A flowering repressor, FLC, has been extensively studied for its role in epigenetic regulation. However, the mechanism of FLC transcription is largely unknown. The authors show that the FRI protein complex recruits basal transcription factors, chromatin modification complexes, and eventually RNA polymerase II to transcribe the FLC gene. This helps to explain the flowering of winter annual Arabidopsis.
Abstract
The flowering of Arabidopsis thaliana winter annuals is delayed until the subsequent spring by the strong floral repressor FLOWERING LOCUS C (FLC). FRIGIDA (FRI) activates the transcription of FLC, but the molecular mechanism remains elusive. The fri mutation causes early flowering with reduced FLC expression similar to frl1, fes1, suf4, and flx, which are mutants of FLC-specific regulators. Here, we report that FRI acts as a scaffold protein interacting with FRL1, FES1, SUF4, and FLX to form a transcription activator complex (FRI-C). Each component of FRI-C has a specialized function. SUF4 binds to a cis-element of the FLC promoter, FLX and FES1 have transcriptional activation potential, and FRL1 and FES1 stabilize the complex. FRI-C recruits a general transcription factor, a TAF14 homolog, and chromatin modification factors, the SWR1 complex and SET2 homolog. Complex formation was confirmed by the immunoprecipitation of FRI-associated proteins followed by mass spectrometric analysis. Our results provide insight into how a specific transcription activator recruits chromatin modifiers to regulate a key flowering gene.
INTRODUCTION
Seasonal flowering is fundamental to the reproductive success and survival of higher plants. Plants have evolved a complex genetic network to control flowering time in response to endogenous cues and environmental factors such as daylength and temperature (Simpson and Dean, 2002). Arabidopsis thaliana is distributed in a wide range of environments, and its flowering behavior has adapted to these local environments (Napp-Zinn, 1979; Gazzani et al., 2003). Most of the Arabidopsis accessions collected from northern areas display winter annual flowering characteristics: late flowering that is accelerated by a long-term winter cold period, called vernalization (Gazzani et al., 2003; Michaels et al., 2003). The genetic analysis of natural variation in the flowering time of Arabidopsis has revealed two major genes conferring such characteristics, FRIGIDA (FRI) and FLOWERING LOCUS C (FLC) (Napp-Zinn, 1979; Clarke and Dean, 1994; Lee et al., 1994; Gazzani et al., 2003). Winter annuals have functional genes of both FRI and FLC, whereas summer annuals (rapid cycling accessions) have mutations in either FRI or FLC or both (Johanson et al., 2000; Gazzani et al., 2003; Michaels et al., 2003). FRI encodes a protein with two coiled-coil motifs and is required to increase the FLC transcript level (Johanson et al., 2000). FLC encodes a MADS box transcription factor that blocks flowering quantitatively by repressing the transcription of downstream floral pathway integrator genes (Michaels and Amasino, 1999; Sheldon et al., 1999). Although the FRI gene was cloned a decade ago (Johanson et al., 2000), its biochemical function remains unknown.
The expression of FLC is regulated by diverse and sophisticated mechanisms. Whereas FRI is required for an increase of the FLC transcript, vernalization completely suppresses FLC expression (Michaels and Amasino, 2001). Such suppression is accomplished through a series of histone modifications by the plant polycomb repressive complex, including VERNALIZATION INSENSITIVE3, VERNALIZATION2 (VRN2), and the subsequent maintenance of the heterochromatin state by VRN1 and LIKE HETEROCHROMATIN PROTEIN1 (Kim et al., 2009). In summer annuals that have a nonfunctional FRI, FLC expression is repressed by the cooperation of the so-called autonomous pathway genes, which are mostly involved in chromatin modifications and RNA processing (Simpson, 2004). The late flowering caused by the mutations in autonomous pathway genes is also suppressed by vernalization (Michaels and Amasino, 2001). Thus, vernalization overrides the function of FRI, and FRI overrides the function of all autonomous pathway genes.
In elucidating the FRI-mediated FLC regulation mechanism, two classes of FLC regulators were revealed: FLC-specific regulators and FLC nonspecific regulators. When mutated, FLC-specific regulators show an early flowering phenotype without any other obvious phenotype, whereas nonspecific regulators show pleiotropic phenotypes such as small size and abnormal morphology (He et al., 2004; Kim et al., 2009). To date, six FLC-specific regulators, FRI, FRIGIDA LIKE1 (FRL1), FRL2, FRIGIDA ESSENTIAL1 (FES1), SUF4, and FLC EXPRESSOR (FLX), have been reported (Michaels et al., 2004; Schmitz et al., 2005; Kim et al., 2006; Andersson et al., 2008). All the mutants for the FLC-specific regulators show reduced FLC expression, and the overexpression of one gene does not induce late flowering in the other mutant backgrounds. These findings suggest that the proteins encoded by the FLC-specific regulators act as the components of the same signaling pathway or the same protein complex rather than acting independently in a dosage-dependent manner. However, protein-to-protein interactions among the FLC specific regulators were not detected by a yeast two-hybrid analysis in previous reports (Michaels et al., 2004; Schmitz et al., 2005; Andersson et al., 2008). Although we have reported that SUF4, which contains two C2H2-type zinc finger motifs and a Pro-rich region, physically interacts with FRI, the existence of a FRI-containing complex (FRI-C) is controversial, and the molecular function remains to be elucidated (Kim et al., 2006).
FLC nonspecific regulators are involved in either chromatin modification or RNA processing. The full activation of FLC by FRI requires chromatin modifiers, including proteins catalyzing histone H2B ubiquitination/deubiquitination, histone H3 lysine 4 (H3K4) methylation, H3K36 methylation, and the PAF1 (RNA polymerase associated factor 1) complex (Kim et al., 2009). In addition, the orthologous components of the yeast ATP-dependent SWR1 chromatin remodeling complex (SWR1-C), such as PHOTOPERIOD INDEPENDENT EARLY FLOWERING1 (PIE1), ACTIN-RELATED PROTEIN6 (ARP6)/SUPPRESSOR OF FRIGIDA3 (SUF3), and SWC6, are also required for incorporation of H2A.Z into FLC chromatin (Noh and Amasino, 2003; Choi et al., 2007; Deal et al., 2007). These chromatin-modifying proteins are well conserved among eukaryotes and are required for the epigenetic marks that are used for active transcription (Li et al., 2007). How these proteins, including SWR1-C, are recruited to the FLC gene remains unclear. Additionally, the RNA-processing factors, HUA2 and SERRATE, and an mRNA cap binding protein, CBP80/ABH1, are also involved in FLC activation (Bezerra et al., 2004; Doyle et al., 2005).
Here, we show that FRI acts as a scaffold protein interacting with known FLC specific regulators and that the resulting protein complex, FRI-C, acts as a specific transcription factor of FLC by recruiting the general transcription machinery and chromatin modification factors.
RESULTS
FRI Forms a Large Protein Complex
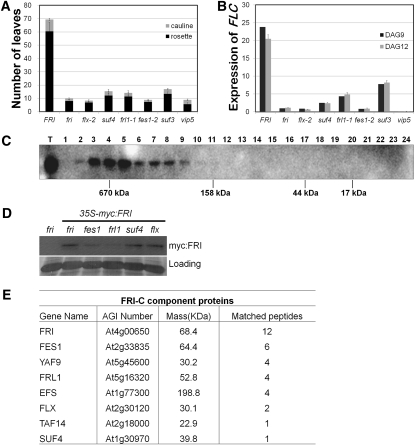
To elucidate the FRI-mediated FLC activation mechanism, we characterized early flowering suf mutants that were generated by fast-neutron mutagenesis; the suf mutations suppress the late flowering phenotype in the FRI-containing line. We isolated a total of 33 early flowering mutants, which were grouped into 12 independent loci (suf1-suf12) based on a complementation analysis (see Supplemental Figure 1A online). All of the suf mutants exhibited reductions in the expression of FLC; some showed only a flowering phenotype, similar to fri, but others showed a pleiotropic phenotype. Thus, we tentatively classified the suf mutants into two groups, one for FLC-specific regulators and the other for FLC nonspecific regulators. The FLC-specific regulators we analyzed were SUF4, SUF5/FLX, and SUF8/FRL1. Among these, the suf4 and frl1 mutations caused slightly later flowering and a higher expression of FLC than did the fri mutation (Figures 1A and 1B).
Figure 1.
The Flowering Phenotype Caused by Mutations in FLC Regulators and the Presence of the FRI Complex.
(A) Flowering time was measured by counting total rosette and cauline leaves at the bolting stage. A higher number of leaves at bolting represents later flowering. Twenty plants per genotype were used for the flowering time measurement. Error bars indicate the sd.
(B) Real-time RT-PCR analysis of the FLC transcript level in the wild type and mutants grown for 9 and 12 d after germination (DAG) under long days. Levels of FLC transcripts were normalized to the levels of ACTIN and presented as a relative value to that of fri. Error bars indicate the sd of triplicate experiments. (C) An immunoblot of the gel filtration fractions from a Superdex 200 column and protein extracts of the 35S-myc:FRI fri transgenic plant. T indicates the total protein as input, and the numbers above the blot represent the fraction order. Calibrated molecular masses are shown below the blot. The myc:FRI protein was detected with a monoclonal myc antibody.
(D) An immunoblot showing the level of myc:FRI protein in fri, fes1, frl1, suf4, and flx backgrounds. Ponceau staining is shown as a loading control.
