This work shows that MCA1, required for the expression of cytochrome f, is degraded by proteolysis upon interaction with unassembled cytochrome f. MCA1 proteolysis appears to be critical for the assembly-dependent regulation of cytochrome f synthesis, known as Control by Epistasy of Synthesis, which tightly couples its expression to that of its assembly partners.
Abstract
Organelle gene expression is characterized by nucleus-encoded trans-acting factors that control posttranscriptional steps in a gene-specific manner. As a typical example, in Chlamydomonas reinhardtii, expression of the chloroplast petA gene encoding cytochrome f, a major subunit of the cytochrome b6f complex, depends on MCA1 and TCA1, required for the accumulation and translation of the petA mRNA. Here, we show that these two proteins associate in high molecular mass complexes that also contain the petA mRNA. We demonstrate that MCA1 is degraded upon interaction with unassembled cytochrome f that transiently accumulates during the biogenesis of the cytochrome b6f complex. Strikingly, this interaction relies on the very same residues that form the repressor motif involved in the Control by Epistasy of cytochrome f Synthesis (CES), a negative feedback mechanism that downregulates cytochrome f synthesis when its assembly within the cytochrome b6f complex is compromised. Based on these new findings, we present a revised picture for the CES regulation of petA mRNA translation that involves proteolysis of the translation enhancer MCA1, triggered by its interaction with unassembled cytochrome f.
INTRODUCTION
In the time since endosymbiosis, most genes of the organelle ancestors have been either lost or transferred to the nucleus of the host cell (Martin et al., 1998; Timmis et al., 2004; Keeling, 2009). In the green lineage, the chloroplast genome has retained <100 protein-encoding genes, out of >3000 in its cyanobacterial progenitor. Their protein products participate mostly in photosynthesis or in the expression of the chloroplast genome (Barkan and Goldschmidt-Clermont, 2000). Mitochondrial genomes have become even smaller, retaining only 8, 13, and 8 open reading frames in the yeast Saccharomyces cerevisiae, in human, and in the green unicellular alga Chlamydomonas reinhardtii, respectively (Anderson et al., 1981; de Zamaroczy and Bernardi, 1986; Foury et al., 1998; Cardol and Remacle, 2009). Consequently, respiratory or photosynthetic protein complexes, with the exception of mitochondrial complex II, result from the stoichiometric assembly of subunits encoded in either the organellar or nuclear genomes. Thus, one expects a need for some coordination in gene expression between these distinct genetic compartments.
The stoichiometric accumulation of the various subunits of energy-transducing protein complexes in organelles relies on a combination of regulatory mechanisms. First, most subunits from a protein complex show a concerted accumulation, such that in the absence of a major subunit; the others are either rapidly degraded or show downregulated synthesis, an assembly-dependent regulation of translation known as the CES process (for Control by Epistasy of Synthesis) (reviewed in Choquet and Wollman, 2009). Secondly, the nucleus tightly controls the expression of organelle genes via a set of trans-acting factors acting at the posttranscriptional level on a single (or a few) specific target mRNA(s) (Fox, 1996; Barkan and Goldschmidt-Clermont, 2000; Choquet and Wollman, 2002; Herrin and Nickelsen, 2004).
Phenotypic studies of photosynthetic or respiratory mutants provided evidence for two major classes of nucleus-encoded factors (Choquet and Wollman, 2002). M (for maturation/stability) factors are required for the stable accumulation of their target mRNA. Typical examples taken from Chlamydomonas studies include the MCA1, NAC2, and MBB1 factors that protect from 5′ to 3′ exonucleolytic degradation the transcripts of the chloroplast genes petA (encoding cytochrome f of the b6f complex; Loiselay et al., 2008), psbD (encoding the D2 subunit of the photosystem II reaction center; Kuchka et al., 1989; Nickelsen et al., 1994), and psbB (encoding the PSII core antenna CP47; Monod et al., 1992; Vaistij et al., 2000a, 2000b), respectively. T factors are required for the translation of a specific transcript, as exemplified in Chlamydomonas by TCA1 for the petA transcript (Wostrikoff et al., 2001; Raynaud et al., 2007) or RBP40 for the psbD transcript (Schwarz et al., 2007).
Expression of these nucleus-encoded factors is critical for organelle biogenesis. Plant and algal cells defective for a chloroplast-targeted trans-acting factor lose their phototrophic properties, whereas mutations in mitochondrial-targeted factors impair respiratory growth in S. cerevisiae (reviewed in Ackerman and Tzagoloff, 2005; Fontanesi et al., 2008). Only a few such factors have been identified in mammals so far, but deficiency in the LRPPRC protein involved in the stabilization and translation of coxI and coxIII mRNAs is associated with severe diseases in human (Xu et al., 2004). Whether M and T factors are merely constitutively required for (i.e., control) mitochondrial or chloroplast gene expression or have true regulatory functions (i.e., regulate) is still a matter of debate. In Chlamydomonas, variations in the abundance of M and T factors with physiological conditions deeply impact the expression of their target gene (Raynaud et al., 2007). Likewise, in S. cerevisiae, the abundance of Pet111p is limiting in the expression level of COXII (Green-Willms et al., 2001), while that of Pet494p, governing the translation of COXIII, is modulated depending on the carbon source and oxygen availability (Steele et al., 1996).
In spite of their physiological importance, relatively little is known regarding the mode of action of these factors. Most of them belong to high molecular weight complexes, some of which contain an RNA moiety as well (Boudreau et al., 2000; Vaistij et al., 2000b; Auchincloss et al., 2002; Dauvillée et al., 2003; Perron et al., 2004). For instance, NAC2 and RBP40 belong to a single high molecular mass complex that also contains the psbD 5′ untranslated region (UTR) (Schwarz et al., 2007).
Most T factors have been shown genetically to target the 5′UTR of the transcripts whose translation they assist, suggesting that they are required for the initiation of translation rather than for its elongation. Accordingly, RBP40, required for the synthesis of the D2 protein, may transiently interact with ribosomes (Schwarz et al., 2007) but is not found in polysomal fractions (Boudreau et al., 2000). However, the molecular events leading to translation initiation remain poorly understood. Some T factors may act by unmasking the initiation codon of their target mRNA, sequestered into a secondary structure (Stampacchia et al., 1997; Klinkert et al., 2006; Schwarz et al., 2007). Alternatively, T factors may recruit the translation machinery, but their affinity for components of this machinery remains to be documented in most cases (however, see McMullin et al., 1990; Haffter et al., 1991; Haffter and Fox, 1992).
According to an emerging consensus, M factors bind to the 5′ or 3′ termini of their target transcripts and stabilize them by acting as a barrier against exonucleases (Drager et al., 1998; Vaistij et al., 2000a; Loiselay et al., 2008; Hattori and Sugita, 2009; Pfalz et al., 2009). Whether, in addition, M factors participate in the translation of their target mRNA is still a matter of debate. In several instances, organelle transcripts in Chlamydomonas and Saccharomyces, even though engineered to accumulate in the absence of their stabilization factor, depend on the latter to be efficiently translated (Drager et al., 1998; Nickelsen et al., 1999; Vaistij et al., 2000a; Islas-Osuna et al., 2002).
To gain deeper insights into the function of M and T factors, we studied how MCA1 and TCA1 participate in the expression of the petA mRNA. We previously provided genetic evidence that these proteins target neighboring but distinct sequences in the very 5′ end of petA 5′UTR, where they display partially overlapping functions in stabilization and translation of the petA mRNA (Loiselay et al., 2008): the MCA1-dependent accumulation of petA mRNA is reduced in the absence of TCA1, whereas a modified petA transcript whose stability does not require the presence of MCA1 shows decreased TCA1-dependent rates of translation in the absence of MCA1 (Loiselay et al., 2008). Thus, MCA1 and TCA1, together with the petA transcript, should be regarded as the petA gene expression system. Here, we used biochemical and gene transformation approaches to provide the molecular basis for the interactions between the three components of the petA gene expression system, MCA1/TCA1/petA mRNA. In particular, we provide new evidence for a critical role of MCA1 in the regulation of petA mRNA translation, which allows us to relate the regulatory function of this M factor to the CES process for cytochrome f synthesis.
RESULTS
MCA1 and TCA1 Are Soluble Proteins
In mitochondria of S. cerevisiae, most trans-acting factors are associated with the inner membrane (Green-Willms et al., 2001; Naithani et al., 2003; Krause et al., 2004). In the green lineage as well, some chloroplast-targeted factors are membrane bound (Perron et al., 1999; Zerges et al., 2002; Balczun et al., 2005; Merendino et al., 2006). Since MCA1 and TCA1 are required for the synthesis of cytochrome f, an integral membrane protein, we first determined whether they behaved as soluble or membrane-bound proteins. To perform these studies, we used strains expressing tagged versions of MCA1 and TCA1, hereafter referred to as mH and tF that allow the immunological detection of these two factors (with apparent molecular masses of ~115 and ~110 kD, respectively) by antibodies directed against the HA or Flag epitopes. These strains (described in Supplemental Figure 1 online and in Raynaud et al., 2007) were obtained by complementation of mca1 and tca1 mutants, with HA- and Flag-tagged versions of MCA1 and TCA1, respectively. As detailed in Methods, in the following, strains are named by their genotype. m (/t) denotes mutated MCA1 (/TCA1) loci, whereas H (/F) indicates that the mca1 (/tca1) mutations had been complemented by the HA-tagged version of MCA1 (/the Flag-tagged version of TCA1).
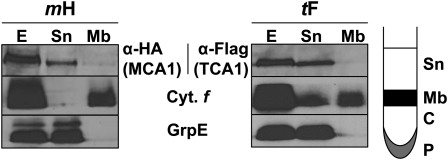
French press lysates of strains mH and tF were layered on top of a 1.5 M sucrose cushion (Figure 1). After ultracentrifugation, MCA1 and TCA1 were found in the supernatant Sn, as was the stromal chaperone GrpE (Schroda et al., 2001), while integral membrane proteins, like cytochrome f, were recovered mainly in the layer at the interface between the supernatant and the sucrose cushion (Mb in Figure 1). Thus, MCA1 and TCA1 behaved as soluble proteins.
Figure 1.
MCA1 and TCA1 Are Soluble Proteins.
Cell extracts (E) from tF and mH strains, overlaid on a 1.5 M sucrose cushion, were separated by ultracentrifugation into the fractions schematically depicted in the right panel. Sn, supernatant; Mb, membrane; C, cushion; P, pellet. Equal volumes of each fraction were immunoreacted with anti-HA or anti-Flag antibodies. In addition, GrpE and cytochrome f were used as markers of soluble and membrane fractions, respectively. In the tF (TCA1) panel, fraction Sn was slightly contaminated by membranes, which explains the presence of cytochrome f (Cyt. f). Fractions C and P are not shown, as they were devoid of significant amounts of MCA1, TCA1, GrpE, or cytochrome f.
Potential Interactions and Functional Domains of MCA1 and TCA1
As detailed in the introduction, MCA1 and TCA1 might synergistically bind to the petA 5′UTR (Loiselay et al., 2008). We tested their ability to interact physically by two-hybrid experiments in the yeast S. cerevisiae.
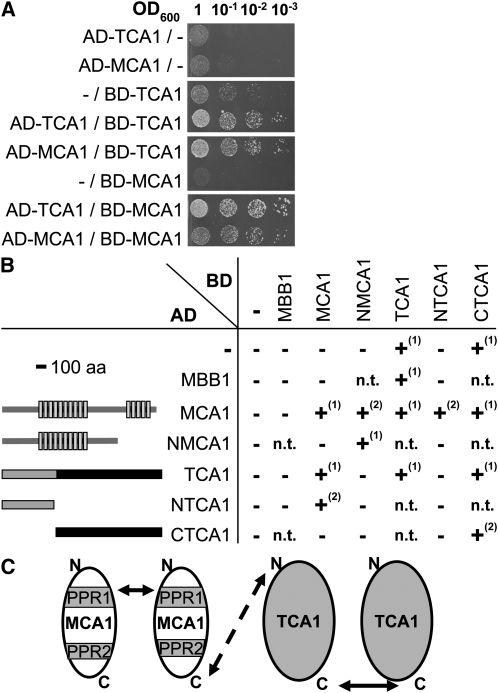
We cloned the whole coding sequences of MCA1 and TCA1 into GAL4-derived vectors, well suited for studying interactions between soluble proteins. In these experiments, interaction between the tested proteins, respectively fused to the GAL4 activating domain (AD) and binding domain (BD), reconstitutes the GAL4 transcription factor that restores prototrophy for His and adenine (Figure 2). Strength of the interaction can be estimated by plating cells on media of increasing stringency (i.e., lacking His and supplemented with 3-amino-triazole [3-AT] or lacking both His and adenine). We observed that MCA1 interacts with itself as well as with TCA1 (Figure 2A). MCA1 homomeric interaction is consistent with our previous observation of a quadratic dependency of petA mRNA levels on MCA1 abundance in vivo (see Figure 3B in Raynaud et al., 2007). However, the BD-TCA1 fusion alone allowed growth of transformed yeast on the less stringent selective medium, which prevented us, in this first set of experiments, to study interaction of TCA1 with itself.
