Abstract
Neuronal activity and energy metabolism are tightly coupled processes. Recently, we found that nuclear respiratory factor 1 (NRF-1) co-regulates all subunits of cytochrome c oxidase (COX, representing oxidative energy metabolism) and glutamatergic neurochemicals, including NR1 (Grin1) and NR2B (Grin2b) of NMDA receptors, GluR2 (Gria2) of AMPA receptors, and neuronal nitric oxide synthase (Nos1). Moreover, all 10 nuclear-encoded COX subunit genes and 3 transcription factor genes for the 3 mitochondrial-encoded COX subunits are transcribed in the same transcription factory. The goal of the present study was to test our hypothesis that genomic loci for Grin1, Grin2b, Gria2, and Nos1 interact with those for COX at the transcriptional level. By means of chromosome conformation capture, interactions were found among all of these genes in neurons, but not in C2C12 muscle cells. COX subunit genes also did not interact with neurochemical genes not regulated by NRF-1, nor with genes for calreticulin, a non-mitochondrial protein. Depolarizing stimulation up-regulated interaction frequencies between COX and neurochemical genes, whereas impulse blockade with TTX or inhibition of COX with KCN down-regulated them in neurons. Thus, an efficient mechanism is in place for coordinating the transcriptional coupling of energy metabolism and glutamatergic neurotransmission at the molecular level in neurons.
Keywords: energy metabolism, excitatory neurochemicals, interactome, KCl depolarization, transcription factory, TTX
Introduction
Transcription factories are discrete sites within the nucleus where gene transcription is known to occur. These sites of active transcription are thought to contain RNA polymerase II molecules surrounded by transcription factors and loops of chromatin encoding actively expressing genes that are transcribed together (Jackson et al. 1998; Osborne et al. 2004; Zhou et al. 2006). Increasing evidence suggests that long-range interactions among the genes can be studied using chromosome conformation capture (3C), which generates novel ligation products between DNA sequences that are closely juxtaposed in vivo (Dekker et al. 2002; Miele and Dekker 2008). 3C employs formaldehyde crosslinking to trap physical interactions between disparate loci within the genome. Cells are then solubilized and chromosomes are digested with a chosen restriction enzyme and religated under conditions that favor intra- and inter-molecular ligation. After the reversal of crosslinking, the DNA is purified and interaction frequencies between specific chromosomal loci are determined by quantifying the amount of corresponding ligation product with polymerase chain reactions (PCR). Previously, co-regulated genes from one or two different chromosomes were found to preferentially cluster at specialized transcription factories (Spilianakis and Flavell 2004; Ling et al. 2006; Lomvardas et al. 2006; Schoenfelder et al. 2010). Recently, however, we reported that all 13 genomic loci of a multisubunit, multichromosomal, bigenomic enzyme, cytochrome c oxidase (COX), and its mitochondrial transcription factors interacted in the same transcription factory in neurons (Dhar et al. 2009a).
In neurons, energy metabolism as reflected by COX activity is tightly coupled to neuronal activity, which consumes the bulk of the energy for repolarizing membrane potentials after excitatory depolarizing activity (Wong-Riley 1989). A major excitatory neurotransmitter in neurons is glutamate (Fonnum 1984). Recently, we found that the same transcription factor, nuclear respiratory factor 1 (NRF-1), co-regulates all subunits of COX from the two genomes (Dhar et al. 2008) as well as critical neurochemicals of glutamatergic synaptic transmission, including NR1 (Grin1) and NR2B (Grin2b) subunits of N-methyl-D-aspartate (NMDA) receptors (Dhar and Wong-Riley 2009), GluR2 (Gria2) subunit of alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors (Dhar et al. 2009b), and neuronal nitric oxide synthase NOS1 (Nos1) (Dhar et al. 2009c). Such co-regulation ensures that energy supply keeps pace with energy demand of neuronal activity. Such coupling also begs the question, are the COX genes and glutamatergic neurochemical genes co-transcribed in the same transcription factory?
The goal of the present study was to test our hypothesis that genes critical for energy metabolism (COX) and glutamatergic transmission (Grin1, Grin2b, Gria2, and Nos1) that are co-regulated by NRF-1 interact in the same transcription factory in neurons. Our 3C results indicate that such interactions of multiple genes from multiple chromosomes did exist in primary neurons, thereby ensuring precise co-regulation of agents of energy metabolism and those of neuronal activity.
