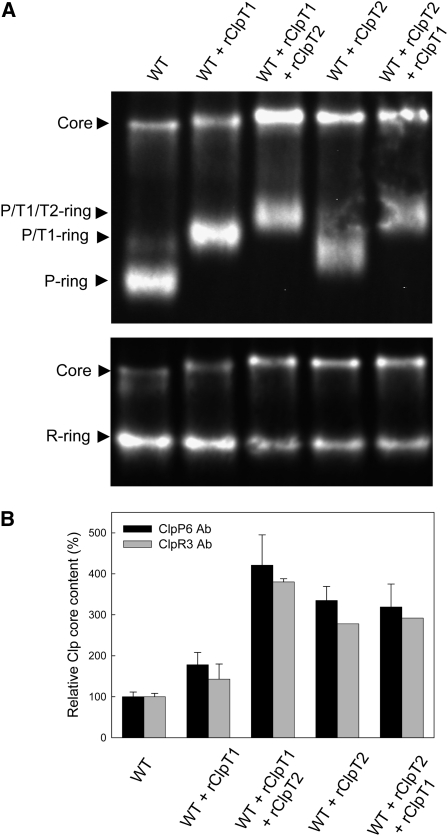
Given that addition of rClpT proteins could not only restore wild-type levels of the Clp core complex in the clpT mutants but also increase the amount further, we tested if the availability of ClpT was in fact limiting the assembly of the Clp core in the wild type (Figure 7A). Since within the assembly pathway of the core complex ClpT1 appears to associate with the P-ring before ClpT2, we first added an excess amount of rClpT1 (1%) to wild-type stroma. Upon addition of rClpT1, all the relatively abundant P-ring was rapidly converted to the P/T1-ring, suggesting the availability of native ClpT1 normally restricts this assembly step. The amount of the Clp core complex also increased but only by ~50% (Figure 7B), suggesting that the level of the P/T1-ring (and thereby ClpT1 itself) also limits the assembly of the proteolytic core.
Figure 7.
Increased Clp Proteolytic Core Content in Wild-Type Arabidopsis by Addition of Recombinant ClpT.
Stroma was extracted from 3-week-old wild-type leaves and then incubated with excess rClpT1 for 1 min at 22°C and then excess rClpT2 for a further 1 min (both ClpT proteins were added at 1% of total stromal protein content). Aliquots of stroma were taken prior to rClpT1 addition (WT), after 1 min incubation with added rClpT1 (WT + rClpT1), and after 1 min incubation with rClpT2 (WT + rClpT1 + rClpT2). The reverse experiment was then performed adding first rClpT2 and then rClpT1 following the same procedure as above. Aliquots were separated by native-PAGE, and complexes of the Clp proteolytic core identified by immunoblotting with specific marker antibodies (ClpP6 and ClpR3) (A) with the relative amount of the Clp proteolytic core then quantified (B). Values shown are averages ± se (n = 3) with the wild-type values set to 100%.
We then added an excess amount of rClpT2 (1%) to the wild-type stroma containing rClpT1. Addition of rClpT2 resulted in a relatively large increase in the amount of the core complex, over 4 times the normal wild-type level (Figure 7B). This shows that the availability of native ClpT2 also regulates the assembly of the core complex. The fact that the P/T1/T2-ring also forms a subcomplex that normally does not accumulate in the wild type suggests that the assembly of additional core complex is restricted by another as yet unknown factor, a conclusion supported by the remaining amounts of R-ring. Indeed, only ~20% of the R-ring is converted to core upon the addition of the two rClpT proteins, whereas ~50% of the P-ring remains in the form of the P/T1/T2-ring. This suggests that the R-ring is relatively more abundant than the P-ring, highlighting again the importance of ClpT regulation on the P-ring for controlling core formation.
