This work describes Pollen Semi-Sterility1, a kinesin-1 like protein that is involved in regulating male meiosis in rice; in pss1 mutants, chromosome segregation is perturbed, causing reduced pollen viability and partial sterility.
Abstract
In flowering plants, male meiosis produces four microspores, which develop into pollen grains and are released by anther dehiscence to pollinate female gametophytes. The molecular and cellular mechanisms regulating male meiosis in rice (Oryza sativa) remain poorly understood. Here, we describe a rice pollen semi-sterility1 (pss1) mutant, which displays reduced spikelet fertility (~40%) primarily caused by reduced pollen viability (~50% viable), and defective anther dehiscence. Map-based molecular cloning revealed that PSS1 encodes a kinesin-1–like protein. PSS1 is broadly expressed in various organs, with highest expression in panicles. Furthermore, PSS1 expression is significantly upregulated during anther development and peaks during male meiosis. The PSS1–green fluorescent protein fusion is predominantly localized in the cytoplasm of rice protoplasts. Substitution of a conserved Arg (Arg-289) to His in the PSS1 motor domain nearly abolishes its microtubule-stimulated ATPase activity. Consistent with this, lagging chromosomes and chromosomal bridges were found at anaphase I and anaphase II of male meiosis in the pss1 mutant. Together, our results suggest that PSS1 defines a novel member of the kinesin-1 family essential for male meiotic chromosomal dynamics, male gametogenesis, and anther dehiscence in rice.
INTRODUCTION
Meiosis is a crucial event in sexual reproduction of eukaryotes. It entails one round of DNA replication and two successive rounds of nuclear division, meiosis I and meiosis II. As a result, diploid somatic cells are converted into haploid gametes needed for fertilization. Meiosis I is a reductional division involving segregation of homologous chromosomes. Meiosis II is an equational division involving segregation of sister chromatids. Both meiosis I and II are further divided into four phases: prophase, metaphase, anaphase, and telophase (Zickler and Kleckner, 1999). During meiosis, homologous chromosome pairing and recombination occur to achieve two contrary purposes: maintenance of genome stability and creation of genetic diversity in offspring. Thus, meiosis plays a critical role in reproduction and evolution of eukaryotes.
Over the last 10 years, we have witnessed great progress in our understanding of the molecular mechanisms controlling meiosis in plants, owing to using forward and reverse genetics screens, availability of genome sequences, and advances in cytological procedures and tools. Approximately 50 genes involved in regulating meiosis have been identified and characterized in plants, including Arabidopsis thaliana, maize (Zea mays), and rice (Oryza sativa; Mercier and Grelon, 2008). These studies have not only demonstrated conservation of gene functions among yeast, plants, and other higher eukaryotes, but also uncovered novel aspects of the molecular mechanisms involved in meiosis in higher plants (Ma, 2006; Ronceret et al., 2008).
Rice is a staple food for nearly half of the world’s population and a model species for monocots developmental studies (Itoh et al., 2005). Development and utilization of hybrid rice have greatly contributed to ensuring sufficient food supply in China and other Asian countries (Virmani, 1996). The genus Oryza consists of more than 20 wild and two cultivated species, indica and japonica. The interspecific F1 hybrids between cultivars and wild-type species often show high sterility, which is partly attributable to defects in homologous pairing at meiosis (Katayama, 1963; Brar and Khush, 1997). Dissecting the meiotic process of Oryza will contribute to reducing reproduction barriers between different cultivars and species. Several genes regulating male meiosis have been identified and functionally characterized in rice (Jenkins et al., 2008). Among them, MEL1 encodes a novel ARGONAUTE family protein essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice (Nonomura et al., 2007). Rice PAIR1, PAIR2, and PAIR3 are essential for homologous chromosome pairing in meiosis I. PAIR2 is an ortholog of Arabidopsis ASY1 proteins (Caryl et al., 2000; Armstrong et al., 2002; Nonomura et al., 2004b, 2006), whereas rice PAIR1 and PAIR3 are putative coiled-coil proteins that do not appear to have close sequence homologs in other organisms (Nonomura et al., 2004a; Yuan et al., 2009). Two homologs of yeast DMC1, rice DMC1A and DMC1B, have also been identified and shown to be required for homologous pairing (Ding et al., 2001; Kathiresan et al., 2002; Deng and Wang, 2007). Rice MER3, similar to its counterparts in yeast and Arabidopsis, encodes a protein required for normal meiotic crossover formation (Wang et al., 2009). Furthermore, genetic analysis also revealed that rice RAD21-4, a homolog of yeast RAD21/SCC1 proteins, is essential for efficient meiosis (Zhang et al., 2006). However, despite the progress, our understanding of the molecular mechanisms regulating meiosis in rice remains fragmentary.
Kinesins are key players of intracellular transport system and thus are essential for many cellular functions and morphogenesis. Kinesins use microtubules as tracks and transport diverse cellular cargos, such as organelles, protein complexes, and mRNAs to specific destinations. Kinesins also participates in chromosomal and spindle movements during mitosis and meiosis (Vale and Fletterick, 1997; Hirokawa et al., 1998; Sharp et al., 2000). All kinesins contain a highly conserved ~360–amino acid globular motor domain (the head or catalytic core) and a long filamentous structure (the stalk/tail). The head domain contains a catalytic pocket for the hydrolysis of ATP and the binding site for microtubules and is responsible for the movement empowered by the hydrolysis of ATP, whereas the stalk/tail domain is responsible for interacting with other subunits of the holoenzyme or with cargo molecules (Miki et al., 2005). Comprehensive genome sequence survey and systematic phylogenetic analyses have grouped kinesins into 14 distinct families. There are 61 kinesins in Arabidopsis (Reddy and Day, 2001) and 41 and 45 kinesins in japonica and indica rice, respectively (Richardson et al., 2006). To date, only one rice kinesin, named DWARF BAMBOO SHOOT1 (DBS1), has been analyzed genetically. The dbs1 mutant exhibits a severe dwarf phenotype due to defective cytokinesis (Sazuka et al., 2005).
In this study, we report a rice semisterile mutant, pollen semi-sterility1 (pss1), which exhibits ~50% pollen viability and ~40% spikelet fertility. The semisterility phenotype is stably inherited through generations. We found that spikelet semisterility was caused jointly by reduced pollen viability and by impairment in anther dehiscence. PSS1 encodes a kinesin-1–like protein and is preferentially expressed in flowers. We observed abnormal chromosome dynamics during male meiosis in the pss1 mutant and in the PSS1 RNA interference (RNAi) transgenic plants. Our results add further evidence to the crucial role of kinesins in regulating meiotic chromosome behavior during male gametogenesis and fertility control in higher plants.
RESULTS
Isolation and Morphological Characterization of the pss1 Mutant
In 1991, a spontaneous semisterile mutant showing ~40% spikelet fertility was found in a japonica rice variety, Nipponbare. The semisterile phenotype was stably inherited, and a semisterile progeny line, w207-2, was selected for further analysis. Genetic analysis indicated that a single recessive nuclear locus was responsible for the mutant phenotype (Yu et al., 2005). The mutant is slightly taller and earlier heading than wild-type plants but is otherwise normal in vegetative and floral development (Figures 1A to 1D). However, anthers of the mutant are shriveled at the basal part of the locules (Figure 1E). The most obvious difference between the mutant and the wild type is fertility. Under normal conditions, pollen viability of wild-type rice is ~97%, and the spikelet fertility is ~94%. In the mutants, pollen viability is ~51%, and spikelet fertility is ~40%. Whole-mount stain-clearing confocal microscopy examination showed that the embryo sac development is normal in the mutant (Figures 1F to 1K). To confirm this finding, we hand-pollinated the mutant with wild-type pollen, and the spikelet fertility reached ~86%. This result indicates that the spikelet semisterility is likely due to defective microspores. Therefore, we named this mutant pollen semi-sterility1 (pss1).
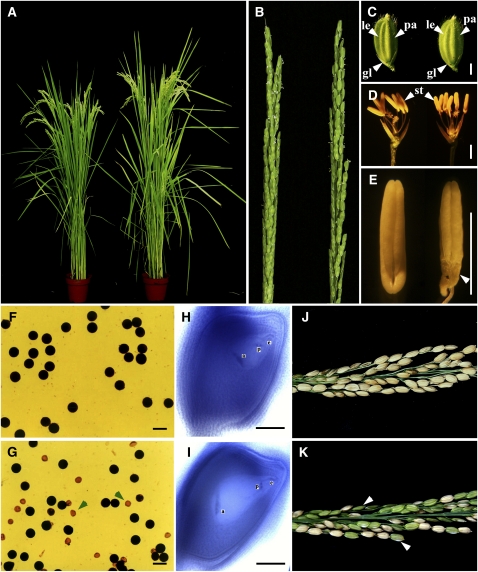
Figure 1.
Phenotypic Analyses of the pss1 Mutant.
(A) The pss1 mutant (right) is almost normal except it is a little taller and earlier heading than the wild-type plant (left) after bolting.
(B) The pss1 mutant has a normal panicle (right) compared with the wild type (left).
(C) The pss1 mutant spikelet (right) appears to be normal compared with the wild type (left) at bolting stage.
(D) Comparison of a wild-type (left) and a pss1 mutant spikelet (right) after removing the lemma and palea.
(E) Comparison of a wild-type (left) and a pss1 mutant anther (right). The basal part of the mutant anther is shriveled (arrowhead).
(F) KI-I2 staining of wild-type pollen.
(G) KI-I2 staining of pss1 mutant pollen. Arrowheads indicate abnormal pollen grains.
(H) and (I) Comparison of a mature wild-type embryo sac (H) and a mature mutant embryo sac (I) observed by the whole-mount stain-clearing confocal laser scanning microscopy method.
(J) and (K) Comparison of a wild-type panicle (J) and a pss1 mutant panicle (K) at the harvest stage. The spikelet fertility of the mutant is ~40%. Arrowheads indicate the sterile spikelets.
le, lemma; pa, palea; gl, glume; st, stamen; a, antipodal cells; p, polar nuclei; e, egg cell. Bars = 2 mm in (C) to (E) and 50 μm in (F) to (I).
To further characterize the difference between the pollen of the wild type and the pss1 mutant, we examined the pollen grains using scanning electron microscopy and transmission electron microscopy. Mature wild-type pollen grains are spherical and plump (Figures 2A and 2B) and contain large numbers of starch granules (Figure 2C). The pollen wall is composed of exine and intine. The exine can be divided into ektexine and endexine. The outside of ektexine is tectum, and the inner side is foot layer. The tectum and foot layer are linked by columella (Figure 2D). In the pss1 mutant, ~50% of the pollen grains have the round and plump shape just like wild-type pollen, while the other ~50% of pollen grains have an irregular and shrunken appearance (Figure 2E). The round pollen grains have a similar shape and internal structure to wild-type pollen grains, with large numbers of starch granules and a normal pollen wall (Figures 2F to 2H). Pollen germination tests (see below) indicated that these pollen grains are viable. By contrast, the shrunken pollen grains have no accumulations of starch granules (Figures 2I and 2J). These irregularly shaped pollen grains also lack intine and have abnormal exine. Their tectum and foot layers are much plumper than the normal ones, and the columella layer tends to degenerate (Figure 2K). Pollen germination tests (see below) indicated that these pollen grains are not viable.
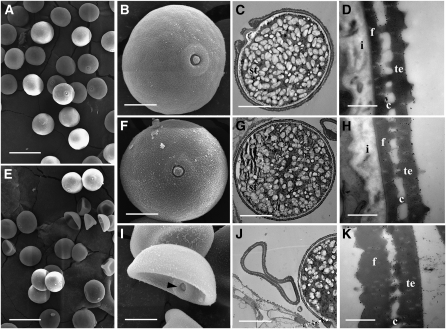
Figure 2.
Scanning Electron Microscopy and Transmission Electron Microscopy Examination of Wild-Type and pss1 Mutant Pollen.
(A) Scanning electron microscopy image of mature wild-type pollen grains.
(B) A higher magnification image of a single pollen grain from (A).
(C) Transmission electron microscopy image showing that the wild-type pollen grain contains a large number of starch granules.
(D) A higher magnification image of the pollen wall from (C).
(E) Scanning electron microscopy image showing the two types of pollen grains in the pss1 mutant.
(F) A higher magnification image of the normal-shaped pollen grain from (E).
(G) Transmission electron microscopy image of a normal-shaped pss1 mutant pollen grain.
(H) A higher magnification image of the pollen wall from (G).