Figure 2.
Interactions between MCA1 and TCA1 Probed by Two-Hybrid Experiments in Yeast.
(A) Interactions between full-length MCA1 and TCA1 proteins. Interactions were assessed between the proteins indicated at the left of the figure, fused either to the AD or to the BD of the Gal4 transcription factor, by spotting serial dilutions of cotransformed yeast cells on selective medium (SD lacking Leu, Trp, and His but supplemented with 5 mM 3-AT).
(B) Interactions observed between the truncated versions of MCA1 and TCA1 schematically depicted at the left of the figure. Light-gray boxes depict the PPR repeats in the MCA1 protein, while the thick black bar indicates the shortest region of TCA1 still able to complement tca1 mutations. Growth of transformed yeast was tested as above on different selective media: –, +(1), and +(2) indicate the absence of growth on any selective media, growth on medium lacking His and supplemented with 3-AT, and growth on a more stringent medium lacking His and adenine. n.t., not tested; aa, amino acids.
(C) Schematic representation of MCA1/TCA1 interactions. PPR1 and 2 represent the two blocks of PPR repeats within MCA1. The dashed arrow points to an interaction suggested, but not fully demonstrated by our experiments, between the corresponding domains.
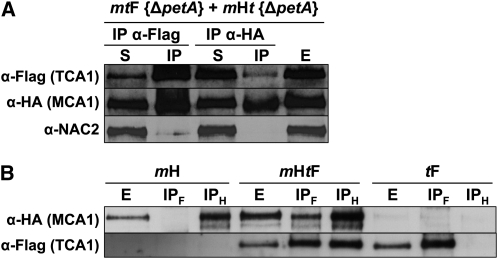
Figure 3.
Interactions among MCA1, TCA1, and petA mRNA Probed by CoIP.
(A) MCA1 and TCA1 interact in the absence of petA mRNA. Equal volumes of mtF {ΔpetA} and mHt {ΔpetA} cultures were mixed and broken with a French press. The soluble extract recovered after centrifugation (E) was immunoprecipitated with anti-HA or anti-Flag antibodies, as indicated. The presence of MCA1/TCA1 in supernatant (S) and immunoprecipitated (IP) fractions was then assessed using the same antibodies, as indicated at the left of the figure. The antibody against the NAC2 protein provided a specificity control.
(B) Interactions between MCA1 and TCA1 in the presence of the petA mRNA. Soluble extracts (E) from strains mH, mHtF, and tF and immunoprecipitates recovered after incubation of the extracts with either anti-HA (IPH) or anti-Flag (IPF) antibodies were analyzed as above for the presence of MCA1 and TCA1.
To characterize further the protein domains responsible for the interactions observed in Figure 2A, we repeated two-hybrid experiments with various truncated versions of TCA1 and MCA1. Indeed, protein interactions are sometimes better identified in the two-hybrid system with cDNAs expressing truncated proteins than with full-length cDNAs. The latter are often poorly expressed in S. cerevisiae, especially when they originate from organisms that display a widely different codon usage, such as C. reinhardtii (Fromont-Racine et al., 1997). We thus generated an N-terminal truncated version of TCA1, lacking the first 391 residues, hereafter referred to as CTCA1, which is still able to complement a tca1-null mutation (see Supplemental Figure 2 online). When fused to Gal4 BD and transformed into yeast, CTCA1 allowed growth on His-depleted but not on the more stringent medium lacking both His and adenine. By contrast, yeast cotransformed with both AD-CTCA1 and BD-CTCA1 constructs grew on this stringent medium (Figure 2B). Thus, TCA1 interacts with itself through its C-terminal domain, at least in S. cerevisiae. We failed to observe an interaction between the C-terminal domain of TCA1 and MCA1 (Figure 2B), suggesting that the N-terminal domain of TCA1 is required for the interaction with MCA1 that we observed in Figure 2A. Indeed, a C-terminal truncated version of TCA1, NTCA1, that retained only the first 391 residues fused to the GAL4-AD interacts with MCA1 (Figure 2B).
We similarly assessed the domains of MCA1 required for interactions with itself or with TCA1. We generated a C-terminal truncated version of MCA1, NMCA1, that lacks the last 290 residues (the last five Pentatrico Peptide Repeats [PPR] and the C-terminal tail). As shown in Figure 2B, NMCA1 still interacted with itself but had lost its ability to bind TCA1, suggesting that the C-terminal moiety of MCA1 is required for TCA1 recognition. Surprisingly, we observed an interaction between the AD-MCA1 and the BD-NMCA1 fusion proteins but not between AD-NMCA1 and BD-MCA1, which may originate from an altered folding of these fusion proteins that may prevent their interaction. The interactions developed by the various versions of MCA1 and TCA1 in the experiments shown in Figure 2 were specific for these two factors: we failed to detect any interaction of either MCA1 or TCA1 with MBB1, another nucleus-encoded factor specifically required for psbB mRNA stability in the chloroplast of C. reinhardtii.
Figure 2C schematically depicts the interactions between TCA1 and MCA1 identified by two-hybrid experiments in yeast. Homomeric interactions rely on protein domains distinct from those involved in hetero-interactions: the C-terminal moiety of MCA1 might interact with the N-terminal domain of TCA1, whereas these proteins interact with themselves through their N-terminal and C-terminal moieties, respectively. Together, these experiments suggested that MCA1 and TCA1 might associate in vivo in high molecular mass complexes.
MCA1 and TCA1 Form Oligomeric Complexes in the Presence and Absence of petA mRNA
Since two-hybrid experiments point to interactions between MCA1 and TCA1 in yeast cells, which are devoid of petA mRNA, we used coimmunoprecipitation (CoIP) to assess their ability to interact in vivo in the absence of the petA transcript.
To this end, we constructed a double mutant strain, mt, lacking both MCA1 and TCA1 (Table 1). We then recovered a strain expressing MCA1-HA in the absence of TCA1, mHt, among the progeny of a cross between strain mH and the mt double mutant. We similarly recovered the mtF strain, expressing TCA1-Fl in the absence of MCA1. The petA gene was then deleted in these two strains by biolistic transformation to generate strains mHt {ΔpetA} and mtF {ΔpetA} that express MCA1-HA or TCA1-Fl, respectively, in the absence of both interacting partners (Table 2). Soluble extracts from these strains were mixed before performing CoIPs with anti-HA or anti-Flag antibodies coupled to sepharose beads. The precipitates were analyzed by protein gel blots probed with anti-HA, anti-Flag, or anti-NAC2 antibodies (Figure 3A). TCA1, but not NAC2, was detected in the pellet after anti-HA precipitation, as was MCA1 after anti-Flag precipitation. Thus, the two proteins have a high affinity for each other, since they specifically interact in the absence of the petA transcript after mixing cell lysates in which preexisting interactions were prevented.
Table 1.
C. reinhardtii Strains Generated by Crosses
| Mating Type− Parent | Mating Type+ Parent | Resulting Strain |
| tca1-8 | mca1-6 | mt |
| mt | mH | mHt |
| mt | tF | mtF |
| mH | {ccsA-B6} | mH {ccsA-B6}a |
Due to the uniparental inheritance of the chloroplast genome from the mt+ parent strain, each member of the resulting tetrads carried the chloroplast mutation ccsA-B6, while the two nuclear loci mca1-8 and MCA1-HA showed independent Mendelian segregation.
Table 2.
C. reinhardtii Strains Generated by Transformation
| Recipient Strain | Transforming Plasmid | Transformed Strains |
| Nuclear transformationa | ||
| mca1-6 | plgMCA1-HA [1]b | mH |
| mtF | plgMCA1-HA [1]b | mHtF |
| tca1-8 | pshTCA1-Fl [1]c | CTCA1 |
| Chloroplast transformationd | ||
| mH | pΔpetA [2] | mH {ΔpetA}e |
| mH | pWFStopK [this study] | mH {petASt}e |
| mH | p5′dAf K [this study] | mH {5′dAf }f |
| mH | pf283St [3] | mH {fSol}eg |
| mH | pf312St [4] | mH {f312St}g |
| mH | pf310St [4] | mH {f310St}g |
| mH | pf307St [4] | mH {f307St}g |
| mH | pf304M [4] | mH {f304M}h |
| mH | pf307S [4] | mH {f307S}h |
| mH | pfΔK [4] | mH {fΔK}h |
| mH | p5′dAf307S [this study] | mH {5′dAf307S}fh |
| mH | pΔpetD [2] | mH {ΔpetD}e |
| mHt | pΔpetA [2] | mHt {ΔpetA}f |
| mHt | p5′dAfK [this study] | mHt {5′dAf}fi |
| mHt | p5′dAf307S [this study] | mHt {5′dAf307S}hi |
| tF | pΔpetA [2] | tF {ΔpetA}e |
| mtF | pΔpetA [2] | mtF {ΔpetA}f |
References are as follows: 1 (Raynaud et al., 2007), 2 (Kuras and Wollman, 1994), 3 (Kuras et al., 1995a); 4 (Choquet et al., 2003), and 5 (Loiselay et al., 2008).
All recipient strains were nonphotosynthetic, and transformants were selected for photoautotrophy on minimum medium (Harris, 1989) under high light (200 μE·m−2·s−1).
Plasmid DNA was linearized with XbaI that cuts upstream of the MCA1 initiation codon before transformation.
Plasmid was cut with XbaI and SfiI, and the 5461-bp fragment that encodes the C-terminal domain of TCA1 was used for transformation after gel purification.
All recipient strains were spectinomycin sensitive. Transformed strains were selected for resistance to spectinomycin (100 μg·mL−1) under low light (5 μE·m−2·s−1) and subcloned in darkness until they reached homoplasmy, unless otherwise indicated.
Homoplasmy was deduced from the loss of photoautotrophic growth capacity.
Homoplasmy was assessed by RNA gel blot experiments.
Homoplasmy was assessed by protein immunoblot analysis for the expression of truncated versions of cytochrome f.
The presence of the f307S substitution was screened by restriction fragment length polymorphism analysis of PCR products amplified with primers Test_petA_For and Test_petA_Rev, as described by Choquet et al. (2003 [see Supplemental Table A]).
Strains were subcloned under dim light (25 μE·m2 ·s−1) as recipient strains were nonphotosynthetic mutants, whereas the transformed strains regained photoautrophic growth capability.
In presence of the petA mRNA, interactions between MCA1 and TCA1 were further confirmed by the same CoIP approach using soluble extracts from a double-complemented mHtF strain that expresses both MCA1-HA and TCA1-Fl, using strains mH and tF as controls (Figure 3B). Antibodies directed against MCA1-HA pulled down TCA1 using extracts from strain mHtF but not from the control strain tF. Similarly, MCA1 was coimmunoprecipitated by antibodies directed against TCA1-Fl in soluble extracts from strain mHtF but not from the control strain mH. Thus, MCA1 and TCA1 do interact in vivo when the petA mRNA is present.
MCA1 and TCA1 Belong in Vivo to Large Complexes That Also Contain petA mRNA
We then used size exclusion chromatography to characterize the protein complexes formed upon association of TCA1 with MCA1. Supernatants recovered after centrifugation of French press lysates from tF and mH cells were fractionated on a Sephacryl S500 column (GE Healthcare), optimal for separating protein complexes in the 100 to 10,000 kD range. As shown in Figure 4 (samples 5 and 12), MCA1 and TCA1 belong to high molecular mass complexes peaking in fractions 10 and 11 with an apparent molecular mass of ~600 kD, a size similar to that of the complexes containing NAC2, used here as a control and internal standard (sample 6; Boudreau et al., 2000).
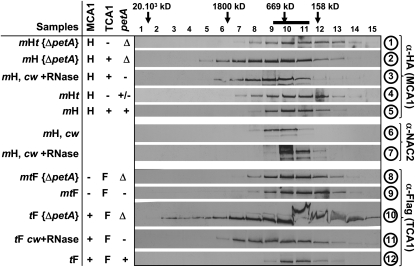
Figure 4.