When adding the rClpT proteins in reverse order, a similar increase in core content was observed. After first adding an excess amount of rClpT2 to wild-type stroma, all the available P-ring was converted to the P/T1-ring, again highlighting the ability of ClpT2 at high concentrations to substitute for ClpT1. The amount of core also increased by ~threefold, confirming that the availability of native ClpT2 restricts core formation. Subsequent addition of excess rClpT1 converted all the P/T1-ring to the P/T1/T2-ring but did not further increase core content. The fact that the rise in core content was less when rClpT2 was added before rClpT1 supports the conclusion from the reconstitution experiments (Figure 6) that ClpT1 associates first with the P-ring followed by ClpT2 during normal core assembly.
DISCUSSION
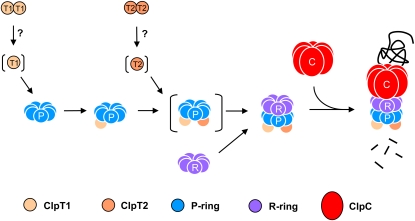
In this study, we have shown that the function of ClpT in chloroplasts of Arabidopsis is essential for plant viability. It now seems clear that ClpT specifically regulates the assembly of the Clp proteolytic core and thereby the Clp protease itself. We propose that this assembly process follows the steps outlined in Figure 8. As shown previously, the core complex consists of two distinct rings: the P- and R-rings (Sjögren et al., 2006). More than 50% of the various ClpP and ClpR subunits within wild-type Arabidopsis accumulate in either of these two ring structures, suggesting their association to form the core complex is restricted in some way. The next step in the assembly pathway is ClpT1 association with the P-ring. The increase in size of the P-ring is consistent with a ClpT1 monomer binding per ring, which is also consistent with previously quantifications of ClpT1 content in the core complex (Peltier et al., 2004). ClpT1 monomers presumably originate from the free ClpT1 dimers and have high affinity for the P-ring since they do not accumulate in the wild type. The amount of the P/T1-ring is relatively small in the wild type, but it does accumulate to observable levels, implying that core complex formation is again restricted at a later step. Formation of the P/T1-ring itself is clearly regulated by the availability of ClpT1, as demonstrated by the complete conversion of the P-ring to the P/T1-ring by adding sufficient rClpT1. However, levels of the P/T1-ring within the wild type limit core assembly to a lesser extent. Even when all the P-ring is converted to the P/T1 form by the addition of excess rClpT1, core content increases by only 50% above wild-type levels.
Figure 8.
Model for the Assembly Pathway of the Chloroplast Clp Protease in Arabidopsis.
The bulk of ClpT1 and ClpT2 in the stroma exists as homogeneous dimers. Prior to association with the Clp proteolytic core, both ClpT dimers undergo monomerization by an unknown mechanism. Both ClpT monomers presumably have high affinity for the P-ring since they do not accumulate within the wild-type stroma (as indicated by parentheses). ClpT1 binds first to the P-ring followed by ClpT2. It is the P/T1/T2-ring that has high affinity for the R-ring to form the intact Clp proteolytic core complex. Later association of the ClpC chaperone partner completes the assembly of the active Clp protease.
[See online article for color version of this figure.]
Association between the P/T1-ring and the R-ring is also clearly regulated by the availability of ClpT2. Like ClpT1, ClpT2 appears to associate with the P-ring but only when ClpT1 is already bound. This P/T1/T2 complex is not observable in wild-type chloroplasts and accumulates only when excessive amounts of rClpT2 are added. The size of the P/T1/T2-ring is again consistent with a ClpT2 monomer per core complex (Peltier et al., 2004). As for ClpT1, ClpT2 monomers almost certainly derive from the abundant free ClpT2 dimers, the monomerization of which is probably regulated in the same way as for ClpT1. It is the P/T1/T2-ring that almost certainly associates with the R-ring to form the core complex, and it is the availability of ClpT2 that clearly limits this interaction. This is highlighted by the large increase in core content (fourfold) by the addition of excess rClpT2 when sufficient P/T1-ring is present. The P/T1/T2-ring therefore appears absent in the wild type since the relatively small amount of it formed immediately associates with the abundant R-ring to form the proteolytic core complex. Since the relative amount of the core complex is lower than that of the chaperone partners, it is the assembly of the core regulated by the availability of ClpT that controls the formation of the Clp protease itself.