(E) The myc-FRI–associated proteins. Plants carrying fully functional myc-FRI protein expressed under the control of the 35S promoter were harvested after 2 weeks of growth under long days and used in the immunoprecipitation purification. The proteins were identified by LC-MS/MS. The fri mutant plants were used as a negative control to determine whether a protein was associated with the myc-FRI protein. The immunoprecipitation and ensuing LC-MS/MS analysis was repeated three times independently, and the representative data are presented. Full details on the peptides and scores from the myc purification and control sample are available in Supplemental Table 1 online.
suf5 was allelic to flx, which was identified in the C24 background (Andersson et al., 2008), and was labeled as flx-2. It showed only early flowering, which was similar to the fri, frl1, fes1, and suf4 mutants (Figures 1A and 1B; see Supplemental Figure 2 online). FLX was highly expressed in the shoot and root apical meristems and vascular tissues (see Supplemental Figure 2F online), where FLC and the FLC positive regulators, FES1, SUF3, SUF4, and SWC6, are expressed (Michaels and Amasino, 2001; Choi et al., 2005, 2007; Schmitz et al., 2005; Kim et al., 2006). In addition, the overexpression of FLX rescued flx-2 but failed to induce late flowering in the Columbia (Col) (fri) background (see Supplemental Figure 2E online). These results suggest that FLX acts in a protein complex with FRI.
To reveal the size of the protein complex that includes FRI, we performed gel filtration analyses using a myc-tagged FRI transgenic line, namely, 35S-myc:FRI fri (Figure 1C). The 35S-myc:FRI fri plants flowered (68 ± 7.6 rosette leaves) as late as the wild type, indicating that the introduced transgene was functional. The size of the protein complex detected by the myc antibody was ~670 kD, ranging from 400 to 1000 kD. This size is much higher than the expected size for a FRI monomer (68.4 kD), indicating that FRI exists mostly as a component of a large protein complex. To determine whether the integrity of the protein complex was affected by the mutations in the FLC-specific regulators, we determined the size of this protein complex in each mutant. The size was not affected by the individual mutation (see Supplemental Figure 3 online). However, fes1 and frl1 consistently showed a decreased steady state level of FRI and the FRI complex (FRI-C) (Figure 1D; see Supplemental Figure 3 online). Taken together, the results suggested that FES1 and FRL1 are required for the stabilization of FRI-C.
To identify the FRI-C components and any additional interacting proteins in vivo, we performed biochemical purifications using 35S-myc:FRI fri seedlings. After isolating a nuclear protein extract from 35S-myc:FRI fri seedlings, the FRI-associated proteins were obtained by immunoprecipitation using c-myc antibody–conjugated agarose beads and then analyzed by liquid chromatography–tandem mass spectrometry (LC-MS/MS). As a negative control, the same procedure was performed using fri seedlings. FRI, FES1, FRL1, FLX, and SUF4 were identified as FRI-associated proteins that were not detected in the fri negative control (Figure 1E). This result indicates that these proteins comprise FRI-C in vivo. In addition, we detected EFS, TAF14, and YAF9 as FRI-associated proteins (Figure 1E; see Supplemental Table 1 online for a full list of the proteins detected), which may explain why the size of the FRI-C was nearly 1 MD. EFS, YAF9, and TAF14 are components of chromatin modification factors and general transcription factors, respectively (see below).
FRI Is a Scaffold Protein Interacting with FLC-Specific Regulators
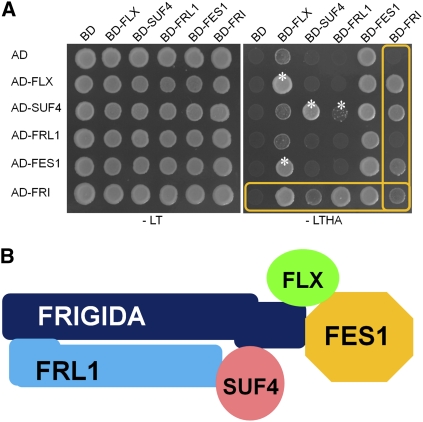
Because the genetic and biochemical data showed that the FLC specific regulators were components of the FRI-C, we carefully performed a yeast two-hybrid (Y2H) interaction analysis with FRI, FRL1, FES1, SUF4, and FLX using activation domain (AD) and binding domain (BD) constructs that contained both the full-length coding sequences and a series of deletions (Figure 2A; see Supplemental Figure 4 online). As expected, FRI interacted with FLX, SUF4, and FES1. The interaction between AD-FRL1 and BD-FRI was not detected when the full coding sequence was used, but we found that the C-terminal deleted FRL1 could interact with FRI (see Supplemental Figure 4 online), indicating either that the AD-fused FRL1 may not be well translated or that the protein structure may impede the interaction in yeast. The interaction between FRI and FRL1 has also been reported in Y2H assays using different yeast strains (Geraldo et al., 2009). Such a result suggests that FRI may act as a scaffold protein to assemble a protein complex that activates FLC. However, any combinations between BD-FES1 and AD-other proteins could not be considered as interaction because the combination of BD-FES1 and AD alone showed good growth even in –LTHA medium due to strong transcriptional activity of FES1 itself. Thus, interactions between BD-FES1 and AD-other proteins require independent confirmation by bimolecular fluorescence complementation (BiFC) as below.
Figure 2.
Y2H Interaction Analyses among FRI, FRL1, FLX, SUF4, and FES1.
(A) Interactions among full-length FRI, FRL1, FLX, SUF4, and FES1. The -LT indicates the synthetic drop (SD) medium lacking Leu and Trp, whereas -LTHA indicates the SD medium without Leu, Trp, His, and Ade. Boxes show the FRI interactions with FLX, SUF4, FRL1, and FES1. White stars mark the interactions with other proteins except FRI.
(B) A schematic drawing showing the interactions among the components of the FRI complex. FRI functions as a scaffold protein interacting with all of the FLC-specific regulators analyzed, FRL1, FES1, SUF4, and FLX. The Y2H analysis using a series of deletion constructs showed that the N terminus of FRI binds to the N terminus of FRL1 and that FRI-CTR interacts with the N terminus of FLX and with the C termini of both SUF4 and FES1. SUF4 interacts with FRL1, and FLX interacts with FES1 (see Supplemental Figure 4 online). All components also form homodimers, but these are not shown.
[See online article for color version of this figure.]
The Y2H analysis performed using a deletion series of FRI and FRL1 showed that the N termini, including the coiled-coil domains, were required for the interaction between the two, and that the C terminus of FRI (FRI-D4, amino acids 459 to 609) was required for the interaction with FLX, SUF4, and FES1 (see Supplemental Figure 4 online). The C-terminal sequence of FRI (FRI-D4, amino acids 459 to 609) is unique and is extended compared with its paralogs, FRL1 and FRL2 (Michaels et al., 2004); this region is Pro rich and has 17 predicted phosphorylation sites (see Supplemental Figure 5 online), the characteristics of which likely provide the scaffolding activity to interact with various proteins, including CBP20 (Geraldo et al., 2009). The deletion series also showed that the N terminus of FLX (amino acids 35 to 78), the C terminus region that includes the Pro-rich region of SUF4, and the C terminus (at least amino acids 459 to 587) of FES1 were required for the interaction with the C terminus of FRI (see Supplemental Figure 4D online). By contrast, two C2H2 zinc finger motifs in SUF4 and the CCCH-type zinc finger motif in FES1 were dispensable for the interaction with FRI. In addition, it was shown that FRI could homodimerize through the C-terminal region, the FRIGIDA domain, and the coiled-coil motif and that FLX, SUF4, FES1, and FRL1 could also homodimerize (Figure 2A; see Supplemental Figure 4D online).