(I) A higher magnification image of an abnormal-shaped pollen grain from (E). Arrowhead indicates the germination pore.
(J) Transmission electron microscopy image showing that there is no accumulations of starch granules in the abnormal-shaped pss1 mutant pollen grain.
(K) A higher magnification transmission electron microscopy image of the pollen wall from (J), which lacks intine and has abnormal exine.
st, starch granules; i, intine; f, foot layer; c, columella; te, tectum. Bars = 50 μm in (A) and (E), 10 μm in (B), (C), (F), (G), (I), and (J), and 500 nm in (D), (H), and (K).
Fertility Analysis of the pss1 Mutant
To determine the underlying reasons for the observed partial spikelet sterility phenotype of the pss1 mutant, we counted the total number of pollen grains per anther. On average, wild-type Nipponbare has 1391 ± 172 pollen grains per anther, while the pss1 mutant has 1084 ± 201 (full plus shrunken pollen). Among them, the regular-shaped and viable pollen grains are ~40% of wild-type pollen (Figure 3A). In vitro germination assays revealed that Nipponbare has a germination rate of ~85% (Figure 3B), while that of the pss1 mutant is ~43% (Figure 3C). This result is generally consistent with the result of iodine-potassium iodide staining.
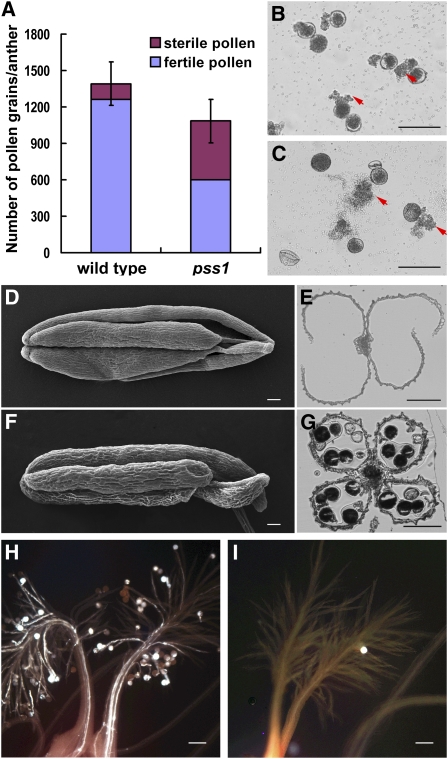
Figure 3.
Fertility Analysis of the pss1 Mutant.
(A) Quantification of the total pollen grain numbers in the wild type and the pss1 mutant anthers. The pollen grain number of the mutant is ~78% of the wild type, and the fertile pollen grain number is ~40% of the wild type. Error bars indicate sd of total pollen grains from five independent samples.
(B) and (C) In vitro germination of the wild-type (B) and the pss1 mutant (C) pollen. The normal-shaped pollen in the mutant can germinate normally. Arrows indicate pollen tubes.
(D) to (G) Comparison of anther dehiscence between the wild type and the pss1 mutant. (D) and (F), scanning electron microscopy images of the wild-type (D) and mutant (F) anthers after anthesis; (E) and (G), cross sections of the wild-type (E) and mutant (G) anthers after anthesis. The mutant anther cannot dehisce normally.
(H) and (I) Comparison of pollen grain number on the stigma between the wild type (H) and the pss1 mutant (I). There are only few pollen grains on the mutant stigma.
Bars = 100 μm.
Even though only half of the pollen grains in the pss1 mutant are viable, there should be enough functional pollen to successfully fertilize each spikelet of the mutant plant. The drastically reduced spikelet fertility suggests that there are likely additional reasons for the observed spikelet sterility. We noticed that before glume opening, pss1 anther shape is similar to the wild type, but after glume closing, the pss1 mutant anther is much thinner than that in wild-type plants, indicating that the pss1 mutant might have a defect in anther dehiscence. At anthesis, wild-type anthers reach the top of the spikelet and begin to dehisce, resulting in release of the pollen grains over the stigma of the pistil. After anthesis, the spikelet begins to close, keeping empty anthers outside the spikelet (Figures 3D and 3E). We found that anthers of the pss1 mutant did not dehisce at the time of spikelet opening or even after its closing (Figures 3F and 3G), suggesting that anther indehiscence of the pss1 mutant might cause a reduction in the number of effective pollen grains shed onto the stigma, resulting in markedly reduced spikelet fertility. To test this hypothesis, we examined the pollen grain number on the stigma 2 h after anthesis. Among 200 wild-type stigmas examined, 90% of wild-type stigmas had more than 20 pollen grains (Figure 3H), while only 8% of pss1 mutant stigmas had more than 20 pollen grains, and up to 75% of pss1 mutant stigmas had fewer than 20 pollen grains (Figure 3I). The other 17% of mutant stigmas did not have any pollen grains at all. These data suggest that the partial spikelet sterility of the pss1 mutant is caused by reduced pollen viability compounded by impairment of anther dehiscence.
Characterization of Male Gametogenesis in the pss1 Mutant
To more precisely determine the timing of the anther developmental defects in the pss1 mutant, we examined anther development at the microscopic level. No significant differences were detected between the wild type and pss1 mutant until the microspore uninucleate stage, when anther morphogenesis has completed. Anthers of both the wild type and the pss1 mutant can form four layers of the anther wall, and the microsporocytes are located at the center of each anther locule, surrounded by four somatic layers, which are, from the surface to interior, the epidermis, the endothecium, the middle layer, and the tapetum layer (Figures 4A, 4B, 4G, and 4H). Moreover, at the postmeiosis stage, anther walls, including the tapetum layer and the middle layer, degenerated normally in the pss1 mutant just like in wild-type anthers (Figures 4C to 4E and 4I to 4K). However, eventually, only ~50% of the microspores showed normal development in the pss1 mutant, whereas the rest of the microspores were degraded (Figures 4K and 4L).
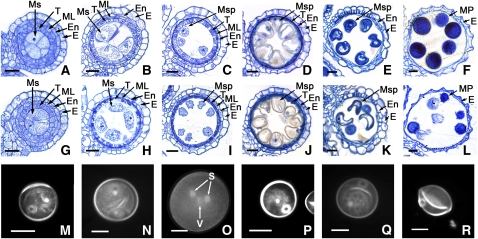
Figure 4.
Comparison of Male Gametogenesis in the Wild Type and the pss1 Mutant.
(A) to (F) and (M) to (O) show the wild type; (G) to (L) and (P) to (R) show the pss1 mutant. The cross sections ([A] to [L]) are stained with 0.25% toluidine blue O. E, epidermis; En, endothecium; ML, middle layer; Ms, microsporocyte; Msp, microspore; MP, mature pollen; S, sperm nuclei; T, tapetum; V, vegetative nuclei. Bars = 15 μm.
(A) and (G) Cross section of single locule at the microspore mother cell stage.
(B) and (H) Cross section of single locule at the meiosis stage.
(C) and (I) Cross section of single locule at the young microspore stage.
(D) and (J) Cross section of single locule at the vacuolated pollen stage.
(E) and (K) Cross section of single locule at the pollen mitosis stage showing two types of pollen grains in the mutant locule.
(F) and (L) Cross section of single locule at the mature pollen stage showing two types of pollen grains in the mutant locule.
(M) and (P) DAPI staining of a uninucleate stage microspore.
(N) and (Q) DAPI staining of a bicellular stage microspore.
(O) and (R) DAPI staining of a tricellular stage microspore.
To characterize the possible male gametogenesis defects in the pss1 mutant in more detail, we examined wild-type and pss1 microspores stained with 4',6-diamidino-2-phenylindole (DAPI). Microscopy observation revealed no striking differences in early-stage microspores. Both the wild type and the mutant can form normal free microspores at the uninucleate stage, each with a brightly stained central nucleus (Figures 4M and 4P).
At the late microspore stage, wild-type pollen undergo polar nuclear migration and asymmetric nuclear division at pollen mitosis I (PM I), giving rise to two daughter nuclei (Figure 4N) and entering the bicellular stage. After pollen mitosis II (PM II) three nuclei are generated, with one dispersed vegetative nucleus and two smaller generative nuclei in the tricellular pollens (Figure 4O). However, in the pss1 mutant, about half of the pollen grains perform PM I and PM II normally, but the other half have one brightly stained nucleus at the bicellular stage and become irregularly shaped and abort at the tricellular stage (Figures 4Q and 4R).
Isolation of the PSS1 Gene
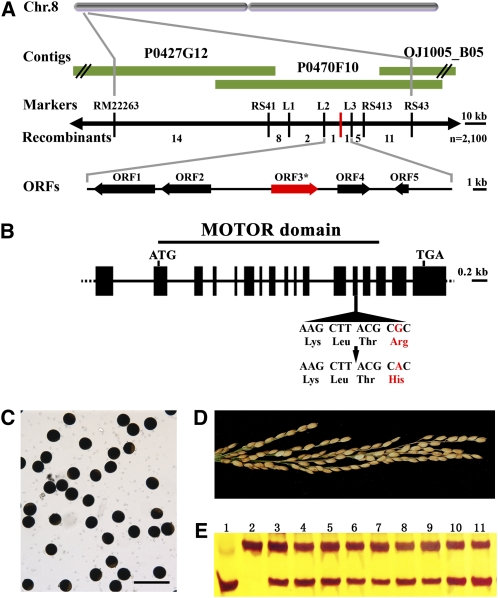
The PSS1 locus was previously mapped to the short arm of rice chromosome 8 between the cleaved-amplified polymorphic sequence marker L2 and the derived cleaved-amplified polymorphic sequence (dCAPS) marker L3, with a physical distance of ~28 kb (Li et al., 2007). There are five predicted open reading frames in this region (Figure 5A). We sequenced the entire region and found that the pss1 mutant carries a single nucleotide substitution of guanine (G) to adenine (A) in the 13th exon of an annotated kinesin-like gene (Os08g02380). This causes an amino acid change from Arg at position 289 (Arg-289) to His (Figure 5B). Sequence analysis of several other cultivars, including Nipponbare, 93-11, Dular, T65, 02428, IR36, N22, and II32B, revealed that they all have the conserved guanine nucleotide and Arg residue at this site.
Figure 5.
Positional Cloning of PSS1.
(A) Fine mapping of the PSS1 gene on chromosome 8. Names of the molecular markers and the number of recombinants are indicated (n = 2100). P0427G12, P0470F10, and OJ1005_B05 are PAC and BAC clones covering this locus. The PSS1 locus is mapped to a 27-kb region between the molecular markers L2 and L3.
(B) A schematic representation of the gene structure of PSS1. The mutant sequence has a single nucleotide change from guanine (G) to adenine (A) in the 13th exon. The motor domain is indicated. ATG and TGA represent the start and stop codons, respectively.
(C) and (D) A 7.6-kb wild-type genomic DNA fragment of PSS1 completely rescues the pollen (C) and spikelet (D) semisterility phenotypes of the pss1 mutant. Bar = 100 μm.
(E) Molecular identification of T0 transgenic plants by a dCAPS marker. The bottom band represents the wild-type allele, and the top band represents the pss1 mutant allele. Lane 1, the wild type; lane 2, pss1 mutant; lanes 3 to 11, T0 transgenic plants.
Kinesins are molecular motors that hydrolyze ATP as they move along microtubules, transporting vesicles and organelles and performing essential roles in chromosome motility and spindle assembly and function (Dagenbach and Endow, 2004; Miki et al., 2005). These functions are highly relevant to male fertility in both animals and plants (Chen et al., 2002; Macho et al., 2002; Yang et al., 2003; Gandhi et al., 2004; Tanaka et al., 2004; Oh et al., 2008). We reasoned that this kinesin gene most likely represents the candidate PSS1. To test this hypothesis, a genomic fragment containing the predicted coding region of Os08g02380 and regulatory sequences was transformed into a pss1 homozygous mutant background. Nine independent transgenic lines showed complete rescue of the semisterility phenotype (Figures 5C and 5D).
To further confirm that disruption of Os08g02380 gene function is responsible for the phenotype observed in the pss1 mutants, we generated RNAi transgenic plants in a japonica rice variety Kitaake background and obtained over 10 independent transgenic lines. Real-time PCR analysis revealed that four of seven transgenic lines tested showed significant reduction in the transcript level of the PSS1 gene compared with wild-type plants (see Supplemental Figure 1A online). These PSS1 knockdown transgenic plants showed reduced pollen viability (ranging from 55 to 69%) and reduced spikelet fertility (ranging from 40 to 60%), while the nontransformed control plants have normal pollen viability (~95%) and spikelet fertility (~93%). Notably, the most severely defective RNAi line R2 (transcript level reduced to ~5% relative to the wild type) only showed ~55% pollen viability and ~40% spikelet fertility (see Supplemental Figures 1B to 1D online). These results confirm that Os08g02380 indeed corresponds to the PSS1 gene.