Interactions between MCA1, TCA1, and the petA mRNA Probed by Size Exclusion Chromatography.
Soluble or stromal (prepared from cw strains lacking cell wall) extracts from strains listed at the left of the figure were fractionated on Sephacryl S500 column and analyzed with the antibodies indicated at the right of the figure. Molecular masses of the complexes found in each fraction were estimated by comparison with standards of the HMW gel filtration calibration kit (GE Healthcare) or with the position of ribulose-1,5-bisphosphate carboxylase/oxygenase (indicated by the thick black bar at the top of the figure). The middle table recapitulates the status of each partner of the petA gene expression system: MCA1 (/TCA1) is absent (−) or expressed from the wild-type (+) or tagged (H/F) gene, while the petA mRNA is either absent in deletion strains (Δ) or in the absence of MCA1 (−), present in reduced amounts in the absence of TCA1 (+/−), or accumulated to wild-type levels (+).
The presence of RNAs, in particular petA mRNA, in these high molecular mass complexes was investigated by two different approaches. First, we RNase treated the soluble extracts from mH and tF strains, prior to size exclusion chromatography (samples 3 and 11). We also analyzed soluble extracts from strains mH {ΔpetA} and tF {ΔpetA}, lacking the petA gene (samples 2 and 10). In all instances, we observed a broadening in the distribution of MCA1 and TCA1, with a shift toward heavier fractions peaking around 1200 kD. By contrast, the distribution of NAC2 (sample 7) shifted toward fractions of lower molecular mass upon RNase treatment, as previously reported (Boudreau et al., 2000). Thus, the presence of petA mRNA is responsible for the typical distribution of complexes containing MCA1 and TCA1 peaking at 600 kD that we observed in strains mH and tF.
MCA1 and TCA1 probably shifted toward fractions of higher molecular mass in the absence of the petA mRNA because they develop homo- and hetero-interactions as we observed by CoIP and two-hybrid experiments in yeast (Figures 2 and 3B). To distinguish homo-oligomers of similar apparent molecular mass containing either MCA1 only or TCA1 only from hetero-oligomers comprising both proteins, we analyzed the distribution of MCA1 in the absence of both TCA1 and petA or of TCA1 in the absence of both MCAl and petA (strains mHt {ΔpetA} and mtF {ΔpetA}, respectively; samples 1 and 8 in Figure 4). In these strains, MCA1 and TCA1 were no longer found in broadly distributed protein complexes centered around 1200 kD but within lighter protein complexes of ~500 kD. Similar protein complexes were found in strains mHt and mtF that express MCA1-HA in the absence of TCA1 or TCA1-Fl in the absence of MCA1 and, consequently, show little or no accumulation of petA mRNA (samples 4 and 9). Thus, the heavy complexes in the 1200 kD range likely correspond to hetero-oligomers of MCA1 and TCA1 that formed in the absence of the petA mRNA.
In summary, the three components of the petA gene expression system (MCA1, TCA1, and the petA transcript), when present all together, are engaged in a ternary complex of ~600 kD, which may accommodate dimers of each protein and one or two copies of the petA 5′UTR. Note that in these experiments, no special care was taken to preserve the integrity of the RNA moiety: the petA mRNA was likely trimmed to the transcript portions protected from degradation by their interaction with nucleus-encoded factors. Thus, RNA fragments retained in the ternary complex in these experiments are most probably restricted to the petA 5′UTR and should account for <100 kD.
In the absence of petA transcripts, MCA1 and TCA1 are likely to form large hetero-oligomeric complexes ranging from several MD to ~100 kD, centered around 1200 kD. This broad distribution suggests variable states of aggregation of these factors, which may also associate with additional proteins. We note that our size exclusion chromatography experiments poorly resolve changes in the distribution of MCA1 and TCA1 in the 500- to 600-kD range between strains mH, tF, mHt, and mtF. These distributions nevertheless corresponded to widely different protein populations since we obtained experimental evidence for a ternary complex in the former two strains as opposed to homo-oligomers, possibly tetramers, in the latter two strains.
Mutual Stabilization of MCA1 and TCA1
We then studied the accumulation of MCA1 and TCA1 in the absence of their partners from the ternary complex. As shown in Figures 3A and 4 (samples 1 and 4), MCA1 still accumulates to detectable levels in the absence of TCA1 and/or petA mRNA. Similarly, TCA1 shows notable accumulation in the absence of MCA1 and/or petA mRNA (Figures 3A and 4, samples 8 and 9). Because MCA1 and TCA1 belong to distinct complexes when expressed alone or together with petA mRNA, their proteolytic susceptibilities and, hence, their accumulation levels may vary depending on these interactions.
We first compared the extent of accumulation of TCA1 in the presence or absence of MCA1. As shown in Figure 5A, TCA1 accumulation was identical in strains tF and mtF, this latter strain also lacking petA mRNA, destabilized in the absence of MCA1. To sort out the consequences of the absence of MCA1 versus defective cytochrome f expression, we analyzed the strain tF {ΔpetA} and observed a 3-fold increased accumulation of TCA1 when compared with strains tF and mtF (Figure 5B). Both strains mtF and tF {ΔpetA} lack petA mRNA, but only strain tF {ΔpetA} expresses MCA1. We conclude that, in absence of petA mRNA, the presence of MCA1 stabilizes TCA1, most probably by allowing the formation of the stable ~1200-kD oligomers that we detected by size exclusion chromatography.
Figure 5.
The Accumulation of MCA1 or TCA1 Depends on the Presence of Their Partners from the petA Gene Expression System.
(A) and (B) Immunoblots showing accumulation of TCA1-Fl in an otherwise wild-type genetic context (strain tF), in progeny expressing TCA1-Fl of one representative tetratype tetrad recovered after crossing mt and tF strains (A), or in the absence of petA mRNA (strain tF {ΔpetA}; [B]). The mtF progeny lacking MCA1 also lacks cytochrome f.
(C) Accumulation of MCA1-HA in strains with an otherwise wild-type genetic context (mH), lacking petA (mH {ΔpetA}), both petA and TCA1 (mHt {ΔpetA}), and in the progeny expressing MCA1-HA of one representative tetratype tetrad from the cross mt × mH. The mHt progeny that inherited the mutant tca1 allele from the mt parent lacks cytochrome f. A dilution series of the mH {ΔpetA} sample is shown for the ease of quantification.
For both panels, expression of cytochrome f (cyt. f) and of the β-subunit of the mitochondrial ATP synthase complex (F1β; loading control) is also shown. The wild-type strain (WT) that does not express MCA1-HA or TCA1-Fl is shown as a control.
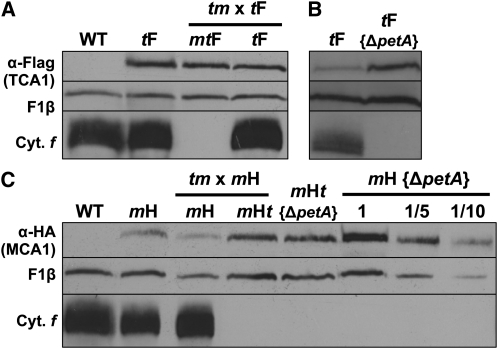
Strikingly, the steady state level of MCA1, as detected in strain mH, increased 10-fold in the absence of petA mRNA (strain mH {ΔpetA}) and 3-fold in the absence of TCA1, irrespective of the presence of petA mRNA (strains mHt and mHt {ΔpetA}; Figure 5C). The much higher accumulation of MCA1 in strain mH {ΔpetA} than in strain mHt {ΔpetA} or mHt, the three of which are defective for cytochrome f expression, suggested that, in the absence of cytochrome f, MCA1 is stabilized by TCA1 most probably again within the ~1200-kD high molecular mass complexes. However, this stabilizing effect of TCA1 was not observed when comparing strains mH and mHt, since the level of MCA1 was 3 times higher in the latter, which lacks TCA1. If one takes into account that cytochrome f is expressed in strain mH, but not in strain mHt, then these contrasting observations can be reconciled by assuming that the steady state level of accumulation of MCA1 results from two opposite effects: (1) a contribution of TCA1 to the stability of MCA1 and (2) a degradation of MCA1 upon cytochrome f expression, which also depends on TCA1. In strain mH, although the two contributions are at work, the expression of cytochrome f would lead to an extensive proteolytic disposal of MCA1.
Cytochrome f Expression Shortens the Half-Life of MCA1
To investigate further the possible role of cytochrome f expression in MCA1 degradation, we transformed the chloroplast genome of strain mH with a modified petA gene that cannot be translated because its initiation codon has been replaced by a Stop codon. In the resulting mH {petASt} strain, despite the accumulation of the untranslatable petA mRNA, MCA1 is 10 times more abundant than in a wild-type genetic context, reaching the same high level as in the absence of petA mRNA (Figure 6A). To confirm that MCA1 is degraded upon cytochrome f expression, we compared the decay of MCA1 in various regimes for petA mRNA translation by immunochase experiments in the presence of cycloheximide that prevents cytosolic translation and, therefore, de novo synthesis of MCA1. As previously reported (Raynaud et al., 2007), MCA1 expressed in a wild-type context is short lived, with a half-life in the hour range (Figure 6B, panel mH). By marked contrast, MCA1 remained stable over 6 h of chase in the absence of cytochrome f expression in strains mH {petASt} or mH {ΔpetA}. Moreover, when the translation-competent mH strain was incubated with cycloheximide and lincomycin to prevent both cytosolic and chloroplast translations (bottom of Figure 6B), the half-life of MCA1 increased markedly, when compared with the control culture treated with cycloheximide alone. Thus, cytochrome f expression targets MCA1 for degradation.
Figure 6.
Half-Life and Accumulation of MCA1 Are Governed by the Expression of Full-Length Cytochrome f.
(A) Accumulation of MCA1-HA in an otherwise wild-type strain (mH), in strains carrying a petA gene deletion (mH {ΔpetA}) or expressing a modified petA gene that (1) cannot be translated (mH {petASt}), (2) is expressed under the control of the atpA 5′UTR (mH {5′dAf}), or (3) encodes a truncated soluble cytochrome f (mH {fSol}). Accumulation of cytochrome f (Cyt. f) in the various strains is shown, while the β-subunit of the mitochondrial ATP synthase complex (F1β) provides a loading control.
(B) Half-life of MCA1 assessed in the same strains by immunochase at the various time points indicated after addition at t = 0 of cycloheximide alone or of cycloheximide plus lincomycin (linco.).
MCA1 Degradation Does Not Result from Its Interaction with the petA 5′UTR
To identify the pathway that leads to MCA1 degradation upon cytochrome f expression, we first investigated whether it required the action of MCA1 on its 5′UTR target on the petA mRNA. We replaced by biolistic transformation the endogenous petA gene in the mH strain by a chimeric version 5′dAf in which the petA coding sequence is fused to the 5′UTR of the unrelated atpA gene (Choquet et al., 1998; Drapier et al., 2007). Since the targets of MCA1 and TCA1 lie within the petA 5′UTR, these trans-acting factors are no longer involved in the expression of the 5′dAf chimera (Wostrikoff et al., 2001; Loiselay et al., 2008). The resulting strain mH {5′dAf} was analyzed for MCA1 content and half-life. When compared with the mH strain, it displayed the same low level (Figure 6A) of short-lived (Figure 6B) MCA1. Thus, MCA1 is still degraded in this strain, although it is no longer involved in cytochrome f synthesis.
Then, we preserved translation of the petA gene but introduced by transformation a premature termination codon in the petA coding sequence. This led to the expression of a soluble variant of cytochrome f, fsol, that lacks the stromal-exposed C-terminal tail and the transmembrane helix anchoring the lumenal domain of cytochrome f into the membrane (Kuras et al., 1995a). In the mH {fsol} strain, the accumulation of MCA1 was much increased with respect to the mH recipient strain and became similar to that detected in the mH {ΔpetA} strain (Figure 6A), as a result of the increased life-time of MCA1 (Figure 6B). Thus, the degradation of MCA1 is not triggered by translation initiation but rather by the expression of the C-terminal domain of cytochrome f.