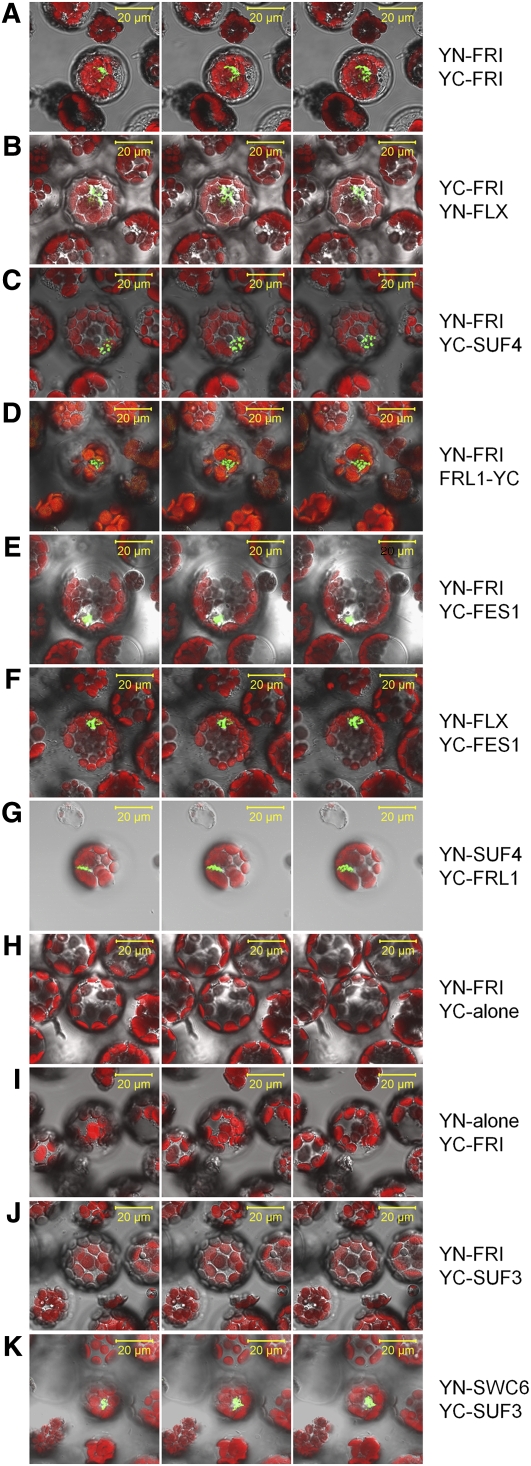
We confirmed the interactions observed in the Y2H assays by BiFC analysis using Arabidopsis protoplasts (Figure 3). The BiFC analysis using YN (amino acids 1 to 173 of yellow fluorescent protein [YFP])-FRI and YC (residues 174 through the end of the YFP sequence)-FRI fusion constructs showed a florescence pattern of nuclear speckles (Figure 3A), which was similar to the previously reported nuclear localization pattern of YFP-FRI (Kim et al., 2006). We observed similar nuclear speckle patterns from the interactions of FRI and FLX, SUF4, FRL1, and FES1 (Figures 3B to 3E). The nuclear fluorescence signals for the combination of FES1 and FRI were relatively weak and disappeared during the period of signal detection using confocal microscopy (Figure 3E). We also confirmed the interactions of SUF4/FRL1 and FLX/FES1 by BiFC because the Y2H data were ambiguous (Figures 3F and 3G). Indeed, the results showed nuclear speckles similar to those of the FRI homodimer, which provided further evidence supporting their interaction. When transfected alone, FLX localized to both the cytosol and the nucleus, as was previously reported (Andersson et al., 2008). However, the BiFC analysis showed that most of the FLX homodimers were present in the cytosol, but the signals of the FLX/FRI and FLX/FES1 heterodimers were strongly detected only in the nucleus (Figures 3B and 3F; see Supplemental Figure 6 online), suggesting that FRI-C recruits FLX to the nucleus. Their physical interactions and similar nuclear speckle patterns supported to the notion that FRI-C is composed (at least partially) of the FLC-specific regulators that we have analyzed.
Figure 3.
In Vivo Interactions among FRI, FRL1, FLX, SUF4, and FES1.
BiFC analyses were performed to confirm the interactions among FRI, FRL1, FLX, SUF4, and FES1. In the YN-/YC-interest or interest-YN/-YC fusion proteins, YN indicates the N-terminal region (amino acids 1 to 173), whereas YC indicates the C-terminal region (from residue 174 to the end of the polypeptide) of YFP. Each image was serially sectioned at intervals of 1 or 1.5 μm and is shown as merged panels of YFP, autofluorescence, and bright-field detections. Shown are the confocal images of BiFC to display the formation of FRI homodimers (A) and the interactions between FRI and FLX (B), FRI and SUF4 (C), FRI and FRL1 (D), FRI and FES1 (E), FLX and FES1 (F), and SUF4 and FRL1 (G). YN-FRI/YC-alone (H), YN-alone/YC-FRI (I), and YN-FRI/YN-SUF3 (J) are shown as negative controls. The previously shown physical interaction between SWC6 and SUF3 is shown as a positive control (K). Bars = 20 μm.
Each Component of the FRI Complex Has a Specialized Function
An obvious function of FRI-C is the transcriptional activation of FLC, such that it overrides the coordinated repression of FLC by autonomous pathway genes. However, it is not known how FRI-C can activate FLC transcription. Thus, we hypothesized that each component of the protein complex has a specialized function and designed experiments to test this hypothesis as below.
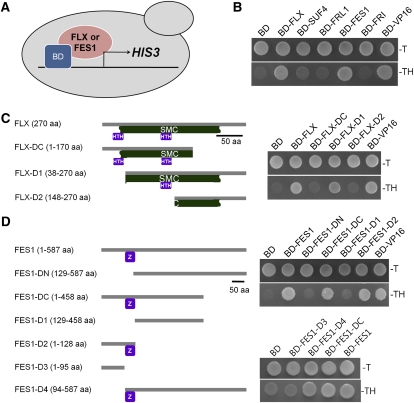
FLX and FES1 Have Transcriptional Activation Potential
Initial evidence supporting the idea of a specialized function was obtained from the Y2H assay of FLX and FES1. Both FLX and FES1 showed intrinsic transcriptional activity when fused with the DNA binding domain of GAL4 (Figure 2A). To confirm this finding, a yeast one-hybrid (Y1H) assay, using a deletion series of FLX and FES1, was performed (Figure 4). The results showed that a specific region was required for the intrinsic transcription activation of each protein in yeast: for FLX, a domain similar to SMC (structural maintenance of chromosomes) was required, whereas the CCCH-type zinc finger motif in FES1 was essential for transcription activation (Figures 4C and 4D). These observations indicated that FLX and FES1 have two separable domains: one for the interaction with FRI and the other for transcription activation. In addition, the observations suggested that FLX and FES1 play roles as transactivators or coactivators for FLC transcription, likely through a conserved mechanism in yeast. It is likely that the specific activation domains of FLX and FES1 may bind to components of general transcription factors and chromatin modifiers in yeast.
Figure 4.
Intrinsic Transcription Activity of FLX and FES1 in Yeast.
(A) A diagram of the Y1H system used to monitor the intrinsic transcription activity. Each protein was fused to the Gal4-DNA BD. When a yeast cell expresses proteins with intrinsic transcription activity, the yeast cell can grow in a medium without TH (Trp and His) by expressing the HIS3 reporter gene.
(B) Intrinsic transcription activity among FLC-specific regulators in yeast. Six independent colonies from each construct were tested, and representative samples are shown. The AD of VP16 (VP16) was fused to BD and used as a positive control. –T and –TH indicate the media lacking Trp only or both Trp and His, respectively.
(C) and (D) Y1H analysis showing that FLX and FES1 contain domains with intrinsic transcription activity. The deletion constructs of FLX (C) and FES1 (D) are shown in the middle of each panel, and the results of the one-hybrid analysis are shown to the right. The SMC region in FLX and the CCCH-type zinc finger region in FES1 were necessary for the intrinsic transcription activity. Boxes under the gray line representing the sequences show the following: SMC, SMC motif in FLX; HTH, HTH domain in FLX; and Z, CCCH-type zinc finger motif in FES1.
[See online article for color version of this figure.]
FES1 is a unique protein, but there are four additional FLX-like proteins in Arabidopsis (see Supplemental Figure 7 and Supplemental Data Set 1 online). The four proteins were also capable of binding to FRI in the Y2H assay, but they had very weak or no intrinsic transcription activity in yeast (see Supplemental Figures 7C and 7D online). Consistently, the T-DNA insertion mutants in three genes (FLX-LIKE1, 2, and 3) did not suppress FRI activity. (The segregation ratios of F2 populations from crosses between FRI-Col and flx like-1, 2, and 3 were approached to three late flowering to one early flowering [61:23, 64:20, 59:25; χ2 = 16.4, 21.7, and 13.4 respectively], indicating that FLX-LIKE genes do not affect flowering time.) Thus, it is very likely that only FLX, which demonstrated strong transcription activity, functions in the FRI-C to activate the transcription of FLC.
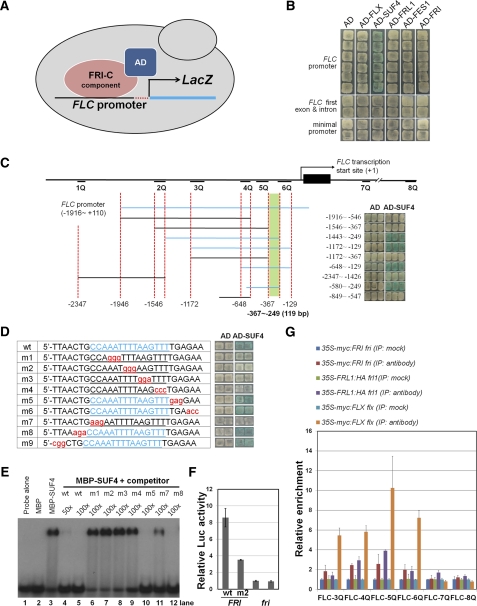
Direct Binding of SUF4 to the FLC Promoter
Next, we hypothesized that at least one of the FRI-C components has DNA binding specificity for the FLC promoter. Previously, chromatin immunoprecipitation (ChIP) analysis showed that SUF4 is enriched at the FLC promoter region; however, direct binding of SUF4 was not demonstrated (Kim et al., 2006). To address this, we performed a Y1H assay using the FLC promoter fused with the lacZ reporter gene (Figure 5). Among the FLC-specific regulators, only SUF4 showed binding to the FLC promoter region but not to the first exon and intron, which is the location of the cis-element for the vernalization response (Figure 5B; Sheldon et al., 2002). To determine the exact binding sequence for SUF4, we analyzed a deletion series of the FLC promoter using the Y1H assay. The results showed that SUF4 binds to a 33-bp sequence in the proximal region (−363 to ~−331 from the transcription start site) of the FLC promoter (Figures 5C; see Supplemental Figure 8A online). We then generated a series of point mutations within the sequence to determine the essential nucleotides for SUF4 binding (Figure 5D), which resulted in a minimal 15-bp sequence (5′-CCAAATTTTAAGTTT-3′). This result was confirmed by an electrophoretic mobility shift assay using a recombinant MBP-SUF4 fusion protein (Figure 5E). The MBP-SUF4 protein bound to the 15-bp sequence, and the wild-type competitor DNA eliminated the binding, whereas the mutant competitor DNA did not.
Figure 5.