Sequence Analysis of PSS1 and Related Proteins
Sequence analysis of amplification products obtained from RT-PCR and rapid amplification of cDNA ends–PCR indicated that the PSS1 cDNA is 1977-bp long and contains an open reading frame of 1434 bp encoding a predicted polypeptide of 477 residues, plus a 270-bp untranslated 5′ region and a 272-bp untranslated 3′ region. Sequence comparison between genomic DNA and cDNA showed that the PSS1 gene contains 17 exons (Figure 5B).
A BLAST search identified a kinesin motor domain in the N terminus of the predicted protein. Interestingly, the mutation in pss1 mutant resides in this region (see Supplemental Figure 2A online). To determine to which family this kinesin belongs, we constructed a kinesin superfamily phylogenetic tree (see Supplemental Figure 3 and Supplemental Data Set 1 online) according to the standardized kinesin nomenclature (Lawrence et al., 2004). In this phylogenetic tree, PSS1 is most closely related to the kinesin-1 family proteins. The kinesin-1 family was previously referred to as conventional kinesin, kinesin heavy chain (KHC), and was reported to function in organelle transport (Brady et al., 1990) or nuclear movement (Holzinger and Lütz-Meindl, 2002). The motor domain of PSS1 and other kinesin-1 family members exhibit high sequence similarities. Notably, the mutated Arg residue in pss1 mutant is strictly conserved among all kinesin-1 family members (see Supplemental Figure 2B online), indicating that this amino acid is likely critical for kinesin-1 protein function.
Spatial and Temporal Expression Pattern of PSS1 Gene and Subcellular Localization of PSS1 Protein
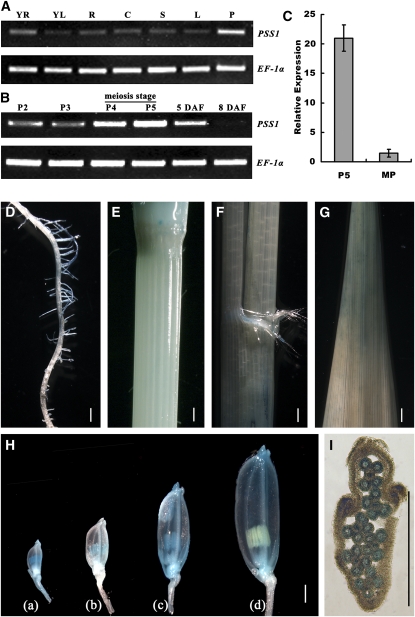
To test whether PSS1 is only expressed in the anther or also in other organs, we analyzed the PSS1 expression pattern. RT-PCR analysis revealed that PSS1 is expressed in a large number of organs, including young root, young leaf, mature root, mature culm, mature sheath, mature leaf, and panicle. The strongest expression was detected in panicles (Figure 6A). We further examined the expression profile of PSS1 at various developmental stages of panicles in more detail. Young panicles were classified into five stages, P1 to P5, according to the method described previously (Nonomura et al., 2003, 2004a): the P1 panicles usually have developed flower primordia (panicle size ~5 mm), the P2 and P3 panicles have developed anthers but have not entered meiosis yet (panicle size ~10 and ~30 mm, respectively), the P4 panicles contain some anthers that have entered meiosis (panicle size ~60 mm), and the P5 panicles have only meiotic or postmeiotic anthers (panicle size ~100 mm). Interestingly, PSS1 shows the highest expression in P4-P5 panicles and then the expression level gradually declines and becomes nearly undetectable at 8 d after heading (Figure 6B). We also used isolated anthers to analyze the expression level of PSS1 at the P5 stage and the mature pollen stage. Using quantitative PCR, we found that the expression level of PSS1 at P5 stage anthers is ~21-fold higher than that in the mature pollen stage anthers (Figure 6C). Thus, we concluded that expression of PSS1 peaks during male meiosis.
Figure 6.
Expression Analysis of the PSS1 Gene.
(A) RT-PCR analysis showing that PSS1 is expressed in various organs. YR, young root; YL, young leaf; R, mature root; C, mature culm; S, mature sheath; L, mature leaf; P, panicle.
(B) RT-PCR analysis showing that PSS1 expression peaks at P4 and P5 stages and decreases afterwards. P2 to P5, different developmental stages of young panicles. 5 DAF, panicles of 5 d after flowering; 8 DAF, panicles of 8 d after flowering.
(C) Real-time PCR analysis showing that the expression of PSS1 in P5 stage is ~21-fold higher than that in the mature pollen stage. Error bars indicate sd of three independent samples.
(D) to (G) Histochemical staining assay of PSS1 promoter–GUS reporter gene. Weak GUS signal is detected in root (D), culm (E), sheath (F), and leave blade (G).
(H) In panicles, the GUS signal was initially limited to the anther, then appears in the other parts of florets (glume, lemma, and palea) as the panicle development progresses. (a) A 2-mm-long spikelet from P2 group panicles; (b) a 3-mm-long spikelet from P3 stage panicles; (c) a 5-mm-long spikelet from P5 stage panicles; (d) postmeiosis spikelet.
(I) Cross section of P5 stage spikelet showing that GUS signal in anthers. Bars = 1 mm in (D) to (I).
To more precisely determine the spatial and temporal expression patterns of PSS1, we cloned the 4.3-kb sequence upstream of the PSS1 ATG start codon and constructed a PSS1 promoter-β-glucuronidase (GUS) construct. The construct was transformed it into the japonica variety Nipponbare. Consistent with the RT-PCR results, we observed GUS signal in various organs, including mature root, mature culm, mature sheath, mature leaf, and panicles of various developmental stages, with the strongest signal found in the panicles (Figures 6D to 6H). At the young microspore stage (1 to 3 mm length spikelet), the GUS signal is largely limited to the anther, and then as the panicle development proceeds, the signal starts to appear in other parts of florets, including glume, lemma, and palea. The strongest GUS staining is detected in spikelet of 4 to 5 mm length (Figure 6H) when meiosis occurs in the anther (Itoh et al., 2005). Cross section of the spikelet reveals that the GUS signal can be detected in both the microsporocyte and the anther walls (Figure 6I). GUS signal disappears in postmeiosis anther but can be still observed in the glume, lemma, and palea (Figure 6H). Together, these results indicate that PSS1 is broadly expressed in various tissues and organs, with the highest expression in panicles, and it peaks during male meiosis.
Previous studies have identified and characterized several rice genes involved in male meiosis control, including MEL1, UGP1, Rad21-4, PAIR1, PAIR2, PAIR3, and MER3 (Nonomura et al., 2004a, 2004b, 2006, 2007; Zhang et al., 2006; Chen et al., 2007; Wang et al., 2009; Yuan et al., 2009). To test whether PSS1 affects expression of these genes, we compared their transcription levels in P5 stage panicles of wild-type and pss1 mutants. We found that none of the tested genes showed any significant difference in expression between the wild type and the mutant (see Supplemental Figure 4A online), suggesting that PSS1 does not regulate their expression.
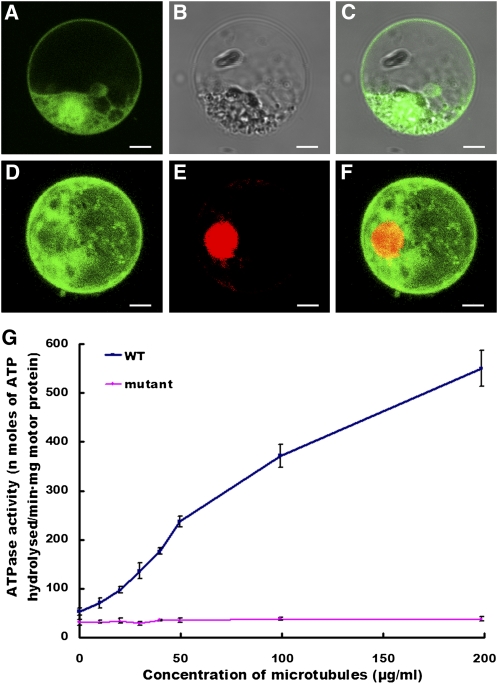
To investigate the subcellular localization of PSS1 protein, we constructed a fusion protein of the full-length PSS1 coding region and the cDNA encoding green fluorescent protein (GFP). The PSS1-GFP fusion construct gene and the GFP alone control, both under the control of the 35S promoter, were introduced into rice protoplast cells. As expected, the free GFP was found in both the nucleoplasm and cytoplasm (Figures 7A to 7C). By contrast, the PSS1-GFP fusion protein is predominantly localized in the cytoplasm (Figures 7D to 7F). Similar observations were made in Arabidopsis protoplast cells (see Supplemental Figure 5 online).
Figure 7.
Subcellular Localization and Microtubule-Stimulated ATPase Activity Assay of the PSS1 Protein.
(A) to (C) GFP alone is detected in both the nucleus and cytoplasm of rice protoplast cells.
(D) PSS1-GFP is predominantly detected in the cytoplasm.
(E) Nuclear localization of RPBF-mCherry (a nuclear marker).
(F) A merged image of (D) and (E). Bars = 5 μm.
(G) Comparison of microtubule-stimulated ATPase activities of the wild type (WT) and PSS1 mutant proteins. Error bars indicate sd of three independent experiments.
The PSS1 Mutant Protein Exhibits a Significant Reduction in Microtubule-Stimulated ATPase Activity
Kinesins are molecular motors that hydrolyze ATP as they move along microtubules, and they exhibit a microtubule stimulated ATPase activity (Vale and Fletterick, 1997; Dagenbach and Endow, 2004). To test whether the microtubule-stimulated ATPase activity is affected in the pss1 mutant, we expressed the N-terminal head domain of both the wild type and the mutant PSS1 proteins in Escherichia coli and conducted an ATPase activity assay using the purified proteins. As shown in Figure 7G, in the absence of microtubules, the basal steady state ATPase activity of the wild-type motor is 52.5 ± 7.4 nmoles of ATP hydrolyzed per min per mg of the motor protein. By contrast, the basal ATPase activity of the mutant PSS1 motor is 31.0 ± 6.0 nmoles of ATP hydrolyzed per min per mg of the motor protein. Moreover, the ATPase activity of the wild-type motor is triggered by microtubules in a concentration-dependent manner, whereas the mutant motor shows only a slight increase of ATPase activity as the concentration of microtubules increases. These results clearly indicate that the mutation in the PSS1 mutant protein diminishes its microtubule-stimulated ATPase activity, suggesting that the R289H amino acid substitution in pss1 mutant likely affects the microtubule interaction activity of the PSS1 protein.
Abnormal Spindle Formation in Male Meiocytes in the pss1 Mutant
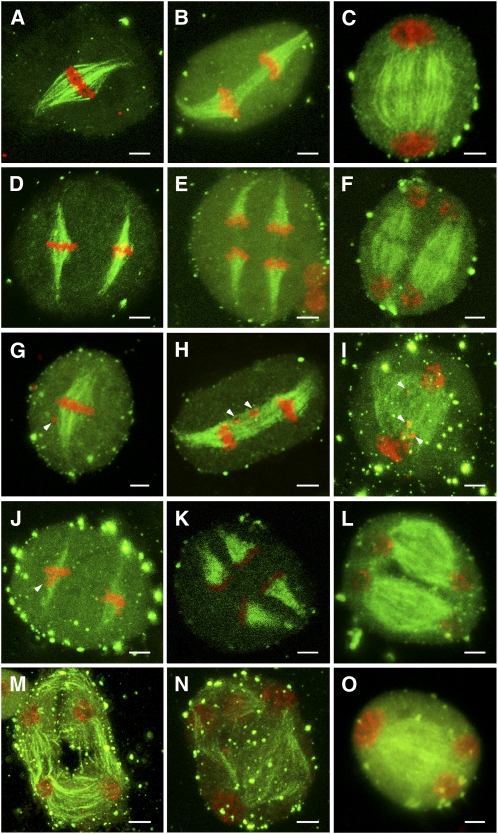
As PSS1 encodes a kinesin-1 like–protein and some kinesins are known to regulate spindle formation during mitosis and meiosis, and our in vitro assay suggested that the PSS1 mutant protein has diminished microtubule interaction ability, we investigated whether meiotic spindle formation is affected in the pss1 mutant. We performed immunofluorescence microscopy with an anti-α-tubulin antibody on male meiocytes in meiosis I and II. At metaphase I, the wild-type meiocytes form typical bipolar spindles, with the 12 rice bivalents arranged at the equator (Figure 8A). Then, the two sets of 12 daughter chromosomes synchronously segregated toward the opposite poles at anaphase I (Figure 8B). At telophase I, the chromosomes become diffuse, and the phragmoplast forms between the two newly generated daughter nuclei (Figure 8C). In metaphase II, two bipolar spindles form (Figure 8D), and chromosomes align on the equatorial plate of each daughter cell. Then, at anaphase II, the sister chromatids separate and are pulled to the opposite poles (Figure 8E). At telophase II, new phragmoplast structures form near the equatorial plate in preparation for meiotic cytokinesis (Figure 8F).