The apparent existence of free P- and R-rings within the stroma raises the question of how their potential proteolytic activity is regulated to prevent inadvertent protein degradation. Such a problem is normally avoided structurally by both narrow entrance apertures to the inner chamber of the core complex and the sequestering of the active sites of the catalytic ClpP subunits within (Wang et al., 1997). Although little is known about the structure of such Clp subcomplexes and how accessible the active site of each ClpP subunit is, evidence that they are likely to be proteolytically inactive comes from recent studies of the cyanobacterial counterpart to the chloroplast Clp protease (Andersson et al., 2009). In the model cyanobacterium Synechococcus, the main Clp proteolytic core consists of a mixture of one catalytic (ClpP3) and one noncatalytic subunit (ClpR). A functional recombinant form of this core complex was able to be purified but only by coexpressing both clpP3 and clpR genes in the same bacterial cell. Earlier purifications of ClpP3 alone showed that it assembled only into a single heptameric ring and was unable to form a stable core complex (Andersson et al., 2009). Despite this, α-casein degradation assays with this purified ClpP3 protein showed no degradation activity, with or without ClpC. This suggests that a single ring of catalytic ClpP subunits is proteolytically inactive, which might explain how free P- and R-rings could exist in the chloroplast.
Although the two ClpT proteins have similar amino acid sequences, they have limited ability to compensate for each other. This was first evident in the single-gene knockout mutants in which the remaining ClpT could only partially substitute for the missing paralog in the assembly of the core complex. This was particularly so for ClpT1, which despite being upregulated in the clpT2 mutant could maintain only 15% of the core complex. Conversely, ClpT2 was better able to replace ClpT1 function, with 30% of the core complex present in the clpT1 mutant. This was also shown by addition of rClpT2 restoring up to 50% wild-type levels of the core complex in stroma from the clpT1 mutant. Interestingly, we have observed that the amount of Clp core complex must drop to 10% of wild-type levels or lower before phenotypic changes are observed in Arabidopsis (Sjögren et al., 2006; Stanne et al., 2009). This threshold might well explain why ClpT1 was induced in the clpT2 mutant but not ClpT2 in the clpT1 mutant.
One seemingly paradoxical feature arising from this study is that even though the amount of ClpT1 and ClpT2 clearly restricts the assembly of the Clp proteolytic core, both proteins are present in the stroma at levels more than sufficient to facilitate this process. Of the total ClpT1 within the stroma, less than half associates with the core complex or the P-ring. The situation is more extreme for ClpT2, with <5% of total ClpT2 bound to the core. Instead, the majority of both proteins exist as homogeneous dimers, suggesting that in this state, ClpT is unavailable for association with the other Clp complexes. If so, then the monomerization of these ClpT dimers might well regulate the availability of ClpT1 and ClpT2 for association with the core complex. This possibility, however, is seemingly contradicted by the results with the rClpT proteins. Both rClpT1 and rClpT2 form a dimer corresponding to that of their native counterparts and yet they were freely available to bind to the P-ring and core complex. This suggests that monomerization of the ClpT dimers per se is not rate limiting for core assembly. Interestingly, the monomerization step itself could well be facilitated by ClpC, given that its bacterial counterpart ClpA is known to possess such a chaperone activity (i.e., the activation of RepA via monomerization of RepA dimers; Wickner et al., 1994). Indeed, ClpC could have been the factor limiting the increase in core assembly when excess rClpT proteins were added to wild-type stroma. We recently estimated that the stromal ClpC content is ~4 times higher than that of the Clp proteolytic core under our standard growth conditions. It might be that once the core increases four- to fivefold by the addition of rClpT1/2 as shown in Figure 7 that all the available ClpC in the stroma is then complexed to the core, and none remains to monomerize additional rClpT for continued core assembly. Despite this, it seems clear that the native ClpT dimers differ from those of the recombinant ones in a way that somehow restricts their monomerization. Such a difference could be some form of posttranslational modification (e.g., phosphorylation) that needs to be removed prior to monomerization, a possibility we are now in the process of examining.