SUF4 Binds to the FLC Promoter and the Elucidation of the cis-Element.
(A) The Y1H system used to determine DNA binding protein at the FLC promoter. Each protein was fused to the AD of B42 and used as the effector protein. When a protein with FLC promoter binding activity is expressed, the reporter gene, bacterial lacZ, is induced, and as a consequence, a blue color appears on the X-gal plate. The red dots just 5′ to the LacZ coding region indicate the minimal promoter from the cytochrome C1 gene fused with the reporter gene.
(B) Y1H analysis of FLX, SUF4, FRL1, FES1, and FRI using the FLC promoter. The FLC promoter (proFLC) construct contains the upstream promoter of FLC from −1916 to +110, before the ATG. Six independent colonies were tested for each FLC-specific regulator. The FLC first exon and intron have a DNA fragment from the translational start ATG (+111) to the first three base pairs of the second exon (+3794). Two independent colonies were tested for each FLC-specific regulator. Vectors containing a minimal promoter (two bottom rows) and the AD (effector) alone were used as negative controls,.
(C) Diagrams of the FLC promoter deletion series and Y1H analysis of SUF4. Blue lines indicate the promoter regions that provided blue colonies on Y1H plates. The green box indicates the minimal region required for SUF4 binding to the FLC promoter. The numbers are the distance from the transcription start site.
(D) A diagram of Y1H analysis to delineate the SUF4 binding sequences. The 28 bp of the DNA fragment containing wild-type or mutated sequences of the FLC promoter, as shown on the left, were fused with the minimal promoter and the lacZ reporter gene and tested for the AD-SUF4 protein activation of the reporters. The 15-bp sequence, 5′-CCAAATTTTAAGTTT-3′, was essential for SUF4 binding.
(E) Electrophoretic mobility shift assay using the MBP-SUF4 recombinant protein and the SUF4 binding elements. Oligoduplex sequences of wild-type probes, the wild type, or cold mutant competitors are shown. Recombinant proteins were mixed with labeled probes and competitors. The 50-fold (lane 4) and 100-fold (lanes 5 to 12) unlabeled wild type or mutant (m1, m2, m3, m4, m5, m7, and m8) oligoduplexes were used as competitors to confirm the specific interaction between SUF4 and the cis-element identified by Y1H analysis.
(F) The FRI complex activates the Luc reporter gene under the control of the wild-type FLC promoter with the first intron (wild type, −1916 to +3794) but not under the mutant promoter (m2) in Arabidopsis protoplasts. The plasmid DNA of two reporters, wild type and m2, was transformed into FRI-Col and Col (fri) protoplasts. Error bars indicate the sd of triplicate experiments. Luc, luciferase. The 35S-GUS gene was cotransformed and used as an internal control.
(G) ChIP–quantitative PCR analysis showing FRI, FRL1, and FLX enriched on the FLC promoter. Epitope-tagged transgenic plants of 35S-myc:FRI fri, 35S-myc:FLX flx, and 35S-FRL1:HA frl1 grown for 12 d under long-day conditions used for ChIP analysis. Mock indicates immunoprecipitation without the specific antibody for MYC or HA. FLC-3Q~8Q indicates primers occupying the region of the FLC promoter, as shown in (C). The TUB2 promoter region was used for the normalization of the qPCR. Error bars indicate the sd of triplicate experiments.
We confirmed the binding of SUF4 to the FLC promoter by an Arabidopsis protoplast transient assay (Figure 5F). For this assay, we introduced either the proFLC-LUC (FLC promoter fused to luciferase) or the proFLCm-LUC (using an FLC promoter with a mutation in the SUF4 binding sequence) construct into wild-type and fri mutant protoplasts. When the wild-type FLC promoter was used, the luciferase activity was approximately 3 times higher than that of the mutant promoter, indicating that this cis-element is necessary for full FLC activation. Furthermore, when the fri mutant plant was used for this assay, the luciferase activity was reduced dramatically, irrespective of the promoter, indicating both that SUF4 bound to the FLC promoter and that the intact FRI-C was required for the activation of FLC.
Because SUF4 is a component of the FRI-C and a DNA binding factor bound to the cis-element in the FLC promoter, we reasoned that all of the components of the FRI-C should bind to this same region of the promoter, either directly or indirectly. Indeed, the ChIP analysis using transgenic plants expressing epitope-tagged proteins driven by a 35S promoter showed that the FRI, FRL1, and FLX proteins were highly enriched near the SUF4 binding region (Figure 5G). When the endogenous promoter of SUF4 was used for the generation of the myc-tagged SUF4 transgenic lines, it also showed the enrichment at the same binding region at FLC-5Q (see Supplemental Figure 8B online). Therefore, our results clearly demonstrated that SUF4 is a sequence-specific DNA binding factor that recruited FRI-C to the FLC promoter.
The FRI Complex Recruits Chromatin Modification Factors
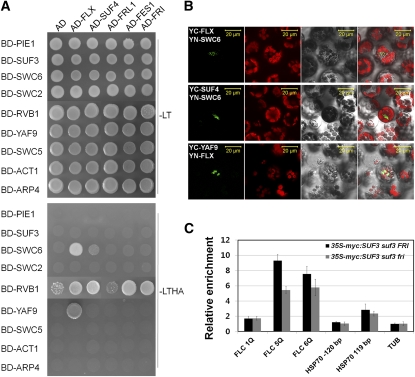
In general, high gene expression depends on physical interactions between sequence-specific activators and general transcription factors or chromatin modification factors. Therefore, we examined whether FRI-C interacts with the previously identified chromatin modification factors regulating FLC expression, such as the SWR1 complex that replaces the histone variant H2A.Z and the PAF1 complex that affects transcriptional elongation and histone methylation (He et al., 2004; Choi et al., 2007). None of the components of the PAF1 complex showed any interaction with FRI-C (data not shown). However, three components (SWC6, YAF9, and RVB1) of SWR1-C bound to components of the FRI-C (Figure 6A). SWC6 bound to both FLX and SUF4, YAF9 interacted with FLX alone, and RVB1 interacted with FLX, SUF4, FES1, and FRI in Y2H analysis (Figure 6A). The interactions of SWC6 and YAF9 with the FRI-C components were confirmed by BiFC analysis (Figure 6B), suggesting a direct link between FRI-C and SWR1-C.
Figure 6.
FRI-C Recruits SWR1-C to the FLC Promoter.
(A) Y2H analysis between components of FRI-C and SWR1-C.
(B) BiFC analysis used to confirm the interactions between FLX and SUF4, and SWC6 and YAF9. From left, fluorescence from YFP, autofluorescence, bright-field, and merged panel, respectively. Bars = 20 μm.
(C) ChIP–quantitative PCR analysis used to show that FRI-C recruits SWR1-C to the FLC promoter. The transgenic plants, 35S-myc:SUF3 suf3 fri and 35S-myc:SUF3 suf3 FRI, were used for ChIP analysis. Enrichment of the ChIP products was normalized based on results using TUB2 promoter primers. The location of 4Q~7Q primers is depicted in Figure 5C. The information of HSP70 primer sets is obtained from previous report (Kumar and Wigge, 2010). Error bars indicate the sd of triplicate experiments.
If FRI-C interacts with SWR1-C, the recruitment of SWR1-C to the FLC promoter may be reduced in the absence of FRI. To address this question, we performed a ChIP assay using the transgenic plants of suf3 fri and suf3 FRI expressing myc-tagged SUF3, which is a subunit of SWR1-C (Figure 6C). As expected, SUF3 was enriched approximately twofold at the proximal region of the FLC promoter when FRI was present, demonstrating that FRI-C enhanced the recruitment of SWR1-C to the FLC promoter. By contrast, SUF3 was not enriched at the promoter of HSP70 by the presence of FRI (Figure 6C), the transcription of which is induced by heat and affected by the activity of the SWR1-C (Kumar and Wigge, 2010). These data support the proposed specificity of FRI-C onto the FLC promoter.
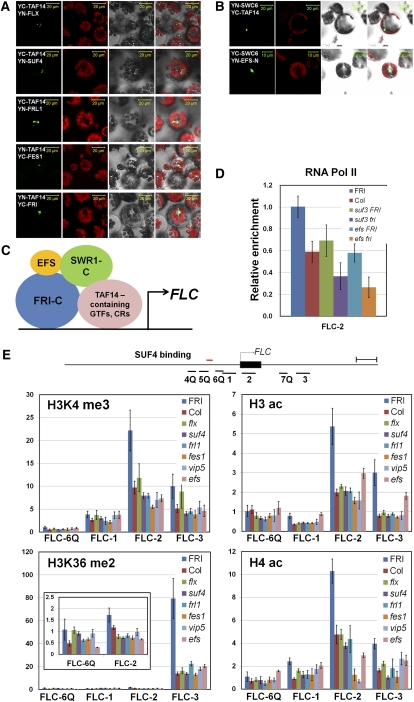
Interestingly, the BiFC analysis showed that SWC6, a subunit of SWR1-C, also binds to EFS-N and TAF14, as well as to FRI-C (Figures 7B). Recently it was reported that EFS interacts with FRI and catalyzes the trimethylation of both H3K4 and H3K36 of FLC chromatin, thus activating transcription (Ko et al., 2010). Therefore, our results suggest that SWC6 plays a critical role in coordinating SWR1-C, FRI-C, EFS, and general transcription factors (Figure 7C).
Figure 7.
Physical Interactions of FRI-C with TAF14 and EFS.