Figure 8.
Immunostaining of Spindles in Wild-Type and pss1 Male Meiocytes.
Chromosomes are stained by propidium iodide (red), and microtubules are labeled by fluorescein isothiocyanate (green). Bars = 5 μm.
(A) to (F) Male meiocytes of the wild type.
(A) Bipolar-oriented spindle at metaphase I.
(B) Two sets of univalents separated by the spindle at anaphase I.
(C) Phragmoplast starts to form between the two daughter pronuclei at telophase I. The weak staining in the midzone indicates the formation of a cytokinetic plane.
(D) Two sets of univalents aligned at the metaphase II plates.
(E) Two sets of univalents are separated to the opposite poles by the anaphase II spindles.
(F) At telophase II, new phragmoplast structure forms near the equatorial plate.
(G) to (O) Male meiocytes of the pss1 mutant.
(G) At metaphase I, the pss1 mutant shows normal-shaped spindles, but one or several chromosomes are delayed (indicated by arrowhead).
(H) Bivalents are pulled by the forces of anaphase I spindles and move away from the equator. Delayed chromosomes are observed (indicated by arrowheads).
(I) The telophase I spindle is somewhat disorganized compared with that of the wild type. In addition, delayed chromosomes are observed (indicated by arrowheads).
(J) The metaphase II spindle has a normal shape but is thinner than that of the wild type. Delayed chromosomes can also be seen (indicated by arrowhead).
(K) The shape and density of the anaphase II spindles are similar to that of the wild type.
(L) Most meiocytes of pss1 mutant show normal shaped spindles at telophase II.
(M) to (O) A small portion of meiocytes has disorganized microtubules and form atypical tetrads.
In the pss1 mutant, most of the bivalents congress to the metaphase plate at metaphase I, but one or several bivalents are delayed in about half of the meiocytes (Figure 8G). At anaphase I and telophase I, the homologous chromosomes move away from the equator toward the poles, but the shape of the telophase spindle is somewhat more disorganized in comparison with the wild-type spindle (Figures 8H and 8I). The metaphase II spindle is thinner than the wild-type spindle (Figure 8J), but the shape and density of the anaphase II and telophase II spindles are similar to the wild-type spindles (Figures 8K and 8L). Among 35 meiocytes examined, we observed 32 meiocytes forming normal microtubule spindles at late telophase II and normal tetrads (Figure 8L). The remaining three meiocytes had disorganized microtubule spindles, and these meiocytes formed atypical tetrads (Figures 8M to 8O). These observations suggest that PSS1 likely plays a minor but not essential role in regulating meiotic spindle formation.
Altered Chromosome Behavior in the pss1 Mutant
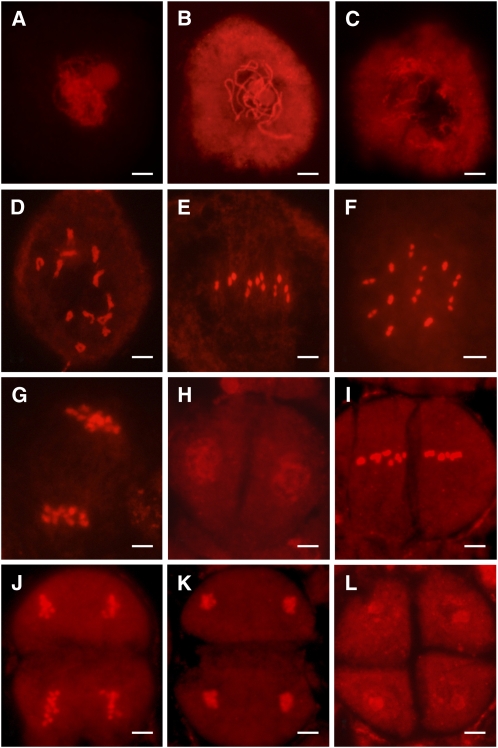
We next examined meiotic chromosomal behavior in meiocytes of the wild type and the pss1 mutants. In the wild type, the chromosomes begin to condense and appear very thin at the leptotene stage, and homologous chromosomes undergo pairing and synapsis at the zygotene stage (Figure 9A). At the pachytene stage (Figure 9B), homologous chromosomes are fully synapsed with the completion of the synaptonemal complex. During diplotene, the synaptonemal complexes are disassembled, and the homologous chromosomes are separated from each other except at the chiasmata (Figure 9C). At diakinesis, chromosomes further condense to produce very short bivalents (Figure 9D). The bivalents align at the equatorial plane at metaphase I (Figures 9E and 9F), after which the homologous chromosomes separate and move to the two poles of the cell at anaphase I (Figure 9G). After meiosis I, two groups of chromosomes align separately at two new division planes. Next, the sister chromatids separate at anaphase II (Figures 9H to 9L).
Figure 9.
Chromosome Dynamics in Wild-Type Male Meiocytes.
Chromosomes are stained with 0.1 mg/mL propidium iodide. Bars = 5 μm.
(A) to (D) Zygotene (A), pachytene (B), diplotene (C), and diakinesis (D).
(E) Metaphase I front view, showing 12 bivalents arranged at the equatorial plate.
(F) Metaphase I polar view, showing 12 completely paired bivalents.
(G) Anaphase I, showing synchronous separation of chromosomes to the opposite poles.
(H) Telophase I.
(I) Metaphase II.
(J) Anaphase II.
(K) Telophase II.
(L) Normal tetrad in wild-type anthers.
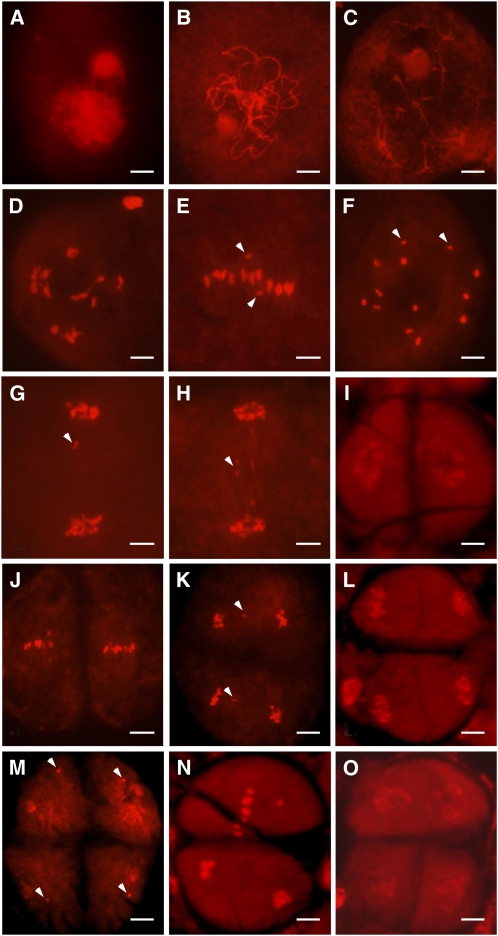
In the pss1 mutant, chromosome behavior seemed generally normal until metaphase I (Figures 10A to 10D), although one pair of univalents could be found in 27 cells of 50 metaphase I cells observed (Figures 10E and 10F). As mentioned above, delayed chromosomes were frequently found at anaphase I (Figure 10G). We also observed anaphase I bridges (Figure 10H). Delayed chromosomes could also be found at anaphase II (Figure 10K), and micronuclei could be detected at the tetrad stage (Figure 10M). In addition, we found that sister chromatids do not separate synchronously in a small portion of anaphase II meiocytes (Figure 10N), thus forming triads instead of tetrads (Figure 10O). The frequency of abnormal chromosomal behavior is summarized in Table 1. These results indicate abnormalities in chromosome congression and segregation in the pss1 mutant.
Figure 10.
Chromosome Dynamics in pss1 Mutant Male Meiocytes.
Chromosomes are stained with 0.1 mg/mL propidium iodide. Bars = 5 μm.
(A) to (D) Zygotene (A), pachytene (B), diplotene (C), and diakinesis (D).
(E) Metaphase I front view, showing only 11 bivalents arranged at the equatorial plate and two isolated univalents (indicated by arrowheads).
(F) Metaphase I polar view, showing 11 bivalents and two univalents (indicated by arrowheads).
(G) Anaphase I. Arrowhead indicates the delayed univalents.
(H) Anaphase I. Arrowhead indicates chromosome bridge.
(I) Telophase I.
(J) Metaphase II.
(K) Anaphase II. Arrowheads indicate the delayed univalents.
(L) Telophase II.
(M) pss1 forms micronuclei (indicated by arrowheads) at the tetrad stage.
(N) Abnormal anaphase II.
(O) Abnormal tetrad in the pss1 mutant.
Table 1.
Statistics of Abnormal Chromosomal Behavior in the pss1 Mutant
| Normal Cells/Frequency | Abnormal Cells | |||||
| Stages | Cells Observed | Chromosome Behavior | Cells/Frequency | Chromosome Behavior | Cells/Frequency | |
| Metaphase I | 50 | 23/46% | Univalents | 27/54% | n.d. | n.d. |
| Anaphase I | 56 | 18/32% | Chromosome bridge | 16/29% | Delayed univalents | 22/39% |
| Anaphase II | 60 | 21/35% | Micronucleus | 17/28% | Delayed univalents | 22/37% |
| Tetrad | 46 | 16/35% | Micronucleus | 25/54% | Triad | 5/11% |
n.d., not detected.
To confirm that the observed chromosome behavior defects in pss1 mutants are due to impairment of the PSS1 gene function, we examined meiocytes in the RNAi transgenic line R2. Similar to the pss1 mutants, univalents, delayed chromosomes, and micronuclei can also be found in PSS1 RNAi plants during male meiosis (see Supplemental Figures 1E to 1H and Supplemental Table 1 online), indicating that the defect in the PSS1 gene is indeed responsible for the abnormal chromosome behavior.
DISCUSSION
In this study, we report the molecular genetic characterization of the spontaneous rice pollen semisterility mutant pss1, positional cloning, temporal, and spatial expression pattern of the PSS1 gene, and subcellular localization of the PSS1 protein. The PSS1 gene encodes a protein belonging to the kinesin-1 family. Substitution of a conserved amino acid (R289H) in the motor domain diminished microtubule-stimulated ATPase activity of the PSS1 protein, resulting in abnormal male meiotic chromosome dynamics in a portion of male meiocytes and causing a partial pollen and spikelet sterility phenotype.