Given the need for ClpT in the assembly of the Clp proteolytic core, the question arises as to why such a regulatory protein is needed for the chloroplast Clp protease in plants but not those in green algae (Majeran et al., 2005) or for the cyanobacterial progenitor (Stanne et al., 2007). One possible explanation might lie in the structural dissimilarity between the different types of core complexes. The main proteolytic core in Synechococcus consists of two identical heptameric rings, each containing three ClpP3 and four ClpR subunits arranged in a defined alternating pattern (Andersson et al., 2009). The two identical rings almost certainly bind symmetrically to each other, with each ClpP and ClpR subunit in one ring aligning with the corresponding subunit in the adjacent ring. Assuming this was the original type of Clp proteolytic core during the evolution of chloroplasts, it has since changed in plants to a more asymmetrical unit in which one ring contains only ClpP subunits and the other all the ClpR subunits. Whatever the underlying reason for this extensive change to the chloroplast core complex, it might well have inadvertently destabilized the association between the two rings. If so, there would have been a strong selection pressure to restabilize core formation, possibly giving rise to ClpT that binds to the P-ring and facilitates its association to the R-ring.
An alternative explanation for the development of ClpT might lie in the specific form of regulation it confers to the assembly of the Clp core complex (this explanation must have in fact been a consequence from the original need to stabilize the core complex). It is clear from this study that the availability of both ClpT1 and ClpT2 affects the amount of core complex that forms and thereby the Clp protease itself. Providing more ClpT protein can rapidly increase the amount of the core complex without the need for new protein synthesis. This regulatory mechanism would therefore enable plant chloroplasts to adjust quickly the levels of Clp protease within the stroma as needed. Such fine-tuning of the plastidic proteolytic activity could be important during plant development, as has been suggested for plants exhibiting leaf variegated phenotypes in which chloroplast protein expression and retrograde signals are somehow disrupted at an early developmental stage (Liu et al., 2010). This form of regulation would also be an obvious advantage to sessile plants that must respond to often fluctuating environmental conditions throughout their lifetime. Indeed, it has been previously shown that the level of almost all of the chloroplast Clp proteins in Arabidopsis remains unchanged during many different types of stresses (Shanklin et al., 1995; Ostersetzer and Adam, 1996; Zheng et al., 2002), whereas the Clp proteins in eubacteria are often highly stress inducible (Porankiewicz et al., 1999). By maintaining excess pools of the ClpT dimers and the various assembly intermediates of the Clp protease in the stroma (i.e., the P- and R-rings and ClpC), the amount of the chloroplast Clp protease can be readily and simply adjusted by the release of available ClpT monomers depending on the required proteolytic activity. Such a regulatory mechanism conferred by ClpT would therefore add a new dimension to the functional importance of chloroplast Clp protease in plants, which has until now been considered exclusively as a constitutive housekeeping enzyme.
METHODS
Plant Growth Conditions
Seeds for Arabidopsis thaliana wild type (ecotype Columbia-0), the clpT1 and clpT2 T-DNA insertion mutants, and the clpP6 antisense line (Sjögren et al., 2006) were sown in a perlite/soil mix (1:5) after vernalization at 4°C for at least 48 h to break dormancy. Plants were cultivated individually in pots or as lawns under the following standard conditions: 8-h photoperiod with white light (~150 μmol photons m−2 s−1), 23/18°C day-night temperatures, and 65% relative air humidity.
Identification of Arabidopsis clpT Null Mutants
Putative clpT mutants were screened by electronic BLAST searches of available populations of Arabidopsis T-DNA insertion mutants using genomic sequences for CLPT1 and CLPT2. One possible clpT1 and one possible clpT2 mutant were identified in the SALK (SALK_05772; Alonso et al., 2003) and SAIL (SAIL_340-A10; Sessions et al., 2002) collections, respectively. Seeds from both lines are obtained and screened using either kanamycin or BASTA resistance as selective marker. Homozygous lines with a single T-DNA insertion were later identified by segregation analysis as previously described (Sjögren et al., 2004).