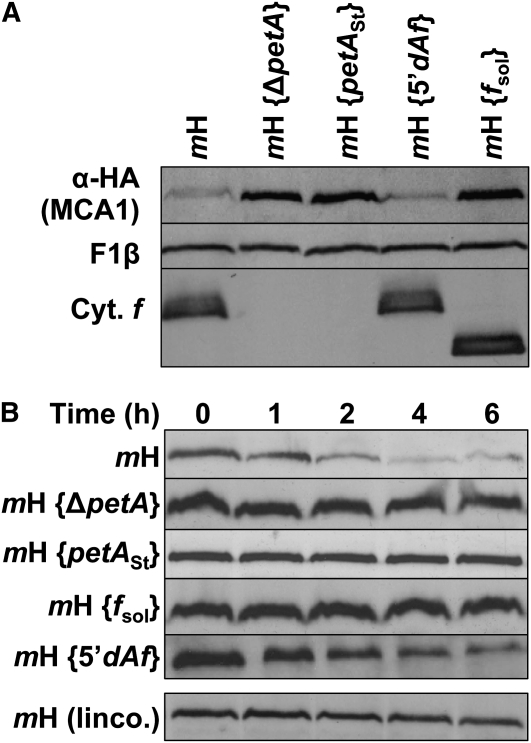
MCA1 Is Degraded upon Interaction with Specific Residues in the C-Terminal Domain of Cytochrome f
To get further insight into the residues of cytochrome f involved in the degradation of MCA1, we took advantage of previously constructed versions of cytochrome f lacking the last 6, 8, or 11 amino acids of the stromal tail (Choquet et al., 2003; Figure 7A). These mutated petA genes were introduced by biolistic transformation into the chloroplast genome of the mH recipient strain to yield strains mH {f312St}, mH {f310St}, and mH {f307St}, respectively. Whereas the shortest truncation f312St did not affect the accumulation of MCA1, which remained similar to that observed in the recipient mH strain, the intermediate truncation, f310St, induced a twofold increase in MCA1 accumulation, and the largest truncation, f307St, led to an ~10-fold higher accumulation of MCA1 (Figure 7B), as observed in the mH {ΔpetA} strain (Figure 6A).
Figure 7.
Disruption of the CES Repressor Domain Stabilizes MCA1.
(A) Schematic representation of the mutations introduced in the stromal-exposed C-terminal tail of cytochrome f in strain mH. Substitutions of the residues shaded in black abolish the cytochrome f CES process, whereas mutations of those shaded in gray attenuate this regulatory process (Choquet et al., 2003).
(B) Accumulation of MCA1-HA in strains with an otherwise wild-type background (strain mH) or carrying the mutations depicted in (A). A dilution series of the strain mH {fsol} is shown for the ease of quantification. Note that strain mH {fΔK} was slightly overloaded in this gel. Cyt., cytochrome.
Strikingly, the very same truncations led to a progressive loss of the CES control of cytochrome f synthesis, a process that couples the rate of cytochrome f synthesis to its assembly into the cytochrome b6f complex. This regulatory process involves the 5′UTR of the petA mRNA (Choquet et al., 1998) and a repressor domain exposed by unassembled cytochrome f, made of a few specific amino acids from the C-terminal domain of the protein. These residues are shown in Figure 7A in black when their substitutions strongly affect the cytochrome f CES process or in gray when substitutions have a significant but more limited effect on this regulatory process (Choquet et al., 2003). Whether these specific residues are also involved in MCA1 degradation was investigated by biolistic transformation of strain mH with petA constructs harboring the corresponding point mutations within the sequence coding for cytochrome f. As shown on Figure 7B, substitution of Lys-304 by a Met, which does not impact the CES process (Choquet et al., 2003), did not alter the accumulation of MCA1 either (compare strain mH {f304M} with strain mH). By contrast, two mutations that compromise the assembly-dependent regulation of cytochrome f synthesis also increased the accumulation of MCA1: substitution of Phe-307 by a Ser led to a 3-fold increase in the accumulation of MCA1 (strain mH {f307S}), whereas deletion of one of the three Lys residues just after the transmembrane helix (strain mH {fΔK}) resulted in the same 10-fold overaccumulation of MCA1 than in strain mH {ΔpetA} (Figure 6A).
We thus conclude that the same C-terminal motif in the sequence of cytochrome f is involved in the CES process and in the degradation of MCA1.
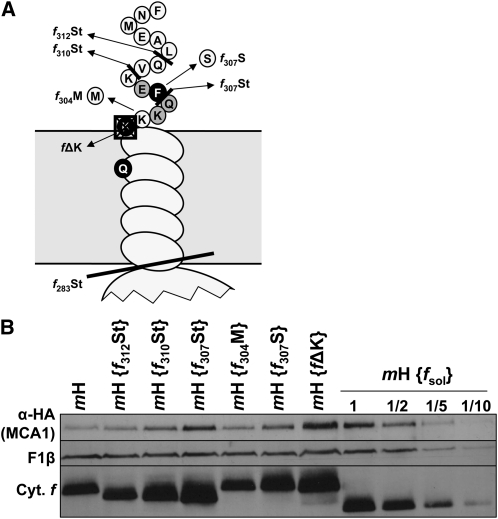
Degradation of MCA1 Depends on the Level of Accumulation of the Unassembled CES Repressor Motif
To determine whether the level of accumulation of the unassembled CES repressor domain or its mere synthesis is responsible for the degradation of MCA1, we compared the level of accumulation of MCA1 under two experimental situations that lead to the most contrasting rates of cytochrome f translation due to extensive changes in the accumulation level of its CES repressor motif.
When holocytochrome f formation is prevented, either because the Cys ligands of the c-type heme are substituted or because mutations inactivate the CCS machinery required for covalent heme binding to apocytochrome f (Kuras et al., 1995b; Xie et al., 1998), the resulting apocytochrome f is rapidly degraded. Consequently, the CES repressor domain accumulates to <0.5% of its level in the wild type, which results in a 3-fold increase in the rate of cytochrome f translation (Kuras et al., 1995b; Choquet et al., 1998). By contrast, in the mH {ΔpetD} strain lacking subunit IV, in which cytochrome f remains unassembled, the repressor domain is particularly long lived, and cytochrome f synthesis is 10 times repressed due to the CES process.
We therefore crossed the mH strain with the mutant strain {ccsA-B6} defective for the CCS pathway because of a premature Stop codon in the chloroplast ccsA gene (Xie et al., 1998) and also deleted the petD gene in the same mH strain by chloroplast transformation. Accumulation of MCA1 was increased in strain mH {ccsA-B6} (Figure 8A) but decreased 2 to 3 times in strain mH {ΔpetD} (Figure 8B), with respect to that in the parental strain mH. Moreover, at variance to its behavior in the mH recipient strain, the half-life of MCA1 was not increased when chloroplast translation was inhibited by addition of lincomycin in strain mH {ΔpetD} (Figure 8C). It appears that the preexisting and long-lived CES repressor domain keeps targeting MCA1 for degradation.
Figure 8.
Accumulation and Half-Life of MCA1 Are Governed by the Accumulation of the CES Repressor Motif.
(A) Accumulation of MCA1-HA and cytochrome f (Cyt. f) in a double mutant mH, {ccsA-B6}, and in the parental strains mH and {ccsA-B6}.
(B) Accumulation of MCA1-HA and cytochrome f in strains mH and mH {ΔpetD}.
In both panels, the accumulation of the β-subunit of the mitochondrial ATP synthase complex (F1β) provides a loading control.
(C) Half-life of MCA1-HA assessed in strains mH and mH {ΔpetD} by immunochase experiments in the presence of cycloheximide (C.) or both cycloheximide and lincomycin (C.+L.).
MCA1-Induced Variations in petA mRNA Abundance
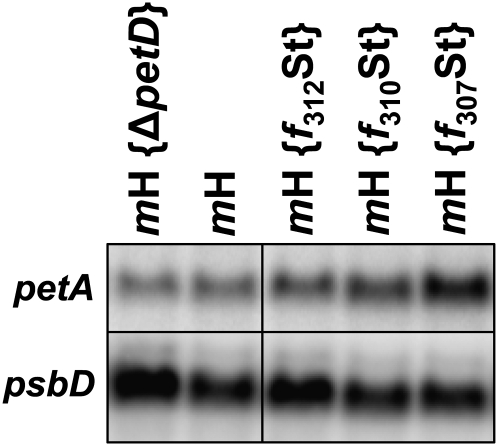
Since the primary function of MCA1 is the stabilization of the petA mRNA, even if it also acts as a translational enhancer (Loiselay et al., 2008), the above-described changes in MCA1 abundance (Figures 7 and 8B) also should cause changes in the accumulation of its target transcript. We thus analyzed by RNA gel blots the accumulation of petA transcripts in diagnostic strains from Figures 7 and 8B. We chose the mH strain as a reference, the mH {ΔpetD} as an archetype of a strain where the synthesis of cytochrome f is repressed because of the CES process, and the three deletion strains, mH {f312St}, mH {f310St}, and mH {f307St} to assess the consequences of a progressive disruption of the CES repressor domain (Figure 9). We observed that the accumulation of the petA mRNA was reduced 2-fold in strain mH {ΔpetD} when compared with the reference strain mH, whereas it increased progressively as truncations extended into the repressor motif, reaching 250% of the wild-type level in strain mH {f307St}. However, we note that in strain mH {ΔpetD}, the 2-fold reduction in petA transcript level cannot alone account for the 10-fold decreased rate of cytochrome f synthesis, as expected for a regulation operating mainly at the translational level. By contrast, in strain mH {f307St}, the 2.5-fold increase in petA mRNA was comparable to the increase in cytochrome f synthesis rate, but we had previously shown that there is little correlation, in an otherwise wild-type genetic context, between the accumulation and the translation of the petA mRNA (Sturm et al., 1994).
Figure 9.
petA Transcript Accumulation in Representative Strains.
Accumulation of petA mRNA in representative strains analyzed in Figures 7B and 8B. A probe specific for the psbD transcript provides a loading control. Note the overloading of sample mH {ΔpetD} and to a lesser extent of sample mH {f312St}.
Interaction between MCA1 and Unassembled Cytochrome f Does Not Require TCA1
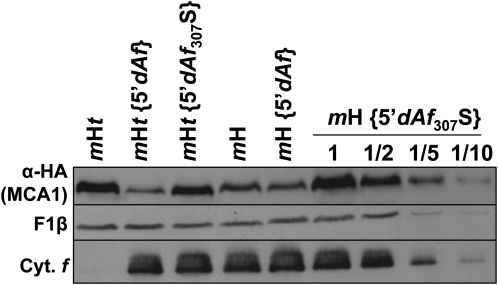
The interaction between unassembled cytochrome f and MCA1 could be restricted to this factor alone, or it could involve a high molecular mass complex that also contains TCA1. We addressed that point using the 5′dAf chimera, which allows the synthesis of cytochrome f in the absence of TCA1. We introduced in the petA coding sequence of this chimera the Phe-307 to Ser substitution that abolishes the CES regulation of endogenous cytochrome f expression and its interaction with MCA1, to yield the 5′dAf307S chimera that was transformed into the chloroplast of strain mH. Compared with the low level of short-lived MCA1 in strain mH {5′dAf} (Figure 6), the immunoblot analysis of the transformed strains revealed a much higher accumulation of MCA1 in strain mH {5′dAf307S} (Figure 10), confirming that MCA1 still interacts with the C-terminal domain of the 5′atpA-driven cytochrome f.
Figure 10.
Destabilization of MCA1 by the Cytochrome f C Terminus Does Not Require the Presence of TCA1.
Accumulation of MCA1-HA, in an otherwise wild-type background (strain mH), in strains where petA coding sequence is expressed under the control of the atpA 5′UTR (strain mH {5′dAf}) and additionally carries a mutation preventing the CES process (strain mH {5′dAf307S}). A dilution series is shown for strain mH {5′dAf307S}. The same chloroplast genotypes were also compared in the mHt nuclear background (i.e., in the absence of TCA1). Accumulations of cytochrome f (Cyt. f) and subunit β of the mitochondrial ATP synthase (F1β, loading control) are also shown in the various strains.
The same 5′dAf and 5′dAf307S constructs were then transformed into strain mHt, lacking TCA1, and the resulting transformants were analyzed by immunoblots. We note that MCA1 was less abundant in strains mHt {5′dAf} and mHt {5′dAf307S} than in strains mH {5′dAf} and mH {5′dAf307S}, respectively, due to the above-described stabilization of MCA1 by TCA1. However, the accumulation of MCA1 was much higher when the CES repressor motif was mutated in strain mHt {5′dAf307S} than when it was preserved in strain mHt {5′dAf} (Figure 10), demonstrating that MCA1 does not require TCA1 for its interaction with the CES repressor motif that regulates its rate of degradation.