PSS1 Is Necessary for Male Meiotic Chromosomal Dynamics and Anther Dehiscence
Male-sterile mutants are valuable for understanding the mechanisms of pollen development. A number of male-sterile mutants have been reported in rice. These mutants can be broadly divided into two groups. One group affects tapetum function as well as meiosis. For example, the msp1 mutant gives rise to an excessive number of male and female sporocytes. The formation of anther wall layers is disordered, and the tapetum layer is completely abolished. In addition, development of pollen mother cells is arrested at various stages of meiotic prophase I, which results in complete male sterility (Nonomura et al., 2003). The udt1 mutant exhibits normal meiosis, but the tapetum fails to differentiate and become vacuolated during the meiotic stage. In addition, meiocytes do not develop into microspores (Jung et al., 2005). The tdr mutant exhibits retardation in the degeneration of the tapetum and the middle layer, resulting in a collapse of microspores and complete male sterility (Li et al., 2006). The other group of male-sterile mutants is primarily defective in various stages of meiosis but with distinct cellular defects. For example, the pollen mother cells of Ugp1-silenced plants begin to degenerate at the early meiosis stage, due to disruption of normal callose deposition, resulting in complete pollen collapse (Chen et al., 2007). In the mel1 mutant, pollen mother cells are arrested at early meiosis I, and meiotic chromosomes frequently exhibited uncondensed morphology. MEL1 encodes a novel ARGONAUTE family protein essential for the progression of the premeiotic mitosis and meiosis in rice (Nonomura et al., 2007), suggesting that RNA-mediated silencing system is involved in sporogenesis in rice. The pair1, pair2, and pair3 rice mutants show 24 completely unpaired univalents at diakinesis and metaphase I, and both male and female meiocytes are affected, causing complete male and female sterility. PAIR2 is a rice ortholog of Arabidopsis ASY1 proteins (Caryl et al., 2000; Armstrong et al., 2002; Nonomura et al., 2004b, 2006). Interestingly, rice PAIR1 and PAIR3 are two putative coiled-coil proteins that do not appear to have close homologs in other organisms (Nonomura et al., 2004a; Yuan et al., 2009). Two homologs of yeast DMC1, DMC1A and DMC1B, have also been shown to be required for homologous pairing in rice (Ding et al., 2001; Kathiresan et al., 2002; Deng and Wang, 2007). Furthermore, genetic analysis also revealed that rice RAD21-4, a homolog of yeast Rec8/RAD21/SCC1 proteins, is essential for meiotic sister chromatid cohesion. The male meiocytes of Rad21-4 RNAi lines show multiple aberrant events at meiotic prophase I, including overcondensation of chromosomes, precocious segregation of homologs, and chromosome fragmentation (Zhang et al., 2006). Rice MER3, similar to its counterparts in yeast and Arabidopsis, encodes a ZMM protein required for normal meiotic crossover formation. Disruption of MER3 function causes defects in meiotic crossover formation, leading to many univalents that are randomly segregated (Wang et al., 2009).
Our analyses showed that the pss1 mutant has normal vegetative and floral development and functional female gametophytes, but both pollen viability and spikelet fertility are greatly reduced (~50 and 40%, respectively). Microscopy examination revealed that unlike msp1, udt1, and tdr, anther wall development in pss1 mutants is normal, and the tapetum and the middle layers degenerate normally. DAPI staining of young microspores revealed that most of the sterile pollen grains abort at the uninucleate stage, and the aborted pollen grains have irregular shapes. Most of them are shrunken with defects in the accumulation of starch granules, but with a clearly visible germination pore, a structure formed at the uninucleate stage. About 50% of the microspores fail to enter PM I, resulting in aborted pollen. Our cytological studies showed that the pss1 mutant displays a range of defects in male meiosis, including univalents at metaphase I, delayed chromosomes and chromosomal bridges at anaphase I and anaphase II, and micronuclei at the tetrad stage. In comparison to other previously reported meiotic mutants, the pss1 mutant does not have detectable defects in chromosome pairing like pair1, pair2, and pair3, with only one pair of univalents in the meiocyte in most cases. The chromosomal behavior in pss1 mutant is similar to that of the Rad21-4 RNAi lines, since both mutants show defective alignment of chromosomes on the equator, lagging chromosomes, and formation of chromosome bridge and asymmetric ‘tetrads’, but the frequency of asymmetric ‘tetrads’ in pss1 mutant is lower than that observed in the Rad21-4 RNAi plants. These observed cellular and subcellular defects together with the transcriptional data suggest that PSS1 most likely acts downstream of MSP1, MEL1, UGP1, UDT1, TDR, PAIR1, PAIR2, PAIR3, MER3, and Rad21-4 in male gametogenesis (see Supplemental Figure 4B online), although their precise functional relationships remain unknown at this stage.
It is worth noting that the pss1 mutants also display several other intriguing phenotypes. First, although PSS1 functions in a sporophytic fashion in male fertility and all microspores of a homozygous mutant have the identical genotype, only about half of the pollen grains are aborted. The underlying reason for the observed partial male sterility of the pss1 mutant is not clear. One possibility is that pss1 may represent a partial loss-of-function mutation, as it carries a missense mutation. However, our results showed that mutation in the pss1 mutant almost completely abolishes the microtubule-stimulated ATPase activity of PSS1, suggesting a near complete loss of function. Alternatively, there may exist functional overlap between PSS1 with kinesins of other families. In line with this possibility, previous studies have shown that Arabidopsis ATK1 and its homologs NCD, KAR3, and KLPA have some functional resemblance as well as distinct functions as revealed by detailed analyses of their mutant phenotypes (Chen et al., 2002).
Second, despite the obvious defects in meiotic chromosome alignment and segregation, the majority of the spindles in pss1 mutants have normal morphology. These observations suggest that there may exist other kinesins sharing an overlapping function in regulating spindle morphogenesis. Alternatively, PSS1 may be not involved in spindle morphology but may participate in regulating chromosome movement. It has been proposed that the flux or slow poleward movement of spindle microtubules with their attached chromosomes, elongation of the spindle, and chromosome movement along spindle fibers probably act together to promote poleward movement of chromosomes in anaphase (Endow, 1999). PSS1 may attach to chromosomes and pull them along the spindle during meiosis, in a fashion similar to mitotic chromokinesins (Mazumdar and Misteli, 2005). The loss of microtubule-stimulated ATPase activity of the mutated PSS1 protein might be responsible for the observed delay in chromosome movement during meiosis.
Another defect in the pss1 mutant is a failure of anthers to dehisce at the time of spikelet opening or even after its closing. Little is known about the genetic control of anther dehiscence in rice and how it is coordinated with other developmental processes that occur in the anther and florets. The rice anther indehiscence1 mutant is defective in anther dehiscence due to a defect in the programmed degradation and breakage of anther wall tissues, which lead to a spikelet sterility phenotype (Zhu et al., 2004). More insights into anther dehiscence have been gained through genetic analysis of Arabidopsis male-sterile mutants. Several Arabidopsis mutants defective in anther dehiscence, such as dde1/opr3 (Sanders et al., 2000; Stintzi and Browse, 2000), coi1 (Feys et al., 1994; Xie et al., 1998), dad1 (Ishiguro et al., 2001), and aos/dde2-2 (Park et al., 2002; von Malek et al., 2002), are all defective in jasmonic acid biosynthesis or signaling. Another anther dehiscence-related gene, MS35/MYB26, is involved in the lignin biosynthesis pathway (Yang et al., 2007). The exact reasons for defective anther dehiscence in pss1 are not known. We speculate that this could be due to reduced mechanical pressure generated in the pollen sacs in the pss1 mutants. In rice, it has been observed that, prior to dehiscence, pollen grains swell rapidly, and it has been proposed that this rapid swelling generates a significant mechanical force that has a role in rupturing the enzymatically weakened septum (Matsui et al., 1999a, 1999b). In pss1 mutants, there are only ~50% of normal pollen, and the other 50% anamorphic pollen grains have no accumulations of starch granules. These aborted pollen grains cannot swell during floret opening; this will reduce the mechanical pressure. Thus, we deduced that the anther indehiscence in pss1 mutants could be a secondary effect of reduced pollen viability, which also contributes to the low panicle fertility of the mutant.
PSS1 Is a Member of the Kinesin-1 Family
In this study, we demonstrated that the rice PSS1 gene encodes a protein belonging to the kinesin-1 family of the kinesin superfamily (Lawrence et al., 2004). The kinesin-1 family is the founding member of the kinesin superfamily, formerly called KHC or conventional kinesin. Based on studies from other eukaryotes, kinesin-1 family proteins are generally known to function as organelle transporters (Brady et al., 1990) and are involved in nuclear movement (Holzinger and Lütz-Meindl, 2002). Little is known about kinesin-1 family proteins in flowering plants. Through computational analysis, we found that PSS1 represents the only kinesin-1 family member in the rice genome and shares highest sequence identity with the product encoded by the Arabidopsis At3g63480 gene (amino acid sequence identity 59.09%). The biological function of At3g63480 is unknown. Our genetic, molecular, and cell biology analyses strongly support a function of PSS1 in chromosome segregation in male meiocytes and male gametogenesis. Thus, our data indicate that the molecular and biological functions of kinesin-1 family members may have diverged in the plant and animal kingdoms.
Several non-kinesin-1 family proteins have been reported to function in regulating diverse cellular processes during male meiosis and gametophyte development in dicotyledonous plants. The recently reported Arabidopsis KIN14a (also known as ATK1 or KATA), a C-terminal kinesin belonging to kinesin-14 family, and KIN14b (formerly ATK5) have evolved partially distinct functions in regulating male meiosis (Quan et al., 2008). Arabidopsis atk1 mutants have greatly reduced male fertility and unfocused spindle poles with disorganized microtubules. Bivalents in the mutant meiocytes fail to align at the metaphase I plate properly, with some bivalents lagging behind in their movement to the metaphase I plate (Chen et al., 2002). Arabidopsis TETRASPORE (TES)/STUD/NACK2 and NACK1/HINKEL are kinesin-like proteins that have overlapping functions essential for regulating meiotic cytokinesis (Tanaka et al., 2004; Oh et al., 2008). PAKRP1/Kinesin-12A and PAKRP1L/Kinesin-12B also jointly play a critical role in the organization of phragmoplast microtubules and cell plate formation during male meiotic cytokinesis (Lee et al., 2007). Intriguingly, mutations in the rice DBS1 gene, a homolog of the Arabidopsis STUD/TES/NACK2 and NACK1/HINKEL genes, cause a severe dwarf phenotype and cell wall stubs in rapidly dividing cells, also reflecting defects in cytokinesis (Sazuka et al., 2005). These observations suggest that there may be some functional overlaps between kinesin-1 family and other kinesins in regulating meiosis and cytokinesis in higher plants.
R289 Plays a Critical Role for the Microtubule Interacting Activity of PSS1
In the pss1 allele, a missense mutation causes substitution of the Arg-289 (R) residue by a His (H) residue (Figure 5B). Arg-289 is located in the conserved region of the motor domain and is highly conserved in all kinesin-1 family members across plant and animal kingdoms. Sequence analysis revealed that Arg-289 of PSS1 corresponds to the Arg-284 residue of human KHC (also known as KIF5B) (see Supplemental Figure 6 online). Human KHC exhibits ubiquitous expression with prominent glial cell expression and plays a significant role in growth and survival of HeLa cells. Depletion of KHC affects lysosomal distribution and stability and induces perinuclear accumulation of autophagosomes in cancer cells (Cardoso et al., 2009). It has been reported that R284 of human KHC is one of the key residues located in the α5 region involved in microtubule interaction (see Supplemental Figure 7 online for the three-dimensional structure), and mutations of this residue decrease affinity for microtubules (Woehlke et al., 1997). Our analyses also showed that the pss1 mutant protein has a drastically reduced microtubule-stimulated ATPase activity, which likely reflects a decrease in its microtubule binding ability. Notably, mutation of the corresponding residue in the Arabidopsis TES protein (Arg-306 to Lys) in the tes-2 allele causes defects in microspore wall separation owing to the failure of the male postmeiotic cytokinesis (Spielman et al., 1997; Yang et al., 2003) (see Supplemental Figure 6 online for amino acid alignment). These findings strongly support the notion that Arg-289 and its corresponding residue in the kinesin motor domain are critical for the biological function of PSS1 and other kinesins, and this residue is most likely involved in microtubule interaction. Future studies will be aimed to more precisely determine the cellular function of PSS1 and its functional relationship with other gene products regulating male meiosis in rice.
METHODS
Plant Materials and Growth Conditions
The pss1 mutant was initially identified as a spontaneous semisterile mutant in the japonica rice (Oryza sativa) variety Nipponbare. Genetic analysis revealed that the semisterility phenotype was stably inherited. A semisterile progeny line, w207-2, was selected for further analysis, which displays ~50% pollen viability and ~40% spikelet fertility. When w207-2 was crossed with the wild type or wide-compatibility varieties, the fertility of F1 plants was normal. F2 plants segregated fertile plants and semisterile plants in a 3:1 ratio, suggesting that the mutant phenotype was controlled by a single recessive nuclear gene (Yu et al., 2005). An F2 mapping population was generated from a cross between the pss1 mutant and the indica variety Dular. Dular carries a suite of neutral alleles: S-7n (Yanagihara et al., 1992), S-5n, S-8n, S-9n, S-16n, and S-17n (Wan et al., 1993, 1996; Wan and Ikehashi, 1995, 1996). All rice materials were grown in the paddy field at the Jiangsu Academy of Agricultural Sciences (Nanjing, China) in the natural growing season.
Evaluation of Pollen, Embryo Sac, and Spikelet Fertility
To estimate the level of pollen viability, three spikelets were collected from each plant shortly before anthesis and fixed in 70% ethanol. Three anthers were sampled at random from each spikelet. Between 100 and 200 stained pollen grains (1% I2-KI solution) were inspected by light microscopy.