Crossing of clpT1 and clpT2 Null Mutants
The confirmed clpT1 and clpT2 null mutants were crossed together using clpT2 as a pollen donor. The F1 progeny were selected on BASTA and then screened by immunoblotting using the ClpT1- and ClpT2-specific antibodies, confirming all as heterozygotes. Immunoblotting was again used to screen 100 individual plants from the F2 population in search of viable clpT1 clpT2 double null mutants. F2 seeds in mature siliques of F1 plants were dissected and visualized by light microscopy (Olympus SZ40).
Production of ClpT2-Specific Antibody
A polyclonal antibody specific to Arabidopsis ClpT2 was generated using a synthetic peptide corresponding to the unique amino acid sequence CELESFASESGFLDE. The peptide was conjugated to BSA and then injected into rabbits intramuscularly and subcutaneously (AgriSera).
Purification of Recombinant ClpT1 and ClpT2
To prepare recombinant forms of Arabidopsis ClpT1 and ClpT2, the mature N terminus of both proteins was first determined using prediction software (TargetP; Emanuelsson et al., 2007), sequence alignment with ClpT orthologs in other plant species, and the available online mass spectrometry database PPDB (http:/ppdb.tc.cornell.edu/) so as to exclude sequences for the chloroplast transit peptide. The Arabidopsis CLPT1 and CLPT2 genes were then amplified from cDNA clones using high-fidelity pfx DNA polymerase and specific primers containing NdeI or KpnI restriction sites to facilitate directional cloning. Also included in the 3′ primer was additional sequence to add His6-tags to the C termini of both ClpT proteins. The primer sequences were as follows: CLPT1 5′ primer, 5′-CATATGTCGGCCAGCACGGTCTTAAACGTC-3′; CLPT1 3′ primer, 5′-GGTACCCTAGTGATGGTGATGGTGATGTTCACCTTGTTTCTTGAAGCTCAAATCTACATC-3′; CLPT2 5′ primer, 5′-CATATGAGCTTACCCACCGCGATTCCAG-3′; CLPT2 3′ primer, 5′-GGTACCCTAGTGATGGTGATGGTGATGTTCATCTAAAAAGCCAGATTCAGAGGCAAAAG-3′. The digested PCR products were ligated into the pCDFDuet-1 vector (Novagen) digested with the same enzymes and transformed into Escherichia coli BL-21 CodonPlus cells (Clontech). The cloned clpT1 and clpT2 genes were sequenced to verify their integrity. Overexpression of ClpT1 and ClpT2 in E. coli and their subsequent purification from cell lysates by sequential Ni2+ affinity and gel filtration chromatography was performed as previously described (Andersson et al., 2006). The purified rClpT proteins were stored in 20 mM Tris-HCl, pH 7.5, 75 mM NaCl, and 20% (w/v) glycerol.
Protein Extractions
Total leaf proteins and intact chloroplasts were isolated from 3-week-old lawns of the wild type, clpT1 and clpT2 null mutants, and clpP6 antisense lines as previously described (Sjögren et al., 2006). Stromal proteins were fractionated from purified intact chloroplasts and the protein concentration determined as detailed earlier (Sjögren et al., 2006).