DISCUSSION
MCA1 and TCA1 Participate with petA mRNA in the Formation of the petA Gene Expression System
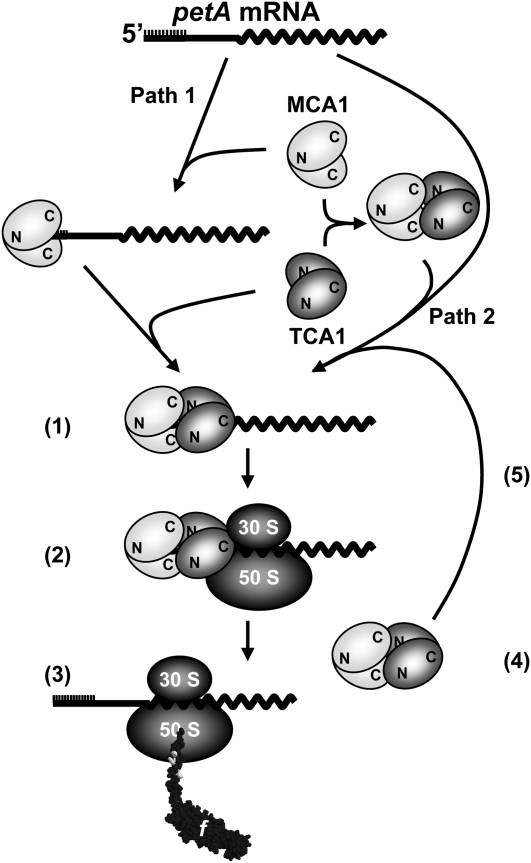
Our previous studies of the nuclear control of cytochrome f expression led to the genetic characterization of two nucleus-encoded factors, MCA1 and TCA1, whose combined actions in Chlamydomonas chloroplasts allow accumulation of the petA mRNA in a translatable form (Loiselay et al., 2008). Here, we further characterized the resulting petA gene expression system by taking advantage of the availability of complemented strains expressing tagged versions of each protein. The transient molecular interactions that may develop during the formation of the petA gene expression system are described in the working model presented in Figure 11.
Figure 11.
Working Model of the Biogenesis of the petA Gene Expression System.
MCA1 and TCA1 likely act in vivo as homo- and hetero-oligomers, whose formation would involve N- and C-terminal protein domains as indicated. MCA1 can bind to petA mRNA in the absence of TCA1 (Path 1) and also can interact with TCA1 in the absence of petA mRNA (Path 2). Both Paths 1 and 2 likely contribute in vivo to the formation of the ternary complex of ~600 kD (1) required for ribosome recruitment (2) and cytochrome f synthesis (3). Once translation is initiated, MCA1 and TCA1 would probably dissociate (4) from the translated mRNA and can be recycled for the formation of a new ternary complex (5) and new rounds of cytochrome f synthesis. Another interaction between MCA1 and unassembled cytochrome f is not shown in this figure but is detailed in Figure 12.
Our biochemical studies suggest that cytochrome f synthesis depends on a ternary complex (~600 kD) comprising MCA1 and TCA1 together with the petA transcript. Size exclusion chromatography experiments suggest that MCA1 interacts with the petA 5′UTR in vivo (Figure 4), even if it remains to be understood whether this interaction is direct or indirect. From its size of ~600 kD, this ternary complex may contain two copies of MCA1 and TCA1, as well as one or two copies of the petA mRNA. Indeed, MCA1 and TCA1 interact through the N-terminal moiety of TCA1 that recognizes MCA1, probably via its PPR-containing C-terminal domain. Since this interaction has been observed in S. cerevisiae by two-hybrid experiments, binding to petA mRNA is not a prerequisite for MCA1/TCA1 recognition; indeed, MCA1 and TCA1 can be coimmunoprecipitated in the absence of petA mRNA. These observations are consistent with the association of TCA1 and MCA1, after RNase treatment or in petA deletion strains, in heavy protein complexes that are probably heterogeneous in size, as shown by their broad distribution around 1200 kD. Moreover, the C-terminal region of TCA1, which does not interact with MCA1 in the yeast two-hybrid system, can still complement tca1 null mutants (see Supplemental Figure 2 online). Thus, MCA1–TCA1 interactions would not be strictly required for TCA1 binding to the petA 5′UTR in vivo nor for translation activation, even though the latter process would be less efficient than in the presence of MCA1.
It is of note that both MCA1 and TCA1 can interact with themselves in yeast: MCA1 develops homomeric interactions via its N-terminal moiety, whereas TCA1 does so through its C-terminal region. In the absence of their other two partners from the petA gene expression system, each of the two proteins is part of a protein complex of ~500 kD. These complexes could accommodate homotetramers of each factor, although the presence of additional proteins cannot be excluded. The ability of other trans-acting factors to oligomerize has been little investigated so far, with the exception of HCF152 that forms homodimers in vitro (Nakamura et al., 2003).
Very few examples of maturation/stability and translation factors acting on the same mRNA have been described to date. The NAC2 and RBP40 proteins that respectively govern the stability and the translation of the psbD transcript in Chlamydomonas interact as well, at least in the presence of the psbD mRNA (Schwarz et al., 2007), but their mode of action differs slightly from those of MCA1 and TCA1 since NAC2 seems to be strictly required for the recruitment of RBP40 onto the psbD 5′UTR and, therefore, for translation initiation (Ossenbühl and Nickelsen, 2000).
Whereas some RNA binding proteins are membrane embedded or show reversible association with the membrane (Zerges and Rochaix, 1998; Trebitsh et al., 2001; Ossenbühl et al., 2002; Zerges et al., 2002; Schult et al., 2007), MCA1 and TCA1, as most trans-acting factors (e.g., CRP1 [Fisk et al., 1999], NAC2 [Boudreau, 2000], MBB1 [Vaistij et al., 2000b], and TAB2 [Dauvillée et al., 2003]), are found in the chloroplast stroma. The functional significance of these contrasting distributions is still unclear but could reflect differences in the mode of action of nucleus-encoded factors: some may remain bound to polysomes, while others bind transiently to their target mRNA but dissociate once translation is initiated. As an example, NAC2 and RBP40 likely associate transiently with monosomes (Schwarz et al., 2007) but are not found in polysome fractions (Boudreau et al., 2000), suggesting that they are released from psbD transcript after translation initiation. We did not find, either, any evidence for the presence of MCA1 (or TCA1) in polysome fractions isolated according to Minai et al. (2006) from strains mH and tF (data not shown).
Unassembled Cytochrome f Induces MCA1 Degradation
In this article, we show that the degradation of MCA1 is regulated by its interaction with cytochrome f, namely, with the CES repressor domain that is located on its stroma-exposed C-terminal domain. This CES repressor domain drives MCA1 degradation when cytochrome f is in its unassembled state, i.e., transiently before cytochrome b6f assembly in a wild-type context or for a longer time period when assembly is prevented, as is the case in a strain lacking SUIV, an assembly partner of cytochrome f. Indeed, MCA1 accumulates to ~10-fold higher levels in the absence of cytochrome f C-terminal domain (strain mH {ΔpetA} or mH {petASt}) than in its presence (strain mH; Figure 6A). In the wild type, MCA1 is unstable, with a half-life in the hour range (Raynaud et al., 2007), but it becomes stable in the absence of cytochrome f (Figure 6B). Moreover, the degradation of MCA1 is triggered by the very same residues in the C-terminal tail that are responsible for CES (Choquet et al., 2003): MCA1 shows no notable degradation when the CES repressor domain is disrupted, either because of truncations (mH {fsol} or mH {f307St}) or point mutations (mH {f307S} or mH {fΔK}; Figure 7B). The CES repressor motif should, however, be exposed by unassembled cytochrome f to promote MCA1 degradation: When its concentration is kept very low, as in strains expressing the unstable apocytochrome f, MCA1 is accumulated to higher levels than in the wild type (Figure 8A). By contrast, in assembly-defective mutants, such as the mH {ΔpetD} strain, that accumulate long-lived unassembled CES repressor domains, MCA1 is less stable (Figure 8C) and hence is less accumulated (Figure 8B) than in the wild type, where the repressor domain is only transiently accessible before being shielded by the other subunits of the cytochrome b6f complex.
Whether the interaction between MCA1 and the unassembled CES repressor domain is direct or indirect remains to be determined. If MCA1 binds to unassembled cytochrome f, it may become accessible to membrane-embedded proteases that have little access to the stromal soluble form of MCA1. This proteolytic disposal when membrane bound would explain why it is poorly found in membrane fractions.
The interaction of MCA1 with unassembled cytochrome f does not depend on its prior association with either TCA1 or the petA mRNA: MCA1 is still degraded, even in the absence of TCA1, when cytochrome f is expressed from a chimeric gene whose 5′UTR is borrowed from the atpA gene. It should also be noted that TCA1, which is long lived in the chloroplast, is limiting for cytochrome f synthesis in complemented strains expressing low levels of this protein (Raynaud et al., 2007) but not in physiological conditions. By contrast, MCA1, which was previously shown to enhance translation (Loiselay et al., 2008), appears as a key player in the CES process for cytochrome f: A modified petA mRNA, even though stable in the absence of MCA1 because of a polyG track in its 5′UTR, is 10-fold less expressed than when MCA1 is present (Loiselay et al., 2008). This activation of translation by TCA1 in the absence of MCA1 is probably born by the C-terminal region of TCA1, which does not interact with MCA1 in the yeast two-hybrid system, but still complements tca1 null mutants.
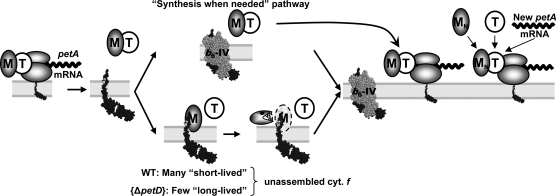
A New Model for the CES Process
Based on these new findings, we revisited our working model for the CES process driving cytochrome f production (Figure 12) to account for the proteolytic regulation of the amount of MCA1 available for cytochrome f translation as an exquisite nuclear control of a chloroplast-based regulation process.
Figure 12.
A Refined Model for the CES Process for Cytochrome f.
The biogenesis of the cytochrome b6f complex in the wild type (WT) “synthesis when needed” pathway; see text. In strains with disrupted CES repressor domain, such as f307St, MCA1 (M) cannot interact with unassembled cytochrome (cyt.) f (f) and is efficiently recycled for new rounds of petA mRNA translation (top part of the figure). In the wild type, free C-terminal tails of cytochrome f accumulate transiently before assembly (“many short-lived” unassembled cytochrome f; bottom part of the figure) and may interact with some MCA1 proteins that are thus targeted for degradation. By contrast, in strains defective for cytochrome b6f complex assembly, such as ΔpetD, C-terminal tails of cytochrome f, although synthesized in reduced amount, are particularly long lived and would target most neighboring MCA1 for degradation (“few long-lived” unassembled cytochrome f; bottom part of the figure). The right side of the figure illustrates the formation of new ternary complexes following import of newly synthesized MCA1 (MN). De novo biogenesis of cytochrome f in assembly-defective cytochrome b6f mutants, such as {ΔpetD}, would mostly rely on these new ternary complexes. For the sake of clarity, MCA1 and TCA1 (T) are drawn as monomers, even if they are likely oligomeric in vivo (Figure 11).
In the wild type, cytochrome f synthesis will obey a “synthesis when needed” mechanism: Most newly synthesized cytochrome f is rapidly assembled into cytochrome b6f complexes, preventing the binding of MCA1. The majority of MCA1 is therefore available for new rounds of translation (top part of the figure). The whole population of MCA1 follows this pathway in strains with disrupted CES repressor domain, such as f307St, in which MCA1 cannot interact with cytochrome f and is very efficiently recycled for new rounds of translation, accounting for the 3-fold increased rate of petA mRNA translation.
In the wild type, a fraction of free C-terminal tail of cytochrome f that transiently accumulates before assembly would nevertheless interact with some MCA1 proteins that thus are targeted for degradation. De novo–synthesized MCA1 imported from the cytosol has to be recruited (right part of the figure) to compensate for MCA1 degradation (bottom part of the figure). This pathway would prevail when the synthesis of cytochrome f exceeds the rate of production of its assembly partners. In particular, when assembly of cytochrome f into the cytochrome b6f complex is fully prevented, as in the {ΔpetD} mutant lacking subunit IV, the long-lived C-terminal tail would target most neighboring MCA1 for degradation. Next rounds of cytochrome f synthesis would thus rely principally on de novo–synthesized MCA1, a process 10 times less efficient than reusing preexisting MCA1 as the wild type does.