A whole-mount stain-clearing laser scanning confocal microscopy method (Zhao et al., 2007) was used to evaluate the structure of mature embryo sacs in the wild type and the pss1 mutant. Between 100 and 150 mature florets from 10 wild-type and mutant plants were fixed in FAA solution (containing an 18:1:1 [by volume] mixture of formalin, 70% ethanol, and acetic acid). The samples were placed in a vacuum for 30 min and incubated for 24 h at room temperature, after which the tissue was stored in 70% ethanol at 4°C. Before staining, the samples were transferred to 70% ethanol; the lemma and palea were removed to expose the ovary to ethanol. The tissue was then processed through an ethanol series (50, 30, and 15%) and finally transferred into distilled water. The whole mature ovary was incubated in 2% aluminum potassium sulfate for 20 min, held in 10 mg/L Eosin Y (Amresco; 0109) water solution for 10 to 12 h, in 2% aluminum potassium sulfate once again for 20 min, washed with distilled water for 24 h, and finally washed three or four times in tap water. The samples were dehydrated by passing through an ethanol series (30, 50, 70, and 100%) and cleared in 100% methyl salicylate three times (for >10 h in the final incubation). The embryo sacs were then examined by confocal laser scanning microscopy (Leica SP2).
Spikelet fertility was calculated as the proportion of fertile spikelets to all spikelets on three panicles for each plant at maturity. Hand-saturated pollination was performed to study the biology of semisterility in pss1 mutant. Ten panicles of the semisterile pss1 mutant were bagged before flowering and then fertilized with wild-type Nipponbare pollen every day for 4 d. One month later, seed set in the bagged spikelets was calculated.
Scanning Electron Microscopy and Transmission Electron Microscopy
For scanning electron microscopy, samples were prepared as described previously (Li et al., 2003) with some modifications. Briefly, rice tissues were excised with a razor and immediately placed in 70% ethanol, 5% acetic acid, and 3.7% formaldehyde for 18 h. Samples were critical point dried, sputter coated with gold in an E-100 ion sputter, and observed with a scanning electron microscope (S3400N; Hitachi).
For transmission electron microscopy, samples were fixed in 3% (w/v) paraformaldehyde and 0.25% glutaraldehyde in 0.2 n sodium phosphate buffer, pH 7.0, and were then postfixed in 2% OsO4 in PBS, pH 7.2. Following ethanol dehydration, samples were embedded in acrylic resin (London Resin Company). Ultrathin sections (50 to 70 nm) were double stained with 2% (w/v) uranyl acetate and 2.6% (w/v) lead citrate aqueous solution and examined with a JEM-1230 transmission electron microscope (JEOL) at 80 kV.
Examination of Pollen Grains on the Stigma
To observe the pollen grains on the stigma, the spikelet of the mutant and wild type were marked when their glume opens. The spikelets were harvested 2 h after anthesis and fixed in FAA fixation (containing an 18:1:1 [by volume] mixture of formalin, 70% ethanol, and acetic acid) for 24 h. The spikelets were then processed through an ethanol series (70, 50, and 30%) and washed with distilled water three times. The spikelets were incubated in 10 n sodium hydroxide for 5 to 8 min at 56°C and then washed with distilled water three times and stained with 0.1% water blue (Fluka; 95290) overnight. Finally, the samples were examined using a fluorescence microscope (Leica DM5000B).
Determination of Total Pollen Grains per Anther
The number of total pollen grains was counted as described previously (Nakamura et al., 2000). Briefly, the number of pollen grains was counted for the spikelets at the specific position on the panicle, which is the 3rd, 4th, and 5th spikelets on the 1st, 2nd, and 3rd primary branches. The spikelets were sampled from six panicles (three panicles from each pot) and fixed with 50% ethanol 1 or 2 d after heading. The pollen grains were counted for two anthers from each of 10 spikelets taken randomly from the sample. The pollen grains were examined by staining with iodine-potassium iodide solution and counted using an optical microscope.
In Vitro Pollen Germination Assay
For pollen germination tests, Brewbaker and Kwach medium (10% sucrose, 100 mg L−1 boric acid, 300 mg L−1 calcium nitrate, 200 mg L−1 magnesium sulfate, and 100 mg L−1 potassium nitrate) was used. Pollen grains were placed on a clean 24 × 50-mm cover glass, and 20 μL of the liquid medium were added. The cover glass was placed on a piece of moist filter paper in a plastic dish. Then, the dish was sealed tightly and incubated for 60 min at 25°C in the dark. The pollen grains were then observed under a microscope. More than 200 pollen grains were examined to determine the percentage of germination. Pollen grains with pollen tube elongated longer than the diameter of the pollen grain were scored as successful germination.
Plastic Sections
Observation of anther development was performed on standard plastic sections as described previously (Li et al., 2006). Spikelets of different developmental stages were collected, based on the length of spikelet, and fixed with 3% (w/v) paraformaldehyde and 0.25% glutaraldehyde in 0.2 n sodium phosphate buffer, pH 7.0, for 20 h at 4°C, rinsed with 0.1 M phosphate buffer, pH 7.0, and dehydrated through an ethanol series. The samples were embedded in Technovit 7100 resin (Hereaus Kulzer) and polymerized at 45°C. Transverse sections of 2 μm were cut using an Ultratome III ultramicrotome (LKB) and stained with 0.25% toluidine blue O (Chroma Gesellshaft Shaud). Images were captured using Leica Application Suite 3.3 and merged and enhanced using Photoshop CS (Adobe).
DAPI Staining
The panicles, including flowers in meiosis, were fixed with 3:1 ethanol:acetic acid and stored at 4°C until observation. Single anthers were dissected from the isolated spikelet using a dissecting microscope (Leica MZ16). Anthers were disrupted on microscope slides using dissecting needles and gently squashed in DAPI staining solution (0.1 M sodium phosphate, pH 7.0, 1 mM EDTA, and 1 mg/mL DAPI) under a cover slip. After staining for 20 min, the stained pollens were monitored under UV light with a fluorescence microscope (Leica DM5000B).
Complementation Test of the pss1 Mutant
For functional complementation, a 7.6-kb genomic DNA fragment containing the entire coding region, 3777-bp upstream sequence, and 368-bp downstream sequence was amplified by PCR with primers named W1f and W1r, using the BAC clone OSJNBa0085H08 as the template. The PCR product was recombined into pDONR-207 (Invitrogen) to generate a Gateway entry clone and finally recombined into the binary Gateway vector pGD625 (Chalfun-Junior et al., 2005) carrying a kanamycin resistance marker to generate the pGD625-PSS1 construct. The pGD625-PSS1 and the control pGD625 binary plasmids were introduced into the Agrobacterium tumefaciens strain EHA105 and used to infect the pss1 mutant calli. Transformed calli were grown in darkness at 25°C for 3 d. Kanamycin-resistant calli were grown in an isolated greenhouse to produce transgenic plants.
For molecular identification of T0 transgenic plants, a dCAPS marker (Neff et al., 1998) was designed to identify the mutation site of pss1 mutant, and a mismatch was made in the antisense primer W2r using dCAPS Finder 2.0 software (Neff et al., 2002) at http://helix.wustl.edu/dcaps/dcaps.html. It generated an additional EcoRI site in the wild-type allele (GAATTC) but not in the mutant allele (GAATTT). The sense primer was W2f. A total of 5 μL of each PCR product was digested with 0.1 units EcoRI (Takara) in a total volume of 20 μL at 37°C overnight. After digestion, 2 μL of each of the digested PCR products was separated on 8% polyacrylamide gel. The positive transgenic plants have bands corresponding to both the wild type and the mutant alleles.
To generate the RNAi construct, two inverted repeat PCR fragments were amplified using the first-strand cDNA derived from rice panicle as the template and the following two pairs of primers named W3f and W3r, and W4f and W4r. The PCR fragments were sequentially cloned into XhoI/KpnI and BamHI/XbaI sites of the pHANNIBAL vector in the sense and antisense orientations. The whole-stem loop fragment was further cloned into pCam23ACT:OCS vector between the rice Actin1 promoter and OCS terminator sequence, yielding the binary PSS1 RNAi vector. The PSS1 RNAi and its empty vector control were introduced into the Agrobacterium strain EHA105 and used to infect the Kitaake (a japonica variety) calli. Transformed calli were grown in darkness at 25°C for 3 d. Kanamycin-resistant calli were grown in an isolated greenhouse to produce transgenic plants. The regenerated plants were further confirmed by PCR using W5f and W5r primers corresponding to the neomycin phosphotransferase II gene. All primer sequences are listed in Supplemental Table 2 online.
RT-PCR and Real-Time PCR Analysis
Total RNA was extracted using a RNA prep pure plant kit (Tiangen) according to the manufacturer’s instructions. The first-strand cDNA was synthesized using oligo(dT) as the primer. For the PCR step, two specific primers, W6f and W6r, within the PSS1 cDNA were used. Rice elongation factor 1α (EF-1α) was used as a control with the primer pair W7f and W7r.
Real-time PCR analysis was conducted using ABI7900HT Fast real-time PCR system with the SYBR Premix Ex Taq (TaKaRa; RR041A) and three biological repeats. For each sample, quantification was made in triplicate. Melt curves were read at the end of each amplification by steps of 0.3°C from 65 to 95°C to ensure that the quantification data were derived from real PCR products and not from primer dimers. Specific gene expression was normalized to the internal control gene Ubiquitin using the primer pair named ubif and ubir. The gene expression value of the wild type was used as a control and set at 1.0. For each specific gene, primer pairs were as follows: for PSS1 gene with primer pair pss1f and pss1r; for MEL1 gene with primer pair mel1f and mel1r; for UGP1 gene with primer pair ugp1f and ugp1r; for Rad21-4 with primer pair osrad21-4f and osrad21-4r; for PAIR1 gene with primer pair pair1f and pair1r; for PAIR2 with pair2f and pair2r; for PAIR3 gene with primer pair pair3f and pair3r; for MER3 gene with primer pair mer3f and mer3r. All primer sequences are listed in Supplemental Table 2 online.
PSS1-GUS Reporter Gene Construction and Analysis
A 4.3-kb promoter genomic fragment upstream of the ATG start codon was amplified by PCR (see PCR primer sequences promoter-f and promoter-r in Supplemental Table 2 online) using the Nipponbare genomic DNA as the template and cloned into the binary vector pCAMBIA1305 to drive the GUS reporter gene expression. Transgenic plants were generated as described above. GUS histochemical staining was performed as described previously (Jefferson, 1987). Images were captured using Leica Application Suite 3.3 and merged and enhanced using Photoshop CS (Adobe).
Subcellular Localization of PSS1 Protein
To investigate the cellular localization of PSS1, the PSS1 cDNA was fused in frame with GFP and inserted between the cauliflower mosaic virus 35S promoter and the NOS terminator in the PA7 vector. The expression construct was transfected into rice and Arabidopsis thaliana protoplasts according to the protocols described previously (Chen et al., 2006; Yoo et al., 2007), with GFP alone as the control. The mCherry-tagged rice prolamin box binding factor (RPBF-mCherry) construct was used as a nuclear marker (Kawakatsu et al., 2009). The samples were observed with a confocal laser scanning microscope (Leica TCS SP5).
Microtubule-Activated ATPase Assay
We cloned cDNA fragments encoding the motor domain of PSS1 (first 350 amino acids; see PCR primer sequences motor-f and motor-r in Supplemental Table 2 online) from Nipponbare or the pss1 mutant into the BamHI and SalI site of pET30a (+) (Novagen) and prepared the fusion proteins following the manufacturer’s protocol. A kinesin enzyme-linked inorganic phosphate assay biochemistry kit (Cytoskeleton) was used to measure the microtubule-activated kinesin ATPase of the motor domain of wild-type and mutantPSS1 proteins. The assay is based on an absorbance shift (330 to 360 nm) that occurs when 2-amino-6-mercapto-7-methylpurine ribonucleoside (MESG) is catalytically converted to 2-amino-6-mercapto-7-methyl purine in the presence of inorganic phosphate (Pi) and purine nucleoside phosphorylase. The reactions were set up in wells of a one-half-area 96-well plate, and each well contained 15 μg wild or mutant PSS1 motor protein, 15 mM PIPES, pH 7.0, 5.0 mM MgCl2, 15 μM paclitaxel, 0.75 mM ATP, 0.2 units purine nucleoside phosphorylase, 0.2 mM MESG reagent, and microtubule concentrations ranging from 0 to 200 μg/mL in a reaction volume of 200 μL. Reactions were started by the addition of ATP and were read every 30 s at 360 nm for a total of 15 min using a monochromatic spectrophotometer (SpectroMax250; Molecular Devices).