PAGE and Immunoblotting
All protein samples were separated under denaturing conditions using the NuPAGE system as previously described (Sjögren et al., 2006). Protein complexes from stromal fractions were separated by native-PAGE using a Tris-borate system as previously described (Sjögren et al., 2006; Stanne et al., 2009). Prior to separation by native-PAGE, the rClpT proteins were diluted in native sample buffer (45 mM Tris-HCl, 45 mM boric acid, 0.002% [w/v] bromphenol blue, and 7% [w/v] glycerol, final concentration). For more accurate size determination of ClpT1 and ClpT2, a modified form of the Tris-borate system was used in which the native protein samples were separated on 7 to 23% polyacrylamide gradient, Tris-borate (45 mM Tris-borate, pH 8.3) gels (16 cm long, 1 mm thick). Gels were electrophoresed at a constant current of 8 mA for up to 69 h at 4°C. Native size markers used were lactalbumin (14 kD), BSA (66-kD monomer, 132-kD dimer), and ferritin (440-kD monomer, 880-kD dimer). Once separated, proteins were either stained with Coomassie Brilliant Blue or transferred to nitrocellulose membranes for immunoblotting (Sjögren et al., 2006). Specific polyclonal antibodies were used to detect the following Clp proteins: ClpC, dilution 1:20,000 (Porankiewicz and Clarke, 1997); ClpD, 1:10,000 (Zheng et al., 2002); ClpP1, 1:1000 (Zheng et al., 2002); ClpP3, 1:5000 (Zheng et al., 2002); ClpP4, 1:3000 (Zheng et al., 2002); ClpP5, 1:1000 (Zheng et al., 2002); ClpP6, 1:10,000 (Zheng et al., 2002); ClpR1, 1:500 (Sjögren et al., 2004); ClpR2, 1:1000 (Sjögren et al., 2004); ClpR3, 1:5000 (Sjögren et al., 2004); ClpR4, 1:4000 (Sjögren et al., 2004); and ClpT1, 1:4000 (Sjögren et al., 2006). Primary antibodies were detected with the horseradish peroxidize–linked anti-rabbit IgG secondary antibody and visualized by enhanced chemiluminescence (GE Healthcare). Chemiluminescent signals were captured and quantified using the ChemiGenius2 imaging system and associated software (Syngene).
Addition of rClpT Proteins to Stroma from clpT Null Mutants
Different amounts (0.03, 0.3, or 3% of total stromal protein content) of rClpT1 or rClpT2 were added to isolated stroma (3.8 μg protein/μL final concentration) from the clpT1 or clpT2 null mutants. As a negative control, an equal volume of the rClpT storage buffer was added to the stromal fractions from the wild type and the two clpT mutants. All samples were incubated at 22°C for 1 min and then placed at 4°C with an equal volume of 2× TB loading buffer (45 mM Tris-HCl, pH 8.3, 45 mM boric acid, 20% glycerol, and 0.02% bromophenol blue) added. Samples were separated by native-PAGE (Sjögren et al., 2006; Stanne et al., 2009) and transferred to nitrocellulose membranes for immunoblotting (Sjögren et al., 2006). The Clp proteolytic core and its various assembly intermediates were detected using specific marker antibodies: ClpP6 antibody for the Clp core, P-ring (ClpP3-6) and P/T1-ring (ClpP3-6 and ClpT1), ClpR3 for the Clp core and R-ring (ClpP1 and ClpR1-4).
Addition of rClpT Proteins to Wild-Type Stroma
Excess amounts of rClpT1 and rClpT2 (1% of total stromal protein content) were added to isolated wild-type stroma stepwise. The rClpT1 protein (0.4 μg) was first added to isolated wild-type stroma (40 μg) and incubated for 1 min at 2°C. The rClpT2 protein (0.4 μg) was then added and the sample incubated for another 1 min at 22°C. The reverse experiment was also performed in which the rClpT2 protein was added before ClpT1 using the same procedure as above. As a negative control, an equal volume of storage buffer was added to a wild-type stroma sample when each rClpT protein was added to the experimental samples. All samples were separated by native-PAGE and the Clp proteolytic core and subcomplexes detected by immunoblotting as described above.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: CLPP1, ATCG00670; CLPP3, At1g66670; CLPP4, At5g45390; CLPP5, At1g02560; CLPP6, At1g11750; CLPR1, At1g49970; CLPR2, At1g12410; CLPR3, At1g09130; CLPR4, At4g17040; CLPT1, At4g25370; and CLPT2, At4g12060.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Size Determination of rClpT1 and rClpT2 by Native-PAGE.
Supplemental Figure 2. Relative Amount of Stromal ClpT1 in Wild-Type Arabidopsis.
Supplementary Material
Acknowledgments
We thank Mats Andersson for technical advice on the crossing of the Arabidopsis clpT mutants and Henrik Antonsson for help with the statistical analysis of the F2 progeny. This work was supported by grants to A.K.C. from The Swedish Research Council for Environment, Agricultural Science, and Spatial Planning (Formas) and from The Swedish Research Council (Vetenskapsrådet).