However, the half-life of MCA1, in the hour range, is much longer than the time it takes for cytochrome f synthesis and assembly, in the minute range. In strain {ΔpetD}, MCA1, after interacting with unassembled cytochrome f, may be transiently trapped, either on the long-lasting unassembled cytochrome f or within oligomeric forms, such as the TCA1-MCA1 complexes observed in our size exclusion chromatography experiments. These forms should be poorly competent for translation initiation, explaining why the steady state concentration of MCA1 in assembly-defective strains is less reduced (2- to 3-fold) than the rate of cytochrome f synthesis (10-fold) when compared with the wild-type situation. It is worth noting that, although 3 times more MCA1 accumulates in tca1 mutants than in the wild type, the accumulation of the petA transcript is nevertheless reduced 5-fold, suggesting that MCA1 is present mostly as homo-oligomers that poorly contribute to petA mRNA stabilization.
Altogether, our results further substantiate the regulatory role of nucleus-encoded factors in the expression of their organelle target genes. The regulatory function of MCA1 in the fine-tuning of cytochrome f synthesis involves a regulated proteolysis, triggered by its interaction with unassembled cytochrome f. Whether this mechanism can be generalized to other factors controlling the expression of other CES proteins remains to be determined.
METHODS
Strains, Growth Conditions, and Crosses
Wild-type (derived from 137c), mutant, and complemented strains of Chlamydomonas reinhardtii were grown in Tris-acetate-phosphate (TAP) medium, pH 7.2 (Harris, 1989), supplemented with 1% sorbitol when growing cw15 (cell wall-less) strains, under continuous light (5 to 10 μE·m−2·s−1) on a rotatory shaker (120 rpm). Strains tca1-8, tca1-8 cw15, and mca1-2 cw15, as well as the complemented derivatives (tF, tF cw15, and mH cw15; the two latter strains used for the isolation of intact chloroplasts and the preparation of stromal extracts) have been described by (Raynaud et al., 2007). The complemented strain expressing TCA1-Fl was the most right-located strain (indicated by an asterisk in Figure 3A from Raynaud et al. [2007]). The strain mca1-6, carrying a new allele of MCA1, is briefly described below and in Supplemental Figure 1 online. It was used as recipient strain for nuclear transformation experiments with plasmid plgMCA1-HA linearized by digestion with XbaI (Raynaud et al., 2007) to generate the complemented strain mH. We also used the chloroplast mutants ccsA-B6 (Xie et al., 1998), f307S (Choquet et al., 2003), and ΔpetD (Kuras and Wollman, 1994).
In this work, strains are named by their genotype. By convention, t indicates a strain carrying the tca1-8 mutant allele at the TCA1 locus, whereas m denotes a mutated MCA1 locus (either the mca1-2 allele in cell wall-less strains carrying the cw15 mutation or the mca1-6 allele in cell-walled strains). F indicates that the tca1-8 mutation had been complemented by a Flag-tagged version of TCA1, whereas H indicates the complementation of either mca1 allele by the HA-tagged version of MCA1. As an example, the mHt strain expresses the tagged version of MCA1 in the absence of TCA1. Chloroplast genotype, when relevant, follows the nuclear genotype and is written between brackets. The construction of the strains used in this study is described in Tables 1 and 2.
Crosses, described in Table 1, were performed according to Harris (1989). We used a genetic strategy to compare the expression of the tagged versions of MCA1 and TCA1 between different strains in Figures 5A and 5B. Indeed, we consistently observed in crosses involving mH or tF strains that the expression of the transgene remains constant (with variations below 15% of the level observed in the parental strain) in progeny with a similar genotype. Compare, for example, the parental strain mH to the mH progeny of the cross mH × mt in Figure 5C or the tF parental strain to the tF progeny from the cross tF × mt in Figure 5A. After transformation, the expression level of a transgene strongly depends on the site of the insertion but appears poorly sensitive to the genetic background, shuffled during the cross.
Description of the mca1-6 Mutation
Strain mca1-6 (CAL015.01.25), kindly provided by Rachel Dent, belongs to a library of mutants generated by random insertion of the ble cassette (Dent et al., 2005). This mutant requires acetate in the medium for growth, lacks accumulation of the petA mRNA (see Supplemental Figure 1B online), and could be complemented by the MCA1 gene, leading to strain mH (see Supplemental Figure 1A online). The mutation is tightly linked to the insertion of the ble cassette conferring resistance to up to 10 μg·mL−1 of zeocin. This allowed following the segregation of the mutation when analyzing the crosses listed in Table 1. The ble cassette is probably inserted in the 3′ part of the MCA1 gene, as no PCR product could be amplified with primers MCA1cod3 and MCA1rev3 (primers used in this work are listed in Supplemental Table 1 online), which respectively hybridize to the downstream third of the coding sequence and to the 3′UTR. The mutation is nevertheless not a deletion, as the reversion frequency, although very low, is not null.
Construction and Nucleic Acid Manipulation
Standard nucleic acid manipulations were performed according to Sambrook et al. (1989).
The petAStop Construct
The pWFStop construct contains a mutated petA gene that cannot be translated since the petA initiation codon was substituted by a Stop codon (TAG). It was created by a two-step PCR procedure (Higuchi, 1990): two pairs of primers, petACodMS/petAStopRev and petAStopCod/petARev2, allowed amplification from the template plasmid pWF (Kuras and Wollman, 1994) of two partially overlapping fragments that were mixed and used as templates in a third PCR with the external primers petACodMS and petARev2. The final amplicon, carrying the Stop codon, was digested by BglII and HindIII, two restriction sites on either side of the mutation, and cloned into plasmid pWF digested with the same enzymes to create plasmid pWFStop. The 5′psaA-aadA-3′rbcL cassette (Wostrikoff et al., 2004) was then inserted at the HincII blunt site, upstream of and in reverse orientation with respect to the petA coding sequence. This plasmid, pWFStop K, was sequenced before transformation in C. reinhardtii.
Construction of 5′atpA-Driven petA Genes
We first associated the 5′dAf chimera with an aadA cassette conferring resistance to the antibiotics spectinomycin and streptomycin to allow selection of transformed strains. The 4214-bp fragment isolated from plasmid pAFFF (Choquet et al., 1998), digested with AccI and ScaI, was ligated with the 5434- and 5565-bp fragments recovered from plasmids pWFB ATG12 (Rimbault et al., 2000) and pf307S (Choquet et al., 2003) digested with the same enzyme to yield plasmids p5′dAfK and p5′dAf307S.
Cloning of MCA1, TCA1, and MBB1 cDNAs in Yeast Two-Hybrid Vectors
MCA1. A 568-bp fragment encompassing the N terminus of MCA1 was amplified with primers MCA1_EcoRI_dir and MCA1_AscI_SalI_rev from the template cDNA clone CL01e04, obtained from the Kazusa DNA Research Institute (http://www.kazusa.or.jp/en/plant/), digested with EcoRI and SalI, and cloned into the corresponding sites of vectors pGAD424 and pGBT-9 (Clontech) to generate the pAD-5′MCA1 and pBD-5′MCA1 plasmids, respectively. Plasmid CL01e04 was then digested with AscI and NsiI to recover the last 2681 bp of the MCA1 cDNA, and this fragment was cloned into vectors pAD-5′MCA1 and pBD-5′MCA1 digested with AscI and PstI to yield plasmids pAD-MCA1 and pBD-MCA1. The NMCA1 constructs were generated by digestion of plasmids pAD-MCA1 and pBD-MCA1 with SalI and BamHI, Klenow treatment, and self-ligation.
TCA1. A 409-bp fragment encompassing nucleotides 814 to 1214 of the TCA1 coding sequence was PCR amplified using primers TCA1_EcoRI_814_dir and TCA1_1212_rev and the cDNA clone HCL001 h11_r, obtained from the Kazusa DNA Research Institute, as a template. The amplicon contained a SfiI site, while the TCA1_EcoRI_814_dir primer introduced an EcoRI site at its 5′ end and was digested by these two enzymes. A 2560-bp fragment of the TCA1 cDNA encompassing the C terminus of the protein was isolated from plasmid HCL001 h11_r digested by SfiI and XhoI. The two fragments were inserted by a trimolecular ligation into vectors pGAD424 and pGBT9 digested with EcoRI and SalI to yield plasmids pAD-CTCA1 and pBD-CTCA1, respectively.
To clone the full-length TCA1 cDNA into pGAD424 and pGBT-9 vectors, the first 1212 bp of TCA1 were amplified with primers TCA1_EcoRV_dir and TCA1_1212_rev and digested with EcoRV and SfiI. The last 2560 bp of the TCA1 cDNA were isolated as above by digestion of plasmid HCL001 h11_r with SfiI and XhoI. These two fragments were coligated into the SmaI and SalI sites of vectors pGAD424 and pGBT9 to generate plasmids pAD-TCA1 and pBD-TCA1, respectively. To create constructs pAD-NTCA1 and pBD-NTCA1 that only contain the first 1173 bp of the TCA1 coding sequence, plasmids pAD-TCA1 and pBD-TCA1 were digested with SfiI and NdeI, blunt-ended with Klenow and T4 DNA polymerase and self-ligated.
MBB1. A 410-bp fragment encompassing the N terminus of the MBB1 gene, PCR amplified with primers MBB1_dir_EcoRI-1 and MBB1_rev_410 from template pKS-MBB1 (Vaistij et al., 2000b) was digested by EcoRI and BamHI. The last 1572 bp of MBB1 were obtained by digesting the same plasmid pKS-MBB1 with BamHI and SalI. These two fragments were cloned between the EcoRI and SalI sites of vectors pGAD424 and pGBT-9 to yield clones pAD-MBB1 and pBD-MBB1, respectively.
Two-Hybrid Assays in Yeast
Yeast two-hybrid assays were performed in the PJ69-a strain (Leu−Trp−Gal4-His, and Gal4-Ade) that expresses the HIS2 and ADE genes under the control of the Gal4-UAS. Interactions between proteins of interest reconstitute a functional Gal4 transcription factor and therefore restore prototrophy for His and adenine. This was tested by assessing growth of double transformants on two media of increasing stringency: minimum medium depleted for His and supplemented with 3-AT (a competitive inhibitor of HIS2) or depleted for both His and adenine.
For yeast transformation, cells were grown overnight in complete YPDA medium. A 1-mL culture was centrifuged at 1000g for 1 min; cells were washed with 1 mL deionized water and resuspended in 20 μL deionized water in the presence of 50 μg of salmon sperm DNA and 1 μg of each plasmid. After addition of 200 μL PEG (PEG-4000 50%, 0.1 M lithium acetate, and 1× TE) and 20 μL DMSO, cells were incubated for 15 min at 30°C with shaking and then at 42°C for 15 min. After addition of 1 mL water, cells were harvested by centrifugation, washed once with 1 mL water, resuspended in 100 μL water, and plated on minimal medium lacking Leu and Trp since cotransformation with both pGAD424 and pGBT-9 derivatives restores the prototrophy for Leu and Trp.
Double transformants were resuspended in water to an OD600 of 1, and 10 μL of serial dilutions were spotted on the various selective media.
Transformation Experiments
Nuclear transformation of mca1-6, tca1-8, and mtF mutant strains was performed by electroporation, as described by Raynaud et al. (2007), with the following parameters: 10 μF/1200 V·cm−1. Transformants were selected for phototrophy on minimum medium (Harris, 1989) under high light (200 μE·m−2·s−1).
Chloroplast transformation experiments, listed in Table 2, were performed by tungsten particle bombardment (Boynton et al., 1988) as described earlier (Kuras and Wollman, 1994). For each transformation, at least four independent transformants were analyzed. Phenotypic variations between independent transformants proved negligible.
RNA Isolation and Analysis
RNA extraction and RNA gel blot analysis were performed as described by Drapier et al. (2002) with probes derived from coding sequences (Eberhard et al., 2002).