Meiotic Chromosome Examination
Young (40- to 60-mm) panicles, including flowers in meiosis, were fixed with 3:1 ethanol:acetic acid and stored at 4°C until examination. Anthers were staged by staining with 1% of Aceto carmine (Sigma-Aldrich Chemical) to visualize chromosomes under an optical microscope. Anthers of appropriate stages were squashed in 40% acetic acid with a cover slip. After freezing in liquid nitrogen for 5 min, the cover slip was removed and the sample dried at room temperature. Then meiocytes were treated with 20 μL of 0.1 mg/mL propidium iodide for ~20 min to stain chromatin. Finally, the male meiocytes were observed using a fluorescence microscope (Leica DM5000B). Images were captured using Leica Application Suite 3.3 and merged and enhanced using Photoshop CS (Adobe).
Immunofluorescence Assay
Observation of the spindle morphology in male meiocytes was performed following a previously described immunofluorescence method (Chan and Cande, 1998). Meiocytes were always used the day the panicles were harvested. Anthers (30 to 40) of appropriate stages were placed into 2 mL of fixative solution: 8% (v/v) paraformaldehyde (Electron Microscopy Sciences) in PHEMS buffer (60 mM PIPES, 25 mM HEPES, 10 mM EGTA, 2 mM MgCl2, and 0.32 M sorbitol, pH 6.8) in a small Petri dish and shaken for 2 h at a medium speed (~100 rpm). Then, the anthers were rinsed with PHEMS buffer without formaldehyde. Using a microblade and forceps, the ends of each anther was cut off, and meiocytes were extruded from the anthers into PHEMS buffer. Aliquots (10 μL) of cell suspension were placed into 1.5-mL microcentrifuge tubes, and 10 μL of molten 3% agarose (ultralow gelling agarose; SeaPrep 15/45, FMC Corporation) in PHEMS buffer were added to each tube. The tubes were cooled to slightly below 15°C, allowing the agarose to solidify. The agarose block in each tube was incubated overnight at room temperature with 100 μL of 1.5% β-glucuronidase (Sigma-Aldrich Chemical; G-0751) in PHEMS buffer to digest the cell walls. The agarose blocks were rinsed with 100 μL PBS (0.14 M NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4, pH 7.2). The agarose block in each tube was then incubated with 50 μL of PBS-diluted (1:1000) mouse monoclonal antibody against α-tubulin (Sigma-Aldrich Chemical; T-9026) overnight at room temperature, rinsed with PBS, and then incubated overnight at room temperature with 30 μL of PBS-diluted (1:30) fluorescein isothiocyanate–conjugated goat-anti-mouse antibody (Sigma-Aldrich Chemical; F-0257). After rinsing with PBS, the blocks were treated with 50 μL of 0.1 mg/mL propidium iodide for ~30 min to stain the chromosomes. The agarose blocks were rinsed with PBS and then placed on glass slides. For each slide, a layer of tape was placed on each side of the slide to raise the cover slip. Approximately 100 μL of 100 mg/mL 1,4-diazobicyclo (2, 2, and 2) octane (DABCO; Sigma-Aldrich Chemical) were placed on each block (100 mg of DABCO were combined with 0.1 mL of PBS and 0.9 mL of glycerin). The slides were heated until the agarose blocks just melted completely. A cover slip was placed onto each slide, and the cover slips were sealed to the slides with fingernail polish. The male meiocytes were observed using confocal laser scanning microscopy (Leica SP2). Five or six photos of optical sections from a meiocyte were merged and enhanced using Photoshop CS (Adobe).
Computational and Database Analysis
The kinesin sequences were downloaded from the kinesin homepage (http://www.cellbio.duke.edu/kinesin/KinesinAlign.html). Multiple sequence alignments were performed using ClustalW (Thompson et al., 1994) with the BLOSUM matrix (Henikoff and Henikoff, 1992). A phylogenetic tree was constructed with the aligned kinesin motor sequences using MEGA software (version 4.0) (http://www.megasoftware.net/index.html) (Tamura et al., 2007) based on the neighbor-joining method with the following parameters: p-distance model, pairwise deletion, and bootstrap (1000 replicates; random seed).
The three-dimensional structure of human KHC (Hs KHC) was downloaded from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov), and figures were generated using Cn3D software (http://www.ncbi.nlm.nih.gov/Structure/CN3D/cn3d.shtml).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession number: PSS1, AK287457; MEL1, AB297928; UGP1, DQ395328; Rad21-4, AY371049; PAIR1, AB158462; PAIR2, AB109238; PAIR3, FJ449712; MER3, FJ008126; and Ubiquitin, AF184279.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. PSS1 RNAi Plants Show a Similar Phenotype to the pss1 Mutant.
Supplemental Figure 2. Sequence Analysis of the Predicted PSS1 and Related Proteins.
Supplemental Figure 3. Neighbor-Joining Tree of the Kinesin Superfamily Proteins.
Supplemental Figure 4. Transcriptional Analysis of Male Meiotic Genes in pss1 Mutant and a Putative Genetic Pathway for Microsporogenesis.
Supplemental Figure 5. Subcellular Localization of PSS1 Protein in Arabidopsis Protoplasts.
Supplemental Figure 6. Amino Acid Sequences Alignment of the Kinesin Motor Domain of Rice PSS1, Human KHC, and Arabidopsis TETRASPORE.
Supplemental Figure 7. A Ribbon Diagram of the Human KHC Motor.
Supplemental Table 1. Statistics of Abnormal Chromosomal Behavior in RNAi Line2.
Supplemental Table 2. List of the Primers Used for Vector Construction, Transgenic Plant Identification, and Gene Expression Analysis.
Supplemental Data Set 1. Text File of Alignment Corresponding to Supplemental Figure 3.
Supplementary Material
Acknowledgments
We thank B. Han (National Center for Gene Research, Chinese Academy of Sciences) for the help in sequencing the candidate gene region and Clemson University Genomics Institute for providing the BAC clone. We also thank X.-F. Cao (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for providing the vector pCam23ACT:OCS and X.-Y. Chen (Institute of Plant Physiology and Ecology, Chinese Academy of Sciences) for the help with the plastic section. We also thank Z.-K. Chen (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) and X.-M. Yang (Institute of Crop Sciences, Chinese Academy of Agricultural Sciences) for advice on meiotic chromosome examination. G.-Q. Liu (China Agricultural University) is also gratefully acknowledged for help with the microtubule-activated ATPase assay. This research was supported by grants from the National High Technology Research and Development Program of China (2006AA10A101 and 2009AA101101), the National Natural Science Foundation of China (30671275 and 30800679), a project from the Ministry of Agriculture of China for transgenic research (2008ZX08009-003 and2009ZX08009-107B), the National Key Basic Research “973” Program of China (2006CB101703), the Jiangsu Science and Technology Development Program (BE2008352), and the 111 Project (B0802).
References
- Armstrong S.J., Caryl A.P., Jones G.H., Franklin F.C. (2002). Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J. Cell Sci. 115: 3645–3655 [DOI] [PubMed] [Google Scholar]
- Brady S.T., Pfister K.K., Bloom G.S. (1990). A monoclonal antibody against kinesin inhibits both anterograde and retrograde fast axonal transport in squid axoplasm. Proc. Natl. Acad. Sci. USA 87: 1061–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar D.S., Khush G.S. (1997). Alien introgression in rice. Plant Mol. Biol. 35: 35–47 [PubMed] [Google Scholar]
- Cardoso C.M., Groth-Pedersen L., Høyer-Hansen M., Kirkegaard T., Corcelle E., Andersen J.S., Jäättelä M., Nylandsted J. (2009). Depletion of kinesin 5B affects lysosomal distribution and stability and induces peri-nuclear accumulation of autophagosomes in cancer cells. PLoS ONE 4: e4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caryl A.P., Armstrong S.J., Jones G.H., Franklin F.C. (2000). A homologue of the yeast HOP1 gene is inactivated in the Arabidopsis meiotic mutant asy1. Chromosoma 109: 62–71 [DOI] [PubMed] [Google Scholar]
- Chalfun-Junior A., Franken J., Mes J.J., Marsch-Martinez N., Pereira A., Angenent G.C. (2005). ASYMMETRIC LEAVES2-LIKE1 gene, a member of the AS2/LOB family, controls proximal-distal patterning in Arabidopsis petals. Plant Mol. Biol. 57: 559–575 [DOI] [PubMed] [Google Scholar]
- Chan A., Cande W.Z. (1998). Maize meiotic spindles assemble around chromatin and do not require paired chromosomes. J. Cell Sci. 111: 3507–3515 [DOI] [PubMed] [Google Scholar]
- Chen C., Marcus A., Li W., Hu Y., Calzada J.P., Grossniklaus U., Cyr R.J., Ma H. (2002). The Arabidopsis ATK1 gene is required for spindle morphogenesis in male meiosis. Development 129: 2401–2409 [DOI] [PubMed] [Google Scholar]
- Chen R., Zhao X., Shao Z., Wei Z., Wang Y., Zhu L., Zhao J., Sun M., He R., He G. (2007). Rice UDP-glucose pyrophosphorylase1 is essential for pollen callose deposition and its cosuppression results in a new type of thermosensitive genic male sterility. Plant Cell 19: 847–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Tao L., Zeng L., Vega-Sanchez M.E., Umemura K., Wang G.L. (2006). A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol. Plant Pathol. 7: 417–427 [DOI] [PubMed] [Google Scholar]
- Dagenbach E.M., Endow S.A. (2004). A new kinesin tree. J. Cell Sci. 117: 3–7 [DOI] [PubMed] [Google Scholar]
- Deng Z.Y., Wang T. (2007). OsDMC1 is required for homologous pairing in Oryza sativa. Plant Mol. Biol. 65: 31–42 [DOI] [PubMed] [Google Scholar]
- Ding Z., Wang T., Chong K., Bai S. (2001). Isolation and characterization of OsDMC1, the rice homologue of the yeast DMC1 gene essential for meiosis. Sex. Plant Reprod. 13: 285–288 [Google Scholar]
- Endow S.A. (1999). Microtubule motors in spindle and chromosome motility. Eur. J. Biochem. 262: 12–18 [DOI] [PubMed] [Google Scholar]
- Feys B., Benedetti C.E., Penfold C.N., Turner J.G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6: 751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi R., Bonaccorsi S., Wentworth D., Doxsey S., Gatti M., Pereira A. (2004). The Drosophila kinesin-like protein KLP67A is essential for mitotic and male meiotic spindle assembly. Mol. Biol. Cell 15: 121–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Henikoff J.G. (1992). Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA 89: 10915–10919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N., Noda Y., Okada Y. (1998). Kinesin and dynein superfamily proteins in organelle transport and cell division. Curr. Opin. Cell Biol. 10: 60–73 [DOI] [PubMed] [Google Scholar]
- Holzinger A., Lütz-Meindl U. (2002). Kinesin-like proteins are involved in postmitotic nuclear migration of the unicellular green alga Micrasterias denticulata. Cell Biol. Int. 26: 689–697 [DOI] [PubMed] [Google Scholar]
- Ishiguro S., Kawai-Oda A., Ueda J., Nishida I., Okada K. (2001). The DEFECTIVE IN ANTHER DEHISCIENCE gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 13: 2191–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh J., Nonomura K., Ikeda K., Yamaki S., Inukai Y., Yamagishi H., Kitano H., Nagato Y. (2005). Rice plant development: from zygote to spikelet. Plant Cell Physiol. 46: 23–47 [DOI] [PubMed] [Google Scholar]
- Jefferson R. (1987). Assaying chimeric genes in plants: The GUS gene fusion system. Plant Mol. Biol. Rep. 5: 387–405 [Google Scholar]
- Jenkins G., Phillips D., Mikhailova E.I., Timofejeva L., Jones R.N. (2008). Meiotic genes and proteins in cereals. Cytogenet. Genome Res. 120: 291–301 [DOI] [PubMed] [Google Scholar]