(A) and (B) BiFC analyses used to show that TAF14 interacts with all of the FRI-C components (A) and that SWC6 interacts with both TAF14 and the N terminus of EFS (B). From left, fluorescence from YFP, autofluorescence, bright-field, and merged panel, respectively. Bars = 20 μm, except bottom panels of (B), where they = 10 μm.
(C) A model of how FRI-C functions for the transcriptional activation of FLC through physical interactions with chromatin modification factors and general transcription factors, such as SWR1-C, EFS, and TAF14.
(D) The relative enrichment of RNA polymerase II at the first exon of FLC in FRI, fri, suf3, efs, suf3 fri, and efs fri. Ten-day-old plants were used for ChIP analysis using the anti-polII antibody. FLC-2 primers for amplifying the region of the first exon of FLC were used. ACTIN was used for the normalization of ChIP-qPCR. Error bars indicate the sd of triplicate experiments.
(E) ChIP and quantitative PCR analyses used to detect the active epigenetic marks of H3K4me3, H3ac, H4ac, and H3K36me2 in FRI-Col, fri, suf4, flx, frl1, fes1, vip5, and efs. The top part of the panel represents the FLC gene structure and the region used for the primers in the ChIP–quantitative PCR analyses. Ten-day-old seedlings were used. Ta3 was used for the normalization of the quantitative PCR analysis. Error bars indicate the sd of triplicate experiments.
The FRI Complex Recruits General Transcription Factors
YAF9, a component of SWR1-C, contains the YEATS (Yaf9/GAS41-ENL-AF9-Taf14-Sas5) domain that has been linked to chromatin modification and transcription (Schulze et al., 2009). The Arabidopsis genome has only two YEATS domain proteins, YAF9 and TAF14 (AT2G18000). TAF14 is a shared component of general transcription factors (TFIID and TFIIF), chromatin remodeling factors (INO80, SWI/SNF, and RSC), and histone H3 acetylation (NuA3) complexes in yeast and humans (Kabani et al., 2005). To determine whether TAF14 also interacts with FRI-C, both Y2H and BiFC analyses were performed (Figure 7; see Supplemental Figure 9 online). In the Y2H assay, interactions with TAF14 were detected with the FRI-C components except for FRL1; however, an interaction with FRL1 was detected in the BiFC analysis. The Y2H analysis, using a truncated version of TAF14 sequence revealed that the FRI-C components bind specifically to the C-terminal half of TAF14, whereas FRI requires the entire TAF14 sequence for interaction (see Supplemental Figure 9C online). It is noteworthy that the sequence of the C terminus of TAF14 is variable among species, in contrast with the conserved YEATS domain at the N terminus (see Supplemental Figures 9A and 9B and Supplemental Data Set 1 online). When single mutants of TAF14 and YAF9 were examined, they showed the same phenotype as the wild type. However, the double mutant showed a pleiotropic phenotype that included small size and abnormal leaf morphology, suggesting that the two genes have functional redundancy (see Supplemental Figure 10 online).
Last, we determined whether FRI-C recruits RNA polymerase II (polII), together with SWR1C and EFS, to the FLC transcription initiation site (Figure 7B). Indeed, RNA polII was enriched approximately twofold higher than the control at the first exon of the FLC when FRI was present. As expected, a mutation in SUF3 or EFS reduced the RNA polII enrichment at the first exon. Moreover, the double mutants, suf3 fri and efs fri, showed an additive effect for RNA polII enrichment compared with the single mutants. These data suggest that SWR1-C, FRI-C, and EFS have partially independent functions in recruiting RNA polII onto the FLC promoter. Taken together, our results indicate that FRI-C mediates FLC transcription through interactions with chromatin modification factors, SWR1-C and EFS, as well as the general transcription factor TAF14, at the proximal region of the FLC promoter (Figure 7D).
The FRI Complex Leads to the Active Chromatin State of the FLC Gene
To define further the function of FRI-C at the molecular level, we monitored the chromatin state of the FLC gene in the mutants for FRI-C components. The epigenetic modifications of H3K4 trimethylation (H3K4me3), H3K36me2, H3 acetylation (H3ac), and H4ac were assessed across the promoter and the first exon and intron of FLC. Typical landscapes of eukaryotic epigenetic marks for active transcription were observed in the wild type: that is, the active epigenetic marks of H3K4me3, H3ac, and H4ac were enriched highly at the 5′ region (the FLC-2 region), and the H3K36me2 level was highest at the first intron region (Figure 7E). By contrast, when mutated in the components of FRI-C, the active epigenetic marks were severely reduced at the 5′ region or the first intron, a result that may reflect the decreased transcription state of FLC in the mutants (Figure 7E). A similar reduction was observed in efs and vip5, a mutant of a PAF1-C component, indicating that the functions of chromatin modification factors and FRI-C are tightly correlated. Taken together, our results support the hypothesis that FRI-C recruits chromatin modification factors to the FLC promoter, resulting in an active chromatin state.
DISCUSSION
FRI is a major determinant of the natural variations in flowering time and vernalization responses observed among Arabidopsis ecotypes. In this report, we show that FRI forms a large protein complex that we call FRI-C, the components of which have specialized functions, such as DNA binding, transcription activation, and maintenance of the complex. In addition, we show the interactions of FRI-C with a diverse range of chromatin modification factors and general transcription factors, which appear to result in the active chromatin state of the FLC gene. Therefore, our results clearly demonstrate that FRI-C acts as a transcription activator complex for FLC expression.
FRI Activates FLC Transcription by Forming a Large Protein Complex
The mutants of the five FLC-specific regulators, fri, frl1, fes1, suf4, and flx, all showed the same phenotype, indicating that they may be components of the same protein complex. As expected, FRI was shown to act as a scaffold protein that assembles a large protein complex with these five regulators. Furthermore, the biochemical and functional analyses of the FRI-C components allowed us to propose a model for the FRI-C–mediated transcriptional activation of FLC, as summarized in Figure 7C. As depicted, SUF4 directly binds to the cis-element (5′-CCAAATTTTAAGTTT-3′) located on the proximal promoter of FLC and forms a transcription complex (FRI-C) with the other FLC-specific regulators. The FRI-C then recruits chromatin modification factors and general transcription factors, such as SWR1-C, EFS, and TAF14. This recruitment promotes the transcription from the basal to the active state and subsequently mediates transcription elongation via the interaction with EFS at the transcribed region. There are many examples of specific transcription factors working in a protein complex with separate DNA binding and transcription activation components (Kidd et al., 2005; Nam et al., 2006; Wilson and Kovall, 2006). However, how such complexes activate transcription is not well known. Our study showing that a plant-specific transcription complex (FRI-C) recruits chromatin modification factors and general transcription factors provides unique insight into the mechanism of this complicated eukaryotic transcription process.
Recruitment of Chromatin Modification Factors and General Transcription Factors
FRI-C interacts with the eukaryotically conserved chromatin modification factors SWR1-C and EFS (Ko et al., 2010). The function of the incorporation of H2A.Z at transcriptional start sites, which is catalyzed by SWR1-C, is highly controversial: it is involved in both gene activation and inactivation (Zlatanova and Thakar, 2008). In Arabidopsis, it has also been implicated in both positive and negative transcriptional regulation. Mutations in the components of SWR1-C cause the derepression of biotic and abiotic plant response genes, such as plant disease resistance genes, heat shock–induced genes, and phosphate starvation response genes, supporting the negative role of H2A.Z (March-Díaz et al., 2008; Kumar and Wigge, 2010; Smith et al., 2010). By contrast, the mutations cause the suppression of the developmentally regulated FLC gene, supporting the positive role of H2A.Z (Martin-Trillo et al., 2006; Choi et al., 2007; Deal et al., 2007; March-Díaz et al., 2007).
The contradictory role of SWR1-C/H2A.Z may be due to changes in the stability of H2A.Z-containing nucleosome particles by the acetylation of H2A.Z and the coincorporation of another histone variant, H3.3, as has been reported (Millar et al., 2006; Jin and Felsenfeld, 2007; Jin et al., 2009). The H2A.Z-containing nucleosomes exhibit a tighter wrapping of their DNA and thus are likely to maintain the inactive state of the genes by preventing the access of RNA polII or specific transcription factors (Thambirajah et al., 2006; Kumar and Wigge, 2010). However, when acetylated or coincorporated with H3.3, the H2A.Z-containing nucleosomes become labile and are evacuated from the promoter region for the onset of active transcription (Millar et al., 2006; Jin et al., 2009). The specific loading of FRI-C onto the proximal promoter of FLC may enhance the replacement of H2A.Z at the transcription start site by a direct interaction with SWR1-C. Thereafter, H2A.Z acetylation and the incorporation of H3.3 may occur at the FLC promoter, resulting in the release of the histone octamer and activation of transcription.
Such a model can explain why the H2A.Z level at the FLC chromatin is negatively correlated with the transcript level of FLC such that the steady state level of H2A.Z is less in FRI than in Col (a fri mutant) (Deal et al., 2007). Recruitment of SWR1-C seems to be specific to the FLC promoter because FRI-C does not affect the enrichment of ARP6 on the HSP70 promoter (Figure 6C), which is one of the targets of H2A.Z incorporation (Kumar and Wigge, 2010). In addition, FRI-C interacts with AtYAF9 and AtTAF14, which are also a shared component of the NuA4 and NuA3 complexes involved in histone acetylation (Lu et al., 2009; Schulze et al., 2009). Thus, FRI-C likely orchestrates the dynamic process in which H2A.Z is reassembled and evacuated for the active transcription of the FLC gene. A similar mechanism has been observed in estrogen receptor signaling in humans, in which H2A.Z incorporation is enhanced by the direct interaction between p400, a component of human SWR1-C, and the estrogen receptor for targeted gene induction (Gévry et al., 2009). Thus, the recruitment of SWR1-C by specific transcription factors seems to be common. It is noteworthy that SWR1-C can also be recruited to the FLC gene independent of FRI because FLC transcription in the fri background is suppressed by the suf3 mutation (Choi et al., 2005). Therefore, FRI-C is not absolutely required, yet it enhances the recruitment of SWR1-C to the FLC promoter.