References
- Adam Z., Adamska I., Nakabayashi K., Ostersetzer O., Haussuhl K., Manuell A., Zheng B., Vallon O., Rodermel S.R., Shinozaki K., Clarke A.K. (2001). Chloroplast and mitochondrial proteases in Arabidopsis. A proposed nomenclature. Plant Physiol. 125: 1912–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam Z., Clarke A.K. (2002). Cutting edge of chloroplast proteolysis. Trends Plant Sci. 7: 451–456 [DOI] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Andersson F.I., Blakytny R., Kirstein J., Turgay K., Bukau B., Mogk A., Clarke A.K. (2006). Cyanobacterial ClpC/HSP100 protein displays intrinsic chaperone activity. J. Biol. Chem. 281: 5468–5475 [DOI] [PubMed] [Google Scholar]
- Andersson F.I., et al. (2009). Structure and function of a novel type of ATP-dependent Clp protease. J. Biol. Chem. 284: 13519–13532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker T.A., Sauer R.T. (2006). ATP-dependent proteases of bacteria: Recognition logic and operating principles. Trends Biochem. Sci. 31: 647–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke A.K., MacDonald T.M., Sjögren L.L.E. (2005). The ATP-dependent Clp protease in chloroplasts of higher plants. Physiol. Plant. 123: 406–412 [Google Scholar]
- Emanuelsson O., Brunak S., von Heijne G., Nielsen H. (2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2: 953–971 [DOI] [PubMed] [Google Scholar]
- Grimaud R., Kessel M., Beuron F., Steven A.C., Maurizi M.R. (1998). Enzymatic and structural similarities between the Escherichia coli ATP-dependent proteases, ClpXP and ClpAP. J. Biol. Chem. 273: 12476–12481 [DOI] [PubMed] [Google Scholar]
- Ishikawa T., Beuron F., Kessel M., Wickner S., Maurizi M.R., Steven A.C. (2001). Translocation pathway of protein substrates in ClpAP protease. Proc. Natl. Acad. Sci. USA 98: 4328–4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Rudella A., Ramirez Rodriguez V., Zybailov B., Olinares P.D.B., van Wijk K.J. (2009). Subunits of the plastid ClpPR protease complex have differential contributions to embryogenesis, plastid biogenesis, and plant development in Arabidopsis. Plant Cell 21: 1669–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.I., Levchenko I., Fraczkowska K., Woodruff R.V., Sauer R.T., Baker T.A. (2001). Molecular determinants of complex formation between Clp/Hsp100 ATPases and the ClpP peptidase. Nat. Struct. Biol. 8: 230–233 [DOI] [PubMed] [Google Scholar]
- Koussevitzky S., Stanne T.M., Peto C.A., Giap T., Sjögren L.L.E., Zhao Y., Clarke A.K., Chory J. (2007). An Arabidopsis thaliana virescent mutant reveals a role for ClpR1 in plastid development. Plant Mol. Biol. 63: 85–96 [DOI] [PubMed] [Google Scholar]
- Kress W., Maglica Z., Weber-Ban E. (2009). Clp chaperone-proteases: Structure and function. Res. Microbiol. 160: 618–628 [DOI] [PubMed] [Google Scholar]
- Kuroda H., Maliga P. (2003). The plastid clpP1 protease gene is essential for plant development. Nature 425: 86–89 [DOI] [PubMed] [Google Scholar]
- Liu X., Yu F., Rodermel S. (2010). An Arabidopsis pentatricopeptide repeat protein, SUPPRESSOR OF VARIEGATION7, is required for FtsH-mediated chloroplast biogenesis. Plant Physiol. 154: 1588–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran W., Friso G., van Wijk K.J., Vallon O. (2005). The chloroplast ClpP complex in Chlamydomonas reinhardtii contains an unusual high molecular mass subunit with a large apical domain. FEBS J. 272: 5558–5571 [DOI] [PubMed] [Google Scholar]