Materials and methods
In silico analysis of mouse COX subunit and neurochemical gene sequences
Genomic sequences, surrounding non-coding sequences, and chromosomal positions of murine genes for nuclear-encoded COX4i1 and COX8a subunits, NMDA receptor subunits Grin1, Grin2b, and Grin3a, AMPA receptor subunits Gria2 and Gria4, and nitric oxide synthase isoforms Nos1 and Nos3 were obtained from mouse genome database (Eppig et al. 2007). COX, Grin1, Grin2b, Grin3a, Gria2, Gria4, Nos1, and Nos3 subunit genes were independently verified by comparing the coding sequences with known cDNA sequences and confirming the presence of intron-exon structures. DNA sequences corresponding to each of these verified genes and its surrounding 10,000 bp region were analyzed for the presence of Bgl II restriction sites.
Primary cortical neuronal cultures and treatment with KCl or tetrodotoxin (TTX)
All experiments were carried out in accordance with the US National Institutes of Health Guide for the Care and Use of Laboratory Animals and the Medical College of Wisconsin regulations. All efforts were made to minimize the number of animals and their suffering.
Primary visual cortical neurons were prepared as described previously (Dhar et al. 2009a). Briefly, 1–2 day old Sprague–Dawley rat pups were anesthetized with CO2 and killed by decapitation. The brains were removed, cleansed of meninges and surface blood vessels, and the cortices were dissected, trypsinized, and triturated to dissociate individual neurons. Cells were plated onto poly-l-lysine- (Sigma, St. Louis, MO, USA) coated 100 mm dish at a density of ~5×106 cells/ml in Dulbecco’s minimal essential medium (DMEM) (Invitrogen, Carlsbad, CA, USA) with 10% fetal bovine serum (Hyclone, Logan, UT, USA) for 3 h. The medium was changed to neurobasal/B27 supplement (Invitrogen) and cultures were maintained at 37°C with 5% CO2 in a humidified incubator. Cytosine arabinoside (Ara-C) (Sigma) was added on the second day of plating neurons to inhibit the replication of non-neuronal cells. Neuronal cultures were maintained by replacing half of the medium every 5 days. Cells were cultured for 7–12 days before exposure to 20 mM KCl for 5h. TTX (Sigma) at a final concentration of 0.4 μM was added to the culture media from the 5th day after plating. Cultures were exposed to TTX for 6 days. All cells were harvested on the same day for each experiment and 3C samples were prepared for each group.
Control cultures were neither depolarized with KCl nor inactivated by TTX. Real time quantitative PCRs (RTqPCR) were carried out in a Cepheid Smart Cycler Detection system (Cepheid, Sunnyvale, CA, USA). iQ SYBR Green Supermix (Bio-Rad, Hercules, CA) was used following the manufacturer’s protocols and as described previously (Dhar et al. 2009a).
Cell culture and KCN treatment
Mouse N2a neuroblastoma cells (ATCC, Manassaa, VA) and C2C12 mouse myoblast cells (gift of Dr. John Lough, Medical College of Wisconsin, Milwaukee, WI) were grown in DMEM (Gibco, Carlsbad CA) supplemented with 10% Fetal Bovine Serum (FBS, Hyclone, Logan, UT). Cells were passaged every 2–3 days and harvested at 80–90% confluency for 3C experiments.
N2a cells were cultured for 3–4 days before exposure to 300 μM KCN for 5 hr. To test for the optimal dosage of KCN treatment, N2a cells were also exposed to 100 and 200 μM KCN for 5 hr, 24 hr, 3 days and 5 days. All cell studies were done in covered Petri dishes to avoid the evaporation of hydrogen cyanide. Control cultures were not treated with KCN. All cells were harvested on the same day for each experiment and 3C samples were prepared for each group.