Protein Preparation, Separation, and Analysis
Protein isolation, separation, and immunoblot analyses were performed on exponentially growing cells (2 × 106 cells·mL−1) as described by Kuras and Wollman (1994). All immunoblots were repeated at least twice and performed on three independent transformants. Cell extracts were loaded on an equal chlorophyll basis, unless otherwise specified. Antibodies against the β-subunit of F1/CF1, the OEE2 subunit from the photosystem II oxygen-evolving complex, cytochrome f, Nac2, and GrpE have been described (Lemaire et al., 1986; de Vitry et al., 1989; Kuras and Wollman, 1994; Boudreau et al., 2000; Schroda et al., 2001). TCA1-Fl and MCA1-HA were detected by ECL using monoclonal antibodies, anti-Flag M2 (Sigma-Aldrich) and anti-HA.11 (Covance), and horseradish peroxidase–conjugated antibody against mouse IgG (Promega). Their accumulation (normalized to that of the F1β subunit from the mitochondrial ATP synthase as an internal standard) was, when required, quantified from scanned films with the Image-Quant software (Molecular Dynamics), according to Raynaud et al. (2007). Cytochrome f accumulation, normalized to that of the β-subunit of the mitochondrial ATP synthase as an internal standard, was quantified from phosphor imager scans of immunoblots revealed with 125I protein A or by ECL, using the ImageQuant software, as described by Choquet et al. (2003) and Raynaud et al. (2007).
Immunochase was performed on whole cells grown in TAP medium (2 × 106 cell·mL−1), supplemented at t = 0 with either cycloheximide (final concentration: 10 μg·mL−1), lincomycin (final concentration 500 μg·mL−1), or both. At the indicated time points, aliquots were removed, briefly chilled on ice, and processed as described above before loading on gels.
Stromal Preparations
Chloroplasts from cell wall–deficient strains were isolated according to Zerges et al. (2002). After limited cell lysis with 1% (w/v) saponin, extracts were loaded on a discontinuous (45/75%) Percoll gradient. Intact chloroplasts were recovered at the interface between the 45 and 75% Percoll solutions and washed once with isotonic buffer (0.3 M sorbitol, 5 mM MgCl2, and 10 mM Tricine, pH 7.8). To prepare stromal fractions, intact chloroplasts were osmotically lysed in hypotonic buffer (10 mM Tricine, pH 7.8; and 5× Roche protease inhibitors) by repeated pipetting. Broken chloroplasts were then centrifuged at 16,000g for 10 min at 4°C, and the concentration of stromal proteins in the supernatant was estimated by the Bradford method (Bio-Rad Quick Start Bradford Dye Reagent).
Gel Filtration Experiments on Stromal or Soluble Extracts
A 600-mL culture at 2 × 106 cells·mL−1 was centrifuged and resuspended in 3 mL of breaking buffer (5 mM HEPES-KOH, pH 7.8, 20 mM KCl, 10% glycerol, 0.5 g·L−1 heparin, and 5× Roche protease inhibitors). Cells were broken with a French press at 6000 p.s.i. and centrifuged at 346,000g for 20 min to pellet the membranes and debris. Two mL of the supernatant or of stromal extracts were loaded on a Sephacryl S500 column (GE Healthcare), and elution was performed at a rate of 700 μL·min−1, at 4°C, with a buffer containing 80 mM Tricine-KOH, pH 7.8, 200 mM KCl, 10 mM EDTA, 20 mM ε-aminocaproic acid, and 0.1× Roche protease inhibitors. Sixteen 5-mL fractions, eluted 40 mL after injection, were collected and concentrated by centrifugation on Amicon Ultra-15 filter units (cutoff: 50 kD) at 4500g for 20 min. Fraction volumes were then adjusted to 500 μL, out of which 80 μL were loaded on 12% acrylamide gels containing 8 M urea. Fraction 16 (smallest molecular mass) lacked any protein of interest and was not loaded on the gels. For RNase treatments, stromal preparations, prepared in breaking buffer lacking heparin, were incubated at 4°C with 2500 units·mL−1 of both RNase I and RNase A for 45 min under gentle and continuous shaking, prior to loading on the column.
Coimmunoprecipitations
A 400-mL culture at 2 × 106 cells·mL−1 was centrifuged and resuspended in 2 mL of breaking buffer (20 mM HEPES-KOH, pH 7.2, 150 mM NaCl, 10 mM KCl, 1 mM MgCl2, 10% glycerol, and 2× Roche protease inhibitors). Cells, broken by a French press at 6000 p.s.i., were centrifuged at 34,000g for 30 min to pellet membranes and debris. Five hundred microliters of the supernatant and 20 μL of beads coupled to anti-HA (HA11 Affinity Matrix; Covance) or anti-Flag (Flag immunoprecipitation kit; Sigma-Aldrich) antibodies were incubated for 1 h at 4°C, after which beads were washed three times with washing buffer (150 mM NaCl, 20 mM HEPES-KOH, pH 7.2, and 10% glycerol supplemented with 1× Roche protease inhibitors) and two more times with 10 mM Tris-HCl, pH 7.5. Bound proteins were detached by incubation for 1 min in 2% SDS at 95°C and analyzed by immunoblots.
Two-Step Centrifugation Procedure
The 400-mL cultures of mH and tF strains at 2 × 106 cells·mL−1 were centrifuged and resuspended in 3 mL of breaking buffer (5 mM HEPES-KOH, pH 7.8, 20 mM KCl, 10% glycerol, 0.5 g/L heparin, and 5× Roche protease inhibitors). Cells, broken by a French press (6000 p.s.i.), were centrifuged at 2100g for 5 min to remove unbroken cells, starch, and large debris. One milliliter of the supernatant (Extract E) was layered on top of a 1-mL 1.5 M sucrose cushion and ultracentrifuged at 272,000g for 30 min, giving rise to supernatant Sn, membrane fraction Mb, sucrose cushion C, and pellet P. All fractions were adjusted to 1 mL with breaking buffer. An aliquot of the whole-cell extract E was loaded on gel, after spectroscopic determination of chlorophyll concentration, together with equal volumes of fractions Sn and Mb, and analyzed by immunoblot. Cytochrome f and GrpE were used as membrane and soluble markers, respectively.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: TCA1, EF503563.1; MCA1, AF330231.1; MBB1, AJ296291.1; NAC2, AJ271460.1; and petA, FJ423446.1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Characterization of the mca1-6 and mH Strains.
Supplemental Figure 2. Accumulation of Cytochrome f, Detected by Immunoblots in the Wild Type and in the CTCA1 Strain (the tca1-8 Mutant Complemented with the C-Terminal Domain of TCA1).
Supplemental Table 1: Oligonucleotides Used in This Work.
Supplementary Material
Acknowledgments
We thank R. Dent for her kind gift of the mca1-6 strain, J.-D. Rochaix for providing antibody against the NAC2 protein and plasmid MBB1-pKS, J. Nickelsen, C. Schwarz, D. Picot, C. Breyton, D. Charvolin, S. Masscheleyn, and B. Miroux for their help with size exclusion chromatography experiments, and B. Bailleul, K. Wostrikoff, D. Drapier, S. Eberhard, and R. Kuras for stimulating discussions and/or critical reading of the manuscript. This work was supported by Centre National de la Recherche Scientifique/Université Pierre et Marie Curie, Unité Mixte de Recherche 7141, and the European Community, SunBioPath contract FP7-KBBE-2009-3-02. C.R. and A.B. were “Attachées Temporaires d’Enseignement et de Recherche” at Université Pierre et Marie Curie.
References
- Ackerman S.H., Tzagoloff A. (2005). Function, structure, and biogenesis of mitochondrial ATP synthase. Prog. Nucleic Acid Res. Mol. Biol. 80: 95–133 [DOI] [PubMed] [Google Scholar]
- Anderson S., et al. (1981). Sequence and organization of the human mitochondrial genome. Nature 290: 457–465 [DOI] [PubMed] [Google Scholar]
- Auchincloss A.H., Zerges W., Perron K., Girard-Bascou J., Rochaix J.-D. (2002). Characterization of Tbc2, a nucleus-encoded factor specifically required for translation of the chloroplast psbC mRNA in Chlamydomonas reinhardtii. J. Cell Biol. 157: 953–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balczun C., Bunse A., Hahn D., Bennoun P., Nickelsen J., Kück U. (2005). Two adjacent nuclear genes are required for functional complementation of a chloroplast trans-splicing mutant from Chlamydomonas reinhardtii. Plant J. 43: 636–648 [DOI] [PubMed] [Google Scholar]
- Barkan A., Goldschmidt-Clermont M. (2000). Participation of nuclear genes in chloroplast gene expression. Biochimie 82: 559–572 [DOI] [PubMed] [Google Scholar]
- Boudreau E., Nickelsen J., Lemaire S.D., Ossenbühl F., Rochaix J.-D. (2000). The Nac2 gene of Chlamydomonas encodes a chloroplast TPR-like protein involved in psbD mRNA stability. EMBO J. 19: 3366–3376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boynton J.E., et al. (1988). Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science 240: 1534–1538 [DOI] [PubMed] [Google Scholar]
- Cardol P., Remacle C. (2009). The mitochondrial genome. The Chlamydomonas Source Book, 2nd ed, Vol. 2, Harris E.E., Stern D.B., Whitman G., (New York, London: Academic Press, Elsevier; ), pp. 445–469 [Google Scholar]
- Choquet Y., Wollman F.-A. (2002). Translational regulations as specific traits of chloroplast gene expression. FEBS Lett. 529: 39–42 [DOI] [PubMed] [Google Scholar]
- Choquet Y., Wollman F.-A. (2009). The CES process. The Chlamydomonas Source Book, 2nd ed, Vol. 2, Harris E.E., Stern D.B., Whitman G., (New York, London, Amsterdam: Academic Press, Elsevier; ), pp. 1027–1064 [Google Scholar]
- Choquet Y., Stern D.B., Wostrikoff K., Kuras R., Girard-Bascou J., Wollman F.-A. (1998). Translation of cytochrome f is autoregulated through the 5′ untranslated region of petA mRNA in Chlamydomonas chloroplasts. Proc. Natl. Acad. Sci. USA 95: 4380–4385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet Y., Zito F., Wostrikoff K., Wollman F.-A. (2003). Cytochrome f translation in Chlamydomonas chloroplast is autoregulated by its carboxyl-terminal domain. Plant Cell 15: 1443–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauvillée D., Stampacchia O., Girard-Bascou J., Rochaix J.-D. (2003). Tab2 is a novel conserved RNA binding protein required for translation of the chloroplast psaB mRNA. EMBO J. 22: 6378–6388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent R.M., Haglund C.M., Chin B.L., Kobayashi M.C., Niyogi K.K. (2005). Functional genomics of eukaryotic photosynthesis using insertional mutagenesis of Chlamydomonas reinhardtii. Plant Physiol. 137: 545–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vitry C., Olive J., Drapier D., Recouvreur M., Wollman F.A. (1989). Posttranslational events leading to the assembly of photosystem II protein complex: A study using photosynthesis mutants from Chlamydomonas reinhardtii. J. Cell Biol. 109: 991–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Zamaroczy M., Bernardi G. (1986). The primary structure of the mitochondrial genome of Saccharomyces cerevisiae—A review. Gene 47: 155–177 [DOI] [PubMed] [Google Scholar]
- Drager R.G., Girard-Bascou J., Choquet Y., Kindle K.L., Stern D.B. (1998). In vivo evidence for 5′-->3′ exoribonuclease degradation of an unstable chloroplast mRNA. Plant J. 13: 85–96 [DOI] [PubMed] [Google Scholar]
- Drapier D., Girard-Bascou J., Stern D.B., Wollman F.-A. (2002). A dominant nuclear mutation in Chlamydomonas identifies a factor controlling chloroplast mRNA stability by acting on the coding region of the atpA transcript. Plant J. 31: 687–697 [DOI] [PubMed] [Google Scholar]
- Drapier D., Rimbault B., Vallon O., Wollman F.-A., Choquet Y. (2007). Intertwined translational regulations set uneven stoichiometry of chloroplast ATP synthase subunits. EMBO J. 26: 3581–3591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard S., Drapier D., Wollman F.-A. (2002). Searching limiting steps in the expression of chloroplast-encoded proteins: relations between gene copy number, transcription, transcript abundance and translation rate in the chloroplast of Chlamydomonas reinhardtii. Plant J. 31: 149–160 [DOI] [PubMed] [Google Scholar]
- Fisk D.G., Walker M.B., Barkan A. (1999). Molecular cloning of the maize gene crp1 reveals similarity between regulators of mitochondrial and chloroplast gene expression. EMBO J. 18: 2621–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanesi F., Soto I.C., Barrientos A. (2008). Cytochrome c oxidase biogenesis: New levels of regulation. IUBMB Life 60: 557–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foury F., Roganti T., Lecrenier N., Purnelle B. (1998). The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae. FEBS Lett. 440: 325–331 [DOI] [PubMed] [Google Scholar]