- Ortega J., Lee H.S., Maurizi M.R., Steven A.C. (2002). Alternating translocation of protein substrates from both ends of ClpXP protease. EMBO J. 21: 4938–4949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostersetzer O., Adam Z. (1996). Effects of light and temperature on expression of ClpC, the regulatory subunit of chloroplastic Clp protease, in pea seedlings. Plant Mol. Biol. 31: 673–676 [DOI] [PubMed] [Google Scholar]
- Peltier J.B., Ripoll D.R., Friso G., Rudella A., Cai Y., Ytterberg J., Giacomelli L., Pillardy J., van Wijk K.J. (2004). Clp protease complexes from photosynthetic and non-photosynthetic plastids and mitochondria of plants, their predicted three-dimensional structures, and functional implications. J. Biol. Chem. 279: 4768–4781 [DOI] [PubMed] [Google Scholar]
- Porankiewicz J., Clarke A.K. (1997). Induction of the heat shock protein ClpB affects cold acclimation in the cyanobacterium Synechococcus sp. strain PCC 7942. J. Bacteriol. 179: 5111–5117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porankiewicz J., Wang J., Clarke A.K. (1999). New insights into the ATP-dependent Clp protease: Escherichia coli and beyond. Mol. Microbiol. 32: 449–458 [DOI] [PubMed] [Google Scholar]
- Sessions A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanklin J., DeWitt N.D., Flanagan J.M. (1995). The stroma of higher plant plastids contain ClpP and ClpC, functional homologs of Escherichia coli ClpP and ClpA: An archetypal two-component ATP-dependent protease. Plant Cell 7: 1713–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai T., Shimizu K., Ueda K., Nishimura Y., Kuroiwa T., Hashimoto T. (2001). The chloroplast clpP gene, encoding a proteolytic subunit of ATP-dependent protease, is indispensable for chloroplast development in tobacco. Plant Cell Physiol. 42: 264–273 [DOI] [PubMed] [Google Scholar]
- Sjögren L.L.E., MacDonald T.M., Sutinen S., Clarke A.K. (2004). Inactivation of the clpC1 gene encoding a chloroplast Hsp100 molecular chaperone causes growth retardation, leaf chlorosis, lower photosynthetic activity, and a specific reduction in photosystem content. Plant Physiol. 136: 4114–4126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjögren L.L.E., Stanne T.M., Zheng B., Sutinen S., Clarke A.K. (2006). Structural and functional insights into the chloroplast ATP-dependent Clp protease in Arabidopsis. Plant Cell 18: 2635–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanne T.M., Pojidaeva E., Andersson F.I., Clarke A.K. (2007). Distinctive types of ATP-dependent Clp proteases in cyanobacteria. J. Biol. Chem. 282: 14394–14402 [DOI] [PubMed] [Google Scholar]
- Stanne T.M., Sjögren L.L.E., Koussevitzky S., Clarke A.K. (2009). Identification of new protein substrates for the chloroplast ATP-dependent Clp protease supports its constitutive role in Arabidopsis. Biochem. J. 417: 257–268 [DOI] [PubMed] [Google Scholar]
- Wang J., Hartling J.A., Flanagan J.M. (1997). The structure of ClpP at 2.3 A resolution suggests a model for ATP-dependent proteolysis. Cell 91: 447–456 [DOI] [PubMed] [Google Scholar]
- Wickner S., Gottesman S., Skowyra D., Hoskins J., McKenney K., Maurizi M.R. (1994). A molecular chaperone, ClpA, functions like DnaK and DnaJ. Proc. Natl. Acad. Sci. USA 91: 12218–12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B., Halperin T., Hruskova-Heidingsfeldova O., Adam Z., Clarke A.K. (2002). Characterization of Chloroplast Clp proteins in Arabidopsis: Localization, tissue specificity and stress responses. Physiol. Plant. 114: 92–101 [DOI] [PubMed] [Google Scholar]
- Zheng B., MacDonald T.M., Sutinen S., Hurry V., Clarke A.K. (2006). A nuclear-encoded ClpP subunit of the chloroplast ATP-dependent Clp protease is essential for early development in Arabidopsis thaliana. Planta 224: 1103–1115 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.