- Jung K.H., Han M.J., Lee Y.S., Kim Y.W., Hwang I., Kim M.J., Kim Y.K., Nahm B.H., An G. (2005). Rice Undeveloped Tapetum1 is a major regulator of early tapetum development. Plant Cell 17: 2705–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama T. (1963). Study on the progenies of autotriploid and asynaptic rice plants. Jpn. J. Breed. 13: 83–87 [Google Scholar]
- Kathiresan A., Khush G., Bennett J. (2002). Two rice DMC1 genes are differentially expressed during meiosis and during haploid and diploid mitosis. Sex. Plant Reprod. 14: 257–267 [Google Scholar]
- Kawakatsu T., Yamamoto M.P., Touno S.M., Yasuda H., Takaiwa F. (2009). Compensation and interaction between RISBZ1 and RPBF during grain filling in rice. Plant J. 59: 908–920 [DOI] [PubMed] [Google Scholar]
- Lawrence C.J., et al. (2004). A standardized kinesin nomenclature. J. Cell Biol. 167: 19–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.R., Li Y., Liu B. (2007). Two Arabidopsis phragmoplast-associated kinesins play a critical role in cytokinesis during male gametogenesis. Plant Cell 19: 2595–2605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., et al. (2006). The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18: 2999–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Jiang L., Zhou S., Wang C., Liu L., Chen L., Ikehashi H., Wan J. (2007). Fine mapping of pss1, a pollen semi-sterile gene in rice (Oryza sativa L.). Theor. Appl. Genet. 114: 939–946 [DOI] [PubMed] [Google Scholar]
- Li Y., Qian Q., Zhou Y., Yan M., Sun L., Zhang M., Fu Z., Wang Y., Han B., Pang X., Chen M., Li J. (2003). BRITTLE CULM1, which encodes a COBRA-like protein, affects the mechanical properties of rice plants. Plant Cell 15: 2020–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H. (June 6, 2006) A molecular portrait of Arabidopsis meiosis. The Arabidopsis Book, Somerville C.R., Meyerowitz E.M., (Rockville, MD: American Society of Plant Biologists; ), doi/10.1199/tab.0095, http://www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho B., Brancorsini S., Fimia G.M., Setou M., Hirokawa N., Sassone-Corsi P. (2002). CREM-dependent transcription in male germ cells controlled by a kinesin. Science 298: 2388–2390 [DOI] [PubMed] [Google Scholar]
- Matsui T., Omasa K., Horie T. (1999a). Mechanism of anther dehiscence in rice (Oryza sativa L.). Ann. Bot. (Lond.) 84: 501–506 [Google Scholar]
- Matsui T., Omasa K., Horie T. (1999b). Rapid swelling of pollen grains in response to floret opening unfolds anther locules in rice (Oryza sativa L.). Plant Prod. Sci. 2: 196–199 [Google Scholar]
- Mazumdar M., Misteli T. (2005). Chromokinesins: Multitalented players in mitosis. Trends Cell Biol. 15: 349–355 [DOI] [PubMed] [Google Scholar]
- Mercier R., Grelon M. (2008). Meiosis in plants: ten years of gene discovery. Cytogenet. Genome Res. 120: 281–290 [DOI] [PubMed] [Google Scholar]
- Miki H., Okada Y., Hirokawa N. (2005). Analysis of the kinesin superfamily: Insights into structure and function. Trends Cell Biol. 15: 467–476 [DOI] [PubMed] [Google Scholar]
- Nakamura T., Chiba M., Koike S., Nishiyama I. (2000). Number of pollen grains in rice cultivars with different cool-weather resistance at the young microspore stage. Plant Prod. Sci. 3: 299–305 [Google Scholar]
- Neff M.M., Neff J.D., Chory J., Pepper A.E. (1998). dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. Plant J. 14: 387–392 [DOI] [PubMed] [Google Scholar]
- Neff M.M., Turk E., Kalishman M. (2002). Web-based primer design for single nucleotide polymorphism analysis. Trends Genet. 18: 613–615 [DOI] [PubMed] [Google Scholar]
- Nonomura K., Miyoshi K., Eiguchi M., Suzuki T., Miyao A., Hirochika H., Kurata N. (2003). The MSP1 gene is necessary to restrict the number of cells entering into male and female sporogenesis and to initiate anther wall formation in rice. Plant Cell 15: 1728–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura K., Morohoshi A., Nakano M., Eiguchi M., Miyao A., Hirochika H., Kurata N. (2007). A germ cell specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice. Plant Cell 19: 2583–2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura K., Nakano M., Eiguchi M., Suzuki T., Kurata N. (2006). PAIR2 is essential for homologous chromosome synapsis in rice meiosis I. J. Cell Sci. 119: 217–225 [DOI] [PubMed] [Google Scholar]
- Nonomura K., Nakano M., Fukuda T., Eiguchi M., Miyao A., Hirochika H., Kurata N. (2004a). The novel gene HOMOLOGOUS PAIRING ABERRATION IN RICE MEIOSIS1 of rice encodes a putative coiled-coil protein required for homologous chromosome pairing in meiosis. Plant Cell 16: 1008–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura K.I., Nakano M., Murata K., Miyoshi K., Eiguchi M., Miyao A., Hirochika H., Kurata N. (2004b). An insertional mutation in the rice PAIR2 gene, the ortholog of Arabidopsis ASY1, results in a defect in homologous chromosome pairing during meiosis. Mol. Genet. Genomics 271: 121–129 [DOI] [PubMed] [Google Scholar]
- Oh S.-A., Bourdon V., Das ’Pal M., Dickinson H., Twell D. (2008). Arabidopsis kinesins HINKEL and TETRASPORE act redundantly to control cell plate expansion during cytokinesis in the male gametophyte. Mol. Plant 1: 794–799 [DOI] [PubMed] [Google Scholar]
- Park J.H., Halitschke R., Kim H.B., Baldwin I.T., Feldmann K.A., Feyereisen R. (2002). A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 31: 1–12 [DOI] [PubMed] [Google Scholar]
- Quan L., Xiao R., Li W., Oh S.A., Kong H., Ambrose J.C., Malcos J.L., Cyr R., Twell D., Ma H. (2008). Functional divergence of the duplicated AtKIN14a and AtKIN14b genes: critical roles in Arabidopsis meiosis and gametophyte development. Plant J. 53: 1013–1026 [DOI] [PubMed] [Google Scholar]
- Reddy A.S., Day I.S. (2001). Kinesins in the Arabidopsis genome: A comparative analysis among eukaryotes. BMC Genomics 2: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson D.N., Simmons M.P., Reddy A.S. (2006). Comprehensive comparative analysis of kinesins in photosynthetic eukaryotes. BMC Genomics 7: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronceret A., Sheehan M., Pawlowski W. (2008). Chromosome dynamics in meiosis. Cell Division Control in Plants, Verma D., Hong Z., (Berlin/Heidelberg: Springer; ), pp. 103–124 [Google Scholar]
- Sanders P.M., Lee P.Y., Biesgen C., Boone J.D., Beals T.P., Weiler E.W., Goldberg R.B. (2000). The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 12: 1041–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazuka T., Aichi I., Kawai T., Matsuo N., Kitano H., Matsuoka M. (2005). The rice mutant dwarf bamboo shoot 1: A leaky mutant of the NACK-type kinesin-like gene can initiate organ primordia but not organ development. Plant Cell Physiol. 46: 1934–1943 [DOI] [PubMed] [Google Scholar]
- Sharp D.J., Rogers G.C., Scholey J.M. (2000). Microtubule motors in mitosis. Nature 407: 41–47 [DOI] [PubMed] [Google Scholar]
- Spielman M., Preuss D., Li F.L., Browne W.E., Scott R.J., Dickinson H.G. (1997). TETRASPORE is required for male meiotic cytokinesis in Arabidopsis thaliana. Development 124: 2645–2657 [DOI] [PubMed] [Google Scholar]
- Stintzi A., Browse J. (2000). The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. USA 97: 10625–10630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tanaka H., Ishikawa M., Kitamura S., Takahashi Y., Soyano T., Machida C., Machida Y. (2004). The AtNACK1/HINKEL and STUD/TETRASPORE/AtNACK2 genes, which encode functionally redundant kinesins, are essential for cytokinesis in Arabidopsis. Genes Cells 9: 1199–1211 [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale R.D., Fletterick R.J. (1997). The design plan of kinesin motors. Annu. Rev. Cell Dev. Biol. 13: 745–777 [DOI] [PubMed] [Google Scholar]
- Virmani S.S. (1996). Hybrid rice. Advances in Agronomy, Donald L.S., (San Diego, CA: Academic Press; ), pp. 377–462 [Google Scholar]
- von Malek B., van der Graaff E., Schneitz K., Keller B. (2002). The Arabidopsis male-sterile mutant dde2-2 is defective in the ALLENE OXIDE SYNTHASE gene encoding one of the key enzymes of the jasmonic acid biosynthesis pathway. Planta 216: 187–192 [DOI] [PubMed] [Google Scholar]
- Wan J., Ikehashi H. (1995). Identification of a new locus S-16 causing hybrid sterility in native rice varieties (Oryza sativa L.) from Tai-hu lake region and Yunnan province, China. Breed. Sci. 45: 461–470 [Google Scholar]
- Wan J., Ikehashi H. (1996). List of hybrid sterility gene loci (HSGLi) in cultivated rice (Oryza sativa L.). Rice Genet. Newsl. 13: 110–113 [DOI] [PubMed] [Google Scholar]
- Wan J., Yamaguchi Y., Kato H., Ikehashi H. (1996). Two new loci for hybrid sterility in cultivated rice (Oryza sativa L.). Theor. Appl. Genet. 92: 183–190 [DOI] [PubMed] [Google Scholar]
- Wan J., Yanagihara S., Kato H., Ikehashi H. (1993). Multiple alleles at a new locus causing hybri sterility between a Korean indica variety and Javanica variety in rice (Oryza sativa L.). Jpn. J. Breed. 43: 507–516 [Google Scholar]
- Wang K., Tang D., Wang M., Lu J., Yu H., Liu J., Qian B., Gong Z., Wang X., Chen J., Gu M., Cheng Z. (2009). MER3 is required for normal meiotic crossover formation, but not for presynaptic alignment in rice. J. Cell Sci. 122: 2055–2063 [DOI] [PubMed] [Google Scholar]
- Woehlke G., Ruby A.K., Hart C.L., Ly B., Hom-Booher N., Vale R.D. (1997). Microtubule interaction site of the kinesin motor. Cell 90: 207–216 [DOI] [PubMed] [Google Scholar]
- Xie D.X., Feys B.F., James S., Nieto-Rostro M., Turner J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]
- Yanagihara S., Kato H., Ikehashi H. (1992). A new locus for multiple alleles causing hybrid sterility between an aus variety and javanica varieties in rice (Oryza sativa L.). Jpn. J. Breed. 42: 793–801 [Google Scholar]
- Yang C., Xu Z., Song J., Conner K., Vizcay Barrena G., Wilson Z.A. (2007). Arabidopsis MYB26/MALE STERILE35 regulates secondary thickening in the endothecium and is essential for anther dehiscence. Plant Cell 19: 534–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C.Y., Spielman M., Coles J.P., Li Y., Ghelani S., Bourdon V., Brown R.C., Lemmon B.E., Scott R.J., Dickinson H.G. (2003). TETRASPORE encodes a kinesin required for male meiotic cytokinesis in Arabidopsis. Plant J. 34: 229–240 [DOI] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yu W.W., Wang C.M., Ikehashi H., Wan J.M. (2005). Mapping of a novel gene for semi-sterility in rice (Oryza sativa L.). Breed. Sci. 55: 15–20 [Google Scholar]
- Yuan W., Li X., Chang Y., Wen R., Chen G., Zhang Q., Wu C. (2009). Mutation of the rice gene PAIR3 results in lack of bivalent formation in meiosis. Plant J. 59: 303–315 [DOI] [PubMed] [Google Scholar]
- Zhang L., Tao J., Wang S., Chong K., Wang T. (2006). The rice OsRad21-4, an orthologue of yeast Rec8 protein, is required for efficient meiosis. Plant Mol. Biol. 60: 533–554 [DOI] [PubMed] [Google Scholar]
- Zhao Z.G., Jiang L., Zhang W.W., Yu C.Y., Zhu S.S., Xie K., Tian H., Liu L.L., Ikehashi H., Wan J.M. (2007). Fine mapping of S31, a gene responsible for hybrid embryo-sac abortion in rice (Oryza sativa L.). Planta 226: 1087–1096 [DOI] [PubMed] [Google Scholar]
- Zhu Q.H., Ramm K., Shivakkumar R., Dennis E.S., Upadhyaya N.M. (2004). The ANTHER INDEHISCENCE1 gene encoding a single MYB domain protein is involved in anther development in rice. Plant Physiol. 135: 1514–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D., Kleckner N. (1999). Meiotic chromosomes: Integrating structure and function. Annu. Rev. Genet. 33: 603–754 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.