Chromosome conformation capture (3C)
3C experiments were performed as described in published methods (Dhar et al. 2009a; Miele and Dekker 2009) with minor modifications. Briefly, N2a cells, primary neurons and C2C12 cells were harvested by trypsinization and resuspended in fresh culture media with 10% FBS. To cross-link chromatin, 1 × 107 N2a cells, 5 × 106 primary neurons, or 1 × 107 C2C12 cells in 10 ml of cell culture media were treated with 2% formaldehyde for 10 min at room temperature (RT). Cross-linking was stopped by quenching with 0.125 M glycine for 5 min at RT. Nuclei were isolated by incubating cells in a lysis buffer (10 mM Tris, pH 8.0, 10 mM NaCl, 0.2% NP-40) on ice with agitation for 30 min. Chromatin was subsequently released by treating isolated nuclei with 0.3% SDS and digested with 400 units of Bgl II (Fermentas, Hanover, MD) overnight. Afterwards, restriction enzyme was heat-inactivated and chromatin was diluted in T4 ligation buffer to achieve a low genomic DNA concentration of 2.5 ng/μl to facilitate intermolecular re-ligation. Chromatin re-ligation was performed by incubating diluted chromatin with 100 Weiss units of T4 ligase (Fermentas) for 4 h at 16°C. Following RNAse A (Roche, Nutley, NJ) digestion to remove RNA contaminant, the 3C chromatin was purified by phenol:chloroform extraction.
Generation of control templates
To generate individual PCR template controls for comparison with 3C product, COX4i1 or COX8a subunit genes were paired with Grin1, Grin2b, Grin3a, Gria2, Gria4, Nos1, and Nos3, respectively, as were the pairing of other genes or gene loci as shown in the controls’ lanes (left columns) of Figs. 1–2. For example, to generate the control template for COX4i1/Grin1, we used PCR to amplify the regions spanning a single Bgl II site of interest with 100–150 bps on either side for each subunit gene. Approximately 30 μg of each PCR product were digested with 300 units of BgI II overnight at 37°C. DNA was extracted using phenol chloroform and ethanol precipitation. Equimolar amounts of digested product (COX4i1 and Grin1) were ligated at a high concentration (~10 ng/μl for each product) to ensure formation of intermolecular cross-linked product. One primer of a given restriction fragment (COX4i1) was combined with a primer of a second (Grin1) in all pairwise combinations of ligated orientations (e.g., forward primer of COX4i1 with forward primer of Grin1; reverse primer of COX4i1 with forward primer of Grin1, etc.). For all other pairing of COX genes, the same steps were used to generate a large number of pairing among subunits and were used as control templates for comparison with 3C samples. The final chosen control template for each pairing had the same orientation as the 3C-generated product. By comparing the signals obtained by quantitative PCR for the 3C cross-linked templates versus the control templates, we corrected for the differences in amplification efficiency between primer sets and also for differences in signal intensities due to the size of the PCR products (as all of the primers were designed to give PCR products of 150–300 bp) (Table 2). All primers were designed to have an annealing temperature of 58–60°C, and they all yielded a product when used with the positive control templates. These control templates and 3C samples for each primer pair were performed during the same PCR run.
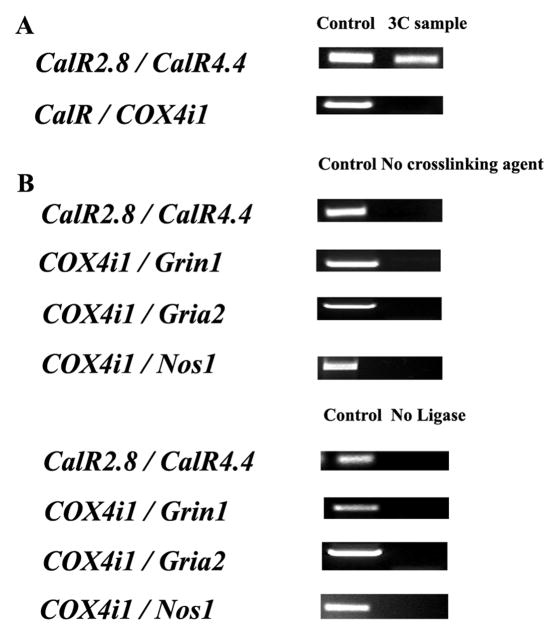
Fig. 1.
Positive and negative controls for chromosome conformation capture. All possible restriction fragments after Bgl II digestions were present in equimolar amounts. A. Two regions of calreticulin gene, CalR2.8 and CalR4.4, were cross-linked and represented a positive control. On the other hand, COX4i1 and CalR were two functionally unrelated genes, and they did not interact in our 3C reactions. B. CalR2.8 and CalR4.4, as well as COX4i1 or COX8a and Grin1, Gria2, or Nos1, respectively, did not interact in the absence of a cross-linking agent (formaldehyde) or T4 ligase. Experiments were performed in triplicates.
Fig. 2.