- Fox T.D. (1996). Genetics of Mitochondrial Translation. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Fromont-Racine M., Rain J.C., Legrain P. (1997). Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat. Genet. 16: 277–282 [DOI] [PubMed] [Google Scholar]
- Green-Willms N.S., Butler C.A., Dunstan H.M., Fox T.D. (2001). Pet111p, an inner membrane-bound translational activator that limits expression of the Saccharomyces cerevisiae mitochondrial gene COX2. J. Biol. Chem. 276: 6392–6397 [DOI] [PubMed] [Google Scholar]
- Haffter P., Fox T.D. (1992). Suppression of carboxy-terminal truncations of the yeast mitochondrial mRNA-specific translational activator PET122 by mutations in two new genes, MRP17 and PET127. Mol. Gen. Genet. 235: 64–73 [DOI] [PubMed] [Google Scholar]
- Haffter P., McMullin T.W., Fox T.D. (1991). Functional interactions among two yeast mitochondrial ribosomal proteins and an mRNA-specific translational activator. Genetics 127: 319–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris E.H. (1989). The Chlamydomonas Source Book: A Comprehensive Guide to Biology and Laboratory Use. (San Diego, CA: Academic Press; ). [DOI] [PubMed] [Google Scholar]
- Hattori M., Sugita M. (2009). A moss pentatricopeptide repeat protein binds to the 3′ end of plastid clpP pre-mRNA and assists with mRNA maturation. FEBS J. 276: 5860–5869 [DOI] [PubMed] [Google Scholar]
- Herrin D.L., Nickelsen J. (2004). Chloroplast RNA processing and stability. Photosynth. Res. 82: 301–314 [DOI] [PubMed] [Google Scholar]
- Higuchi R. (1990). Recombinant PCR. PCR Protocols: A Guide to Methods and Applications, Gelfand D.H., Innis M.A., Sninsky J.J., White T.J., (New York/London/Amsterdam: Academic Press, Elsevier; ), pp. 177–183 [Google Scholar]
- Islas-Osuna M.A., Ellis T.P., Marnell L.L., Mittelmeier T.M., Dieckmann C.L. (2002). Cbp1 is required for translation of the mitochondrial cytochrome b mRNA of Saccharomyces cerevisiae. J. Biol. Chem. 277: 37987–37990 [DOI] [PubMed] [Google Scholar]
- Keeling P.J. (2009). Role of horizontal gene transfer in the evolution of photosynthetic eukaryotes and their plastids. Methods Mol. Biol. 532: 501–515 [DOI] [PubMed] [Google Scholar]
- Klinkert B., Elles I., Nickelsen J. (2006). Translation of chloroplast psbD mRNA in Chlamydomonas is controlled by a secondary RNA structure blocking the AUG start codon. Nucleic Acids Res. 34: 386–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause K., Lopes de Souza R., Roberts D.G., Dieckmann C.L. (2004). The mitochondrial message-specific mRNA protectors Cbp1 and Pet309 are associated in a high-molecular weight complex. Mol. Biol. Cell 15: 2674–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchka M.R., Goldschmidt-Clermont M., van Dillewijn J., Rochaix J.-D. (1989). Mutation at the Chlamydomonas nuclear NAC2 locus specifically affects stability of the chloroplast psbD transcript encoding polypeptide D2 of PS II. Cell 58: 869–876 [DOI] [PubMed] [Google Scholar]
- Kuras R., Büschlen S., Wollman F.-A. (1995b). Maturation of pre-apocytochrome f in vivo. A site-directed mutagenesis study in Chlamydomonas reinhardtii. J. Biol. Chem. 270: 27797–27803 [DOI] [PubMed] [Google Scholar]
- Kuras R., Wollman F.-A. (1994). The assembly of cytochrome b6/f complexes: An approach using genetic transformation of the green alga Chlamydomonas reinhardtii. EMBO J. 13: 1019–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras R., Wollman F.-A., Joliot P. (1995a). Conversion of cytochrome f to a soluble form in vivo in Chlamydomonas reinhardtii. Biochemistry 34: 7468–7475 [DOI] [PubMed] [Google Scholar]
- Lemaire C., Girard-Bascou J., Wollman F.-A., Bennoun P. (1986). Studies on the cytochrome b6f complex. I. Characterization of the complex subunits in Chlamydomonas reinhardtii. Biochim. Biophys. Acta 851: 229–238 [Google Scholar]
- Loiselay C., Gumpel N.J., Girard-Bascou J., Watson A.T., Purton S., Wollman F.-A., Choquet Y. (2008). Molecular identification and function of cis- and trans-acting determinants for petA transcript stability in Chlamydomonas reinhardtii chloroplasts. Mol. Cell. Biol. 28: 5529–5542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullin T.W., Haffter P., Fox T.D. (1990). A novel small-subunit ribosomal protein of yeast mitochondria that interacts functionally with an mRNA-specific translational activator. Mol. Cell. Biol. 10: 4590–4595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W., Stoebe B., Goremykin V., Hapsmann S., Hasegawa M., Kowallik K.V. (1998). Gene transfer to the nucleus and the evolution of chloroplasts. Nature 393: 162–165 [DOI] [PubMed] [Google Scholar]
- Merendino L., Perron K., Rahire M., Howald I., Rochaix J.-D., Goldschmidt-Clermont M. (2006). A novel multifunctional factor involved in trans-splicing of chloroplast introns in Chlamydomonas. Nucleic Acids Res. 34: 262–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minai L., Wostrikoff K., Wollman F.A., Choquet Y. (2006). Chloroplast biogenesis of photosystem II cores involves a series of assembly-controlled steps that regulate translation. Plant Cell 18: 159–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monod C., Goldschmidt-Clermont M., Rochaix J.-D. (1992). Accumulation of chloroplast psbB RNA requires a nuclear factor in Chlamydomonas reinhardtii. Mol. Gen. Genet. 231: 449–459 [DOI] [PubMed] [Google Scholar]
- Nakamura T., Meierhoff K., Westhoff P., Schuster G. (2003). RNA-binding properties of HCF152, an Arabidopsis PPR protein involved in the processing of chloroplast RNA. Eur. J. Biochem. 270: 4070–4081 [DOI] [PubMed] [Google Scholar]
- Naithani S., Saracco S.A., Butler C.A., Fox T.D. (2003). Interactions among COX1, COX2, and COX3 mRNA-specific translational activator proteins on the inner surface of the mitochondrial inner membrane of Saccharomyces cerevisiae. Mol. Biol. Cell 14: 324–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickelsen J., Fleischmann M., Boudreau E., Rahire M., Rochaix J.-D. (1999). Identification of cis-acting RNA leader elements required for chloroplast psbD gene expression in Chlamydomonas. Plant Cell 11: 957–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickelsen J., van Dillewijn J., Rahire M., Rochaix J.-D. (1994). Determinants for stability of the chloroplast psbD RNA are located within its short leader region in Chlamydomonas reinhardtii. EMBO J. 13: 3182–3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossenbühl F., Hartmann K., Nickelsen J. (2002). A chloroplast RNA binding protein from stromal thylakoid membranes specifically binds to the 5′ untranslated region of the psbA mRNA. Eur. J. Biochem. 269: 3912–3919 [DOI] [PubMed] [Google Scholar]
- Ossenbühl F., Nickelsen J. (2000). cis- and trans-acting determinants for translation of psbD mRNA in Chlamydomonas reinhardtii. Mol. Cell. Biol. 20: 8134–8142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perron K., Goldschmidt-Clermont M., Rochaix J.-D. (1999). A factor related to pseudouridine synthases is required for chloroplast group II intron trans-splicing in Chlamydomonas reinhardtii. EMBO J. 18: 6481–6490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perron K., Goldschmidt-Clermont M., Rochaix J.-D. (2004). A multiprotein complex involved in chloroplast group II intron splicing. RNA 10: 704–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J., Bayraktar O.A., Prikryl J., Barkan A. (2009). Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J. 28: 2042–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud C., Loiselay C., Wostrikoff K., Kuras R., Girard-Bascou J., Wollman F.-A., Choquet Y. (2007). Evidence for regulatory function of nucleus-encoded factors on mRNA stabilization and translation in the chloroplast. Proc. Natl. Acad. Sci. USA 104: 9093–9098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimbault B., Esposito D., Drapier D., Choquet Y., Stern D., Wollman F.-A. (2000). Identification of the initiation codon for the atpB gene in Chlamydomonas chloroplasts excludes translation of a precursor form of the β subunit of the ATP synthase. Mol. Gen. Genet. 264: 486–491 [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. (1989). Molecular Cloning. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Schroda M., Vallon O., Whitelegge J.P., Beck C.F., Wollman F.-A. (2001). The chloroplastic GrpE homolog of Chlamydomonas: Two isoforms generated by differential splicing. Plant Cell 13: 2823–2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schult K., Meierhoff K., Paradies S., Töller T., Wolff P., Westhoff P. (2007). The nuclear-encoded factor HCF173 is involved in the initiation of translation of the psbA mRNA in Arabidopsis thaliana. Plant Cell 19: 1329–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz C., Elles I., Kortmann J., Piotrowski M., Nickelsen J. (2007). Synthesis of the D2 protein of photosystem II in Chlamydomonas is controlled by a high molecular mass complex containing the RNA stabilization factor Nac2 and the translational activator RBP40. Plant Cell 19: 3627–3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stampacchia O., Girard-Bascou J., Zanasco J.L., Zerges W., Bennoun P., Rochaix J.-D. (1997). A nuclear-encoded function essential for translation of the chloroplast psaB mRNA in chlamydomonas. Plant Cell 9: 773–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele D.F., Butler C.A., Fox T.D. (1996). Expression of a recoded nuclear gene inserted into yeast mitochondrial DNA is limited by mRNA-specific translational activation. Proc. Natl. Acad. Sci. USA 93: 5253–5257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm N.R., Kuras R., Büschlen S., Sakamoto W., Kindle K.L., Stern D.B., Wollman F.A. (1994). The petD gene is transcribed by functionally redundant promoters in Chlamydomonas reinhardtii chloroplasts. Mol. Cell. Biol. 14: 6171–6179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmis J.N., Ayliffe M.A., Huang C.Y., Martin W. (2004). Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 5: 123–135 [DOI] [PubMed] [Google Scholar]
- Trebitsh T., Meiri E., Ostersetzer O., Adam Z., Danon A. (2001). The protein disulfide isomerase-like RB60 is partitioned between stroma and thylakoids in Chlamydomonas reinhardtii chloroplasts. J. Biol. Chem. 276: 4564–4569 [DOI] [PubMed] [Google Scholar]
- Vaistij F.E., Boudreau E., Lemaire S.D., Goldschmidt-Clermont M., Rochaix J.-D. (2000b). Characterization of Mbb1, a nucleus-encoded tetratricopeptide-like repeat protein required for expression of the chloroplast psbB/psbT/psbH gene cluster in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 97: 14813–14818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaistij F.E., Goldschmidt-Clermont M., Wostrikoff K., Rochaix J.-D. (2000a). Stability determinants in the chloroplast psbB/T/H mRNAs of Chlamydomonas reinhardtii. Plant J. 21: 469–482 [DOI] [PubMed] [Google Scholar]
- Wostrikoff K., Choquet Y., Wollman F.-A., Girard-Bascou J. (2001). TCA1, a single nuclear-encoded translational activator specific for petA mRNA in Chlamydomonas reinhardtii chloroplast. Genetics 159: 119–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wostrikoff K., Girard-Bascou J., Wollman F.-A., Choquet Y. (2004). Biogenesis of PSI involves a cascade of translational autoregulation in the chloroplast of Chlamydomonas. EMBO J. 23: 2696–2705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Culler D., Dreyfuss B.W., Kuras R., Wollman F.-A., Girard-Bascou J., Merchant S. (1998). Genetic analysis of chloroplast c-type cytochrome assembly in Chlamydomonas reinhardtii: One chloroplast locus and at least four nuclear loci are required for heme attachment. Genetics 148: 681–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F., Morin C., Mitchell G., Ackerley C., Robinson B.H. (2004). The role of the LRPPRC (leucine-rich pentatricopeptide repeat cassette) gene in cytochrome oxidase assembly: Mutation causes lowered levels of COX (cytochrome c oxidase) I and COX III mRNA. Biochem. J. 382: 331–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerges W., Rochaix J.-D. (1998). Low density membranes are associated with RNA-binding proteins and thylakoids in the chloroplast of Chlamydomonas reinhardtii. J. Cell Biol. 140: 101–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerges W., Wang S., Rochaix J.-D. (2002). Light activates binding of membrane proteins to chloroplast RNAs in Chlamydomonas reinhardtii. Plant Mol. Biol. 50: 573–585 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.