The interaction of FRI-C with EFS has recently been reported, and EFS has a dual catalytic activity for the trimethylation of both H3K4 and H3K36, which are active epigenetic marks for transcription initiation and elongation (He et al., 2004; Sims et al., 2004; Ko et al., 2010). In agreement, there is a report showing that Set2, an EFS homolog, and H3K36 methylation affect the recruitment of RNA polII at the promoter as well (Krogan et al., 2003b). Therefore, likely through EFS, FRI-C may enhance both the initiation and the elongation steps of transcription. Prerecruitment of EFS to the promoter of FLC by FRI-C may help the dynamic transcription progress from preinitiation to initiation and to productive elongation for FLC transcription. Together with the fact that FRI-C also interacts with general transcription factors and the mRNA cap binding protein, CBP20 (Geraldo et al., 2009), our results support the concept of a unified model of gene expression in which each step regulating gene expression is physically and functionally connected to the next (Orphanides and Reinberg, 2002).
Conservation and Specification of the FRI Complex
Molecular genetic analyses in Arabidopsis have revealed two flowering pathways that respond to environmental signals: the photoperiod and vernalization pathways. Although the CO-FT regulatory module that perceives daylength in the photoperiod pathway seems to be well conserved among both monocots and dicots (Izawa et al., 2003; Hayama and Coupland, 2004; Böhlenius et al., 2006), the FRI-FLC regulatory module in the vernalization pathway seems not to be conserved in monocots; FLC-like genes have not been identified in monocots, and vernalization in wheat (Triticum aestivum) suppresses the expression of a flowering repressor that is distinct from FLC, Ta VRN2, which encodes a B box zinc finger protein (Goff et al., 2002; Yan et al., 2004). In addition, Ta VRN2 expression is not suppressed by vernalization under short days, a normal photoperiod during the winter season, suggesting that the molecular mechanism of vernalization in monocots is quite dissimilar with that in Arabidopsis (Trevaskis et al., 2007). Interestingly, we could detect the homologs of the components of FRI-C by database searching in both monocots and dicots (see Supplemental Figure 11 and Supplemental Data Set 3 online). However, the amino acid sequence identities, compared with rice (Oryza sativa) homologs, were as low as 25% with the exception of SUF4. In addition, the C terminus (amino acids 459 to 609) of FRI, where the majority of the FRI-C components interact, is very unique: no homologous sequence was detected outside the Brassicaceae family. Therefore, it is likely that the C terminus of FRI has evolved recently and provides a function as a scaffold for FRI-C in this Brassicaceae family. It is tempting to propose that FRI underwent neofunctionalization for generating FRI-C to regulate the flowering repressor FLC. Recently, the FLC homolog in sugar beet (Beta vulgaris), which is outside the Brassicaceae family, has been reported to play a critical role in the vernalization pathway (Reeves et al., 2007). It may be interesting to ascertain whether the FRI-FLC regulatory module is also conserved in the plant family that includes the sugar beet.
By contrast, SUF4 is relatively highly conserved in both sequence (60% identity) and biological function: the overexpression of the rice SUF4 homolog (EEE70223) successfully rescued the suf4 mutant (see Supplemental Figures 12 and 13 online). Although the obvious function of all of the components of FRI-C is to regulate FLC, the higher conservation of SUF4 may indicate that it has an additional function regulating the transcription of other genes in a subtle way. Indeed, when double mutants for the components of FRI-C were generated in Arabidopsis, the suf4 mutation caused an increase in the floral organ number (in suf4 flx and suf4 fes1) and the retardation of root growth (in suf4 fes1). Such an additional function may constrain the evolutionary change of SUF4.
The genetic variation in flowering time among Arabidopsis ecotypes is mainly caused by aberrations in FRI and FLC, although a mutation in any of the components of FRI-C causes a similar flowering phenotype (Gazzani et al., 2003; Michaels et al., 2003; Lempe et al., 2005; Werner et al., 2005). This suggests that evolutionary constraints may exist in the other components, such that they have other functions in addition to the regulation of FLC. The phenotype regulated by these additional functions may be expressed in natural environments, which then leads to different outcomes of genotypes compared with standard laboratory conditions (Wilczek et al., 2009).
METHODS
Plant Materials and Growth Conditions
The wild type, Col:FRISF2 (FRI-Col), and all the flowering time mutants used in this study have been previously described (Michaels et al., 2004; Choi et al., 2005; Schmitz et al., 2005; Kim et al., 2006). Seed for SALK insertion lines (see accession numbers section) were obtained from the ABRC. Plants were grown and the flowering time was measured by counting the number of rosette leaves from at least 10 plants.
Plasmid Construction
The sequence information of the primer pairs used for PCR amplification is presented in Supplemental Table 2 online. To generate the constructs for the plant transformation, the full open reading frames (ORFs) of FLX, FRI, and FRL1 were cloned into pCGN18, myc-pBA, or the pGWB14 binary gateway vector.
For the Y2H analysis, the ORFs of FLX, FES1, and other FLC regulators were amplified, cloned into the pGEM-T Easy vector obtained from Promega, sequenced, and finally subcloned into the proper restriction sites of the pGBKT7 and pGADT7 vectors obtained from Clontech (Matchmaker GAL4 Yeast Two-Hybrid System 3). To generate the truncated constructs, proper restriction sites within the genes and vectors were used, or amplified PCR fragments including the truncated region were cloned into the Y2H vectors.
For the reporter constructions used in the Y1H analysis, various FLC promoter fragments, the first intron of FLC, and 28-bp oligomer duplexes were cloned into the SmaI site of the pLacZi2μ vector (kindly provided by H. Wang). For the effector constructions, the ORFs of FRI, FRL1 (SmaI and BamHI), FES1 (BglII), SUF4, and FLX (BamHI), within the pGBKT7 vectors, were removed by restriction enzyme digestion, and the overhanging ends were filled in using DNA polymerase Klenow fragment. The inserts were cloned into the EcoRI site of the pJG4-5 vector, whose ends had been blunted using Klenow.
To produce the constructs for the BiFC analysis or the cyan fluorescent protein (CFP) and YFP localization analyses using the protoplast transient assay, the ORFs or the ORFs without the stop codon of FRI, FRL1, FES1, SUF4, and FLX were cloned into the BamHI site of E3081/pSAT4-nEYFP-C1, E3082/pSAT4-cEYFP-C1, E3083/pSAT4-nEYFP-N1, E3084/pSAT4-cEYFP-N1 (http://www.bio.purdue.edu/people/faculty/gelvim/nsf/index.htm), the YFP vector (Kim et al., 2006), or the CFP vector, which was modified from the YFP vector by exchanging the ORF of YFP (NcoI-BamHI) with the ORF of CFP.
Y2H Analysis
The vectors and yeast strains (Matchmaker GAL4 Two-Hybrid System 3) were obtained from Clontech. The Y2H assay was performed according to the manufacturer's instructions. After 3 d of incubation at 30°C, yeast cells were spotted on the selection plates containing SD medium lacking Leu, Trp, Ade, and His. These plates were further incubated at 22 or 30°C until the yeast cells formed colonies.
Y1H Assay
To detect intrinsic transcriptional activity, plasmids with GAL4 DNA binding domain fusions were transformed into the yeast strain AH109 (Matchmaker GAL4 Yeast Two-Hybrid System 3 from Clontech) using standard transformation techniques. To find the DNA binding proteins, plasmids for the AD fusions were cotransformed into the yeast strain EGY48, with the LacZ reporter gene driven by various FLC promoter fragments.
Protoplast Transient Assay
Rosette leaves of Col plants that had been grown for 4 weeks on short days (8 h light/16 h dark) were sampled for the isolation and transformation of protoplasts as described (Yoo et al., 2007). All of the plasmid DNA for the protoplast transformation was prepared by the CsCl gradient method. After ~12 h of transformation, the protoplasts were observed as previously described (Choi et al., 2005) or used for luciferase activity assays and ChIP analysis.
Gel Filtration Assays
Two hundred micrograms of total protein extracts from 35S-myc:FRI fri transgenic plants grown for 10 d was injected on a Superdex 200 10/300 GL column (Amersham Biosciences). After fractionation by the AKTA fast protein liquid chromatography system (Amersham Biosciences), 24 tubes containing 0.5 mL were collected. Proteins in each fraction were concentrated using 10 μL of Strataresin (Stratagene), electrophoresed, blotted, and identified using monoclonal anti-c-myc antibodies (Sigma-Aldrich M5546).
Purification of the Components of FRI-C
Purification of the FRI complexes was performed following a protocol previously described (Cho et al., 2006). Two-week-old plants of 35S:myc-FRI (50 g fresh weight), which fully rescues the fri mutant, were used. As a negative control, the fri mutant was used while following the same procedure. Nuclear extracts were obtained as previously described (Lee et al., 2007), incubated with 300 μL of anti-c-myc affinity agarose in TBS (50 mM Tri-HCl, pH 7.4, and 150 mM NaCl) overnight, and then washed and eluted with 100 μL sample buffer. The eluates were analyzed by LC-MS/MS (ProteomeX LTQ 2D/MS/MS mass spectrometer). The analysis was processed according to the manufacturer’s procedures. Full details of the data on the matching peptides and scores from the purification of the protein complexes and control samples are included in Supplemental Table 1 online.
Electrophoretic Mobility Shift Assays
The MBP and MBP-SUF4 recombinant fusion proteins were expressed in Escherichia coli BL21 strain and purified using amylose resin. The single-stranded oligonucleotides were labeled with [α-32P]ddATP using terminal transferase (Roche 03333566001) and then annealed. One hundred nanograms of protein and ~40,000 cpm of 32P-labeled probe were incubated in 20 μL of binding mixtures, including 10 mM Tris-HCl, pH 7.5, 100 mM KCl, 1 mM EDTA, 0.1 μg/μL BSA, 100 μM ZnCl2, 6% glycerol, and 1 mM DTT at 4°C for 1 h and separated on a 6% polyacrylamide gel in Tris-acetate-EDTA buffer (40 mM Tris and 2.5 mM EDTA, pH 7.8) at 10 V/m for 2 h. For the competition assay, excess unlabeled probes (50× and 100×) were added to the binding mixture.
ChIP Analysis
All ChIP procedures were followed according to a previous report (Choi et al., 2007). Whole plants grown for 10 d under long-day conditions and antibodies against acetyl H4K5/8/12/16 (Upstate 06-866), acetyl H3K9/14 (Upstate 06-599), trimethyl H3K4 (Upstate 17-614), dimethyl H3K36 (Upstate 07-274), and the RNA polymerase II C-terminal domain (Abcam ab817) were used for immunoprecipitation. To determine the enrichment in the epitope-tagged transgenic plants, monoclonal anti-c-myc (Sigma-Aldrich M5546) and monoclonal anti-HA (Sigma-Aldrich H9658) were used. Information on the primer pairs for the ChIP–quantitative PCR is presented in Supplemental Table 2 online.
Phylogenetic Analysis
Multiple alignments of amino acid sequences were performed using the ClustalX2.1 program (http://www.clustal.org/download/current/). For phylogenetic analysis, the PHYLIP program (version 3.69) was used (http://evolution.genetics.washington.edu/phylip.html). In the PHYLIP software, SEQBOOT, PROTDIST, NEIGHBOR, and CONSENSE programs were used to draw phylogenetic trees (unrooted) and obtain bootstrap values. For the phylogenetic tree, JTT model, neighbor joining, and bootstraps with 1000 trial options were used. The phylogenetic tree was drawn with the TreeView program (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html).
Quantitative PCR
For real-time quantitative PCR, total RNA was isolated using the RNeasy plant mini kit (Sigma-Aldrich 74904). Four micrograms of total RNA was treated with recombinant DNaseI (TaKaRa 2270A) to eliminate genomic DNA. cDNA was generated using RNA with reverse transcriptase (Fermentas EP0441) and oligo(dT). Quantitative PCR was performed using the 2× SYBR Green SuperMix (Bio-Rad 170-8882) and monitored by the CFX96 real-time PCR detection system (Kim et al., 2006). The relative transcript level and the ChIP enrichment level were normalized with actin7 and tubulin2 genes according to the 2-ΔΔCt method (Livak and Schmittgen, 2001).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL database or the Arabidopsis Genome Initiative database under the following accession numbers: ACT1 (AT2G37620), ARP4 (AT1G18450), ARP6 (AT3G33520), EFS (AT1G77300), FES1 (AT2G33835), FLX (AT2G30120), FRI (AT4G00650), FRL1 (AT5G16320), HSP70 (AT3G12580), PIE1 (AT3G12810), SUF4 (AT1G30970), SWC2 (AT2G36740), SWC4 (AT2G47210), SWC5 (AT5G30490), SWC6 (AT5G37055), RVB1 (AT5G22330), TAF14 (AT2G18000), TUB (AT5G62690), VP16 (U8963), and YAF9 (AT5G45600). SALK insertion lines from ABRC are as follows: SALK_002678 (flx like 1), SAIL_535_H09 (flx like 2), SAIL_156_B10 (flx like 3), and SALK_075203 (yaf9).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Grouping of suf Mutants and FLC Expression .
Supplemental Figure 2. Characterization of flx-2 and the expression patterns of FLX.
Supplemental Figure 3. Gel Filtration Analysis of myc:FRI Proteins Expressed in the fes1, frl1, suf4, and flx Mutant Backgrounds .
Supplemental Figure 4. Yeast Two-Hybrid Interaction Analyses among the Deletion Forms of FRI, FRL1, FLX, SUF4, and FES1 .
Supplemental Figure 5. Characteristics of the C-Terminal 150 Amino Acids of FRI .
Supplemental Figure 6. Localization of FLX, FRL1, and FES1 .
Supplemental Figure 7. FLX-Like Genes in Arabidopsis thaliana.
Supplemental Figure 8. SUF4 Binding to the FLC Promoter in Vitro and in Vivo .
Supplemental Figure 9. YEATS-Containing Proteins in Different Organisms .
Supplemental Figure 10. Phenotype of atyaf9 and attaf14-kd Plants .
Supplemental Figure 11. Phylogenetic Trees of FRI-C Components .
Supplemental Figure 12. Alignment of SUF4 Homologs from Different Plant Species .
Supplemental Figure 13. The Overexpression of OsSUF4 Rescued the suf4 Mutant Phenotype .
Supplemental Table 1. LC-MS/MS Results for the myc-FRI IP and the Control Sample .
Supplemental Table 2. Primers for Plasmid Construction and PCR .
Supplemental Data Set 1. Text File of Sequences Used for the Phylogenetic Analysis in Supplemental Figure 7A.
Supplemental Data Set 2. Text File of Sequences Used for the Phylogenetic Analysis in Supplemental Figure 9A.
Supplemental Data Set 3. Text File of Sequences Used for the Phylogenetic Analysis in Supplemental Figure 11.
Supplementary Material
Acknowledgments
We thank the ABRC for SALK_002678 (flx-like1), SAIL_535_H09 (flx-like2), SAIL_156_B10 (flx-like3), and SALK_075203 (yaf9); S. Michaels for providing the frl1-1 and fes1-2 seeds; H. Wang for the pLacZi2μ vector; N. Baek for the BiFC vectors; Y. Noh for FLAG vector; and H. Park for the pJG4-5 vector and the yeast EGY48 strain. This work was supported partially by the Korea Ministry of Science and Technology under the National Research Laboratory Program (2006-01952) and the Global Research Laboratory Program (2006-03870) and by a grant (Code 20070301034011) from the BioGreen 21 Program, Rural Development Administration. J.K. and H.-J.H. were supported by the Brain Korea 21 Program.
References
- Andersson C.R., Helliwell C.A., Bagnall D.J., Hughes T.P., Finnegan E.J., Peacock W.J., Dennis E.S. (2008). The FLX gene of Arabidopsis is required for FRI-dependent activation of FLC expression. Plant Cell Physiol. 49: 191–200 [DOI] [PubMed] [Google Scholar]
- Bezerra I.C., Michaels S.D., Schomburg F.M., Amasino R.M. (2004). Lesions in the mRNA cap-binding gene ABA HYPERSENSITIVE 1 suppress FRIGIDA-mediated delayed flowering in Arabidopsis. Plant J. 40: 112–119 [DOI] [PubMed] [Google Scholar]
- Böhlenius H., Huang T., Charbonnel-Campaa L., Brunner A.M., Jansson S., Strauss S.H., Nilsson O. (2006). CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312: 1040–1043 [DOI] [PubMed] [Google Scholar]
- Cho Y.H., Yoo S.D., Sheen J. (2006). Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell 127: 579–589 [DOI] [PubMed] [Google Scholar]
- Choi K., Kim S., Kim S.Y., Kim M., Hyun Y., Lee H., Choe S., Kim S.G., Michaels S., Lee I. (2005). SUPPRESSOR OF FRIGIDA3 encodes a nuclear ACTIN-RELATED PROTEIN6 required for floral repression in Arabidopsis. Plant Cell 17: 2647–2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K., Park C., Lee J., Oh M., Noh B., Lee I. (2007). Arabidopsis homologs of components of the SWR1 complex regulate flowering and plant development. Development 134: 1931–1941 [DOI] [PubMed] [Google Scholar]
- Clarke J.H., Dean C. (1994). Mapping FRI, a locus controlling flowering time and vernalization response in Arabidopsis thaliana. Mol. Gen. Genet. 242: 81–89 [DOI] [PubMed] [Google Scholar]
- Deal R.B., Topp C.N., McKinney E.C., Meagher R.B. (2007). Repression of flowering in Arabidopsis requires activation of FLOWERING LOCUS C expression by the histone variant H2A.Z. Plant Cell 19: 74–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle M.R., Bizzell C.M., Keller M.R., Michaels S.D., Song J.D., Noh Y.S., Amasino R.M. (2005). HUA2 is required for the expression of floral repressors in Arabidopsis thaliana. Plant J. 41: 376–385 [DOI] [PubMed] [Google Scholar]
- Gazzani S., Gendall A.R., Lister C., Dean C. (2003). Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiol. 132: 1107–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldo N., Bäurle I., Kidou S., Hu X.Y., Dean C. (2009). FRIGIDA delays flowering in Arabidopsis via a cotranscriptional mechanism involving direct interaction with the nuclear cap-binding complex. Plant Physiol. 150: 1611–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gévry N., Hardy S., Jacques P.E., Laflamme L., Svotelis A., Robert F., Gaudreau L. (2009). Histone H2A.Z is essential for estrogen receptor signaling. Genes Dev. 23: 1522–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff S.A., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92–100 [DOI] [PubMed] [Google Scholar]
- Hayama R., Coupland G. (2004). The molecular basis of diversity in the photoperiodic flowering responses of Arabidopsis and rice. Plant Physiol. 135: 677–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y.H., Doyle M.R., Amasino R.M. (2004). PAF1-complex-mediated histone methylation of FLOWERING LOCUS C chromatin is required for the vernalization-responsive, winter-annual habit in Arabidopsis. Genes Dev. 18: 2774–2784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa T., Takahashi Y., Yano M. (2003). Comparative biology comes into bloom: Genomic and genetic comparison of flowering pathways in rice and Arabidopsis. Curr. Opin. Plant Biol. 6: 113–120 [DOI] [PubMed] [Google Scholar]
- Jin C.Y., Felsenfeld G. (2007). Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 21: 1519–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C.Y., Zang C.Z., Wei G., Cui K.R., Peng W.Q., Zhao K.J., Felsenfeld G. (2009). H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat. Genet. 41: 941–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson U., West J., Lister C., Michaels S., Amasino R., Dean C. (2000). Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290: 344–347 [DOI] [PubMed] [Google Scholar]
- Kabani M., Michot K., Boschiero C., Werner M. (2005). Anc1 interacts with the catalytic subunits of the general transcription factors TFIID and TFIIF, the chromatin remodeling complexes RSC and INO80, and the histone acetyltransferase complex NuA3. Biochem. Biophys. Res. Commun. 332: 398–403 [DOI] [PubMed] [Google Scholar]
- Kidd A.R., III, Miskowski J.A., Siegfried K.R., Sawa H., Kimble J. (2005). A beta-catenin identified by functional rather than sequence criteria and its role in Wnt/MAPK signaling. Cell 121: 761–772 [DOI] [PubMed] [Google Scholar]
- Kim D.H., Doyle M.R., Sung S., Amasino R.M. (2009). Vernalization: Winter and the timing of flowering in plants. Annu. Rev. Cell Dev. Biol. 25: 277–299 [DOI] [PubMed] [Google Scholar]
- Kim S., Choi K., Park C., Hwang H.J., Lee I. (2006). SUPPRESSOR OF FRIGIDA4, encoding a C2H2-Type zinc finger protein, represses flowering by transcriptional activation of Arabidopsis FLOWERING LOCUS C. Plant Cell 18: 2985–2998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J.H., Mitina I., Tamada Y., Hyun Y., Choi Y., Amasino R.M., Noh B., Noh Y.S. (2010). Growth habit determination by the balance of histone methylation activities in Arabidopsis. EMBO J. 29: 3208–3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan N.J., et al. (2003b). Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 23: 4207–4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S.V., Wigge P.A. (2010). H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140: 136–147 [DOI] [PubMed] [Google Scholar]
- Lee I., Michaels S.D., Masshardt A.S., Amasino R.M. (1994). The late-flowering phenotype of FRIGIDA and mutations in LUMINIDEPENDENS is suppressed in the landsberg erecta strain of Arabidopsis. Plant J. 6: 903–909 [Google Scholar]
- Lee J., He K., Stolc V., Lee H., Figueroa P., Gao Y., Tongprasit W., Zhao H.Y., Lee I., Deng X.W. (2007). Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lempe J., Balasubramanian S., Sureshkumar S., Singh A., Schmid M., Weigel D. (2005). Diversity of flowering responses in wild Arabidopsis thaliana strains. PLoS Genet. 1: 109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Carey M., Workman J.L. (2007). The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lu P.Y.T., Lévesque N., Kobor M.S. (2009). NuA4 and SWR1-C: Two chromatin-modifying complexes with overlapping functions and components. Biochem. Cell Biol. 87: 799–815 [DOI] [PubMed] [Google Scholar]
- March-Díaz R., García-Domínguez M., Florencio F.J., Reyes J.C. (2007). SEF, a new protein required for flowering repression in Arabidopsis, interacts with PIE1 and ARP6. Plant Physiol. 143: 893–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- March-Díaz R., García-Domínguez M., Lozano-Juste J., León J., Florencio F.J., Reyes J.C. (2008). Histone H2A.Z and homologues of components of the SWR1 complex are required to control immunity in Arabidopsis. Plant J. 53: 475–487 [DOI] [PubMed] [Google Scholar]
- Martin-Trillo M., Lázaro A., Poethig R.S., Gómez-Mena C., Piñeiro M.A., Martinez-Zapater J.M., Jarillo J.A. (2006). EARLY IN SHORT DAYS 1 (ESD1) encodes ACTIN-RELATED PROTEIN 6 (AtARP6), a putative component of chromatin remodelling complexes that positively regulates FLC accumulation in Arabidopsis. Development 133: 1241–1252 [DOI] [PubMed] [Google Scholar]
- Michaels S.D., Amasino R.M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S.D., Amasino R.M. (2001). Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 13: 935–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S.D., Bezerra I.C., Amasino R.M. (2004). FRIGIDA-related genes are required for the winter-annual habit in Arabidopsis. Proc. Natl. Acad. Sci. USA 101: 3281–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S.D., He Y.H., Scortecci K.C., Amasino R.M. (2003). Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis. Proc. Natl. Acad. Sci. USA 100: 10102–10107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar C.B., Xu F., Zhang K.L., Grunstein M. (2006). Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes Dev. 20: 711–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam Y., Sliz P., Song L.Y., Aster J.C., Blacklow S.C. (2006). Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell 124: 973–983 [DOI] [PubMed] [Google Scholar]
- Napp-Zinn K. (1979). On the genetical basis of vernalization requirement in Arabidopsis thaliana (L.) Heynh. (Paris: La Physiologie de la Floraison, Colloques Internationaux. Centre National de la Recherche Scientifique) [Google Scholar]
- Noh Y.S., Amasino R.M. (2003). PIE1, an ISWI family gene, is required for FLC activation and floral repression in Arabidopsis. Plant Cell 15: 1671–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G., Reinberg D. (2002). A unified theory of gene expression. Cell 108: 439–451 [DOI] [PubMed] [Google Scholar]
- Reeves P.A., He Y.H., Schmitz R.J., Amasino R.M., Panella L.W., Richards C.M. (2007). Evolutionary conservation of the FLOWERING LOCUS C-mediated vernalization response: Evidence from the sugar beet (Beta vulgaris). Genetics 176: 295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz R.J., Hong L., Michaels S., Amasino R.M. (2005). FRIGIDA-ESSENTIAL 1 interacts genetically with FRIGIDA and FRIGIDA-LIKE 1 to promote the winter-annual habit of Arabidopsis thaliana. Development 132: 5471–5478 [DOI] [PubMed] [Google Scholar]
- Schulze J.M., Wang A.Y., Kobor M.S. (2009). YEATS domain proteins: A diverse family with many links to chromatin modification and transcription. Biochem. Cell Biol. 87: 65–75 [DOI] [PubMed] [Google Scholar]
- Sheldon C.C., Burn J.E., Perez P.P., Metzger J., Edwards J.A., Peacock W.J., Dennis E.S. (1999). The FLF MADS box gene: A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11: 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon C.C., Conn A.B., Dennis E.S., Peacock W.J. (2002). Different regulatory regions are required for the vernalization-induced repression of FLOWERING LOCUS C and for the epigenetic maintenance of repression. Plant Cell 14: 2527–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson G.G. (2004). The autonomous pathway: Epigenetic and post-transcriptional gene regulation in the control of Arabidopsis flowering time. Curr. Opin. Plant Biol. 7: 570–574 [DOI] [PubMed] [Google Scholar]
- Simpson G.G., Dean C. (2002). Arabidopsis, the Rosetta stone of flowering time? Science 296: 285–289 [DOI] [PubMed] [Google Scholar]
- Smith A.P., Jain A., Deal R.B., Nagarajan V.K., Poling M.D., Raghothama K.G., Meagher R.B. (2010). Histone H2A.Z regulates the expression of several classes of phosphate starvation response genes but not as a transcriptional activator. Plant Physiol. 152: 217–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thambirajah A.A., Dryhurst D., Ishibashi T., Li A., Maffey A.H., Ausió J. (2006). H2A.Z stabilizes chromatin in a way that is dependent on core histone acetylation. J. Biol. Chem. 281: 20036–20044 [DOI] [PubMed] [Google Scholar]
- Trevaskis B., Hemming M.N., Dennis E.S., Peacock W.J. (2007). The molecular basis of vernalization-induced flowering in cereals. Trends Plant Sci. 12: 352–357 [DOI] [PubMed] [Google Scholar]
- Werner J.D., Borevitz J.O., Uhlenhaut N.H., Ecker J.R., Chory J., Weigel D. (2005). FRIGIDA-independent variation in flowering time of natural Arabidopsis thaliana accessions. Genetics 170: 1197–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilczek A.M., et al. (2009). Effects of genetic perturbation on seasonal life history plasticity. Science 323: 930–934 [DOI] [PubMed] [Google Scholar]
- Wilson J.J., Kovall R.A. (2006). Crystal structure of the CSL-Notch-Mastermind ternary complex bound to DNA. Cell 124: 985–996 [DOI] [PubMed] [Google Scholar]
- Yan L.L., Loukoianov A., Blechl A., Tranquilli G., Ramakrishna W., SanMiguel P., Bennetzen J.L., Echenique V., Dubcovsky J. (2004). The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303: 1640–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zlatanova J., Thakar A. (2008). H2A.Z: View from the top. Structure 16: 166–179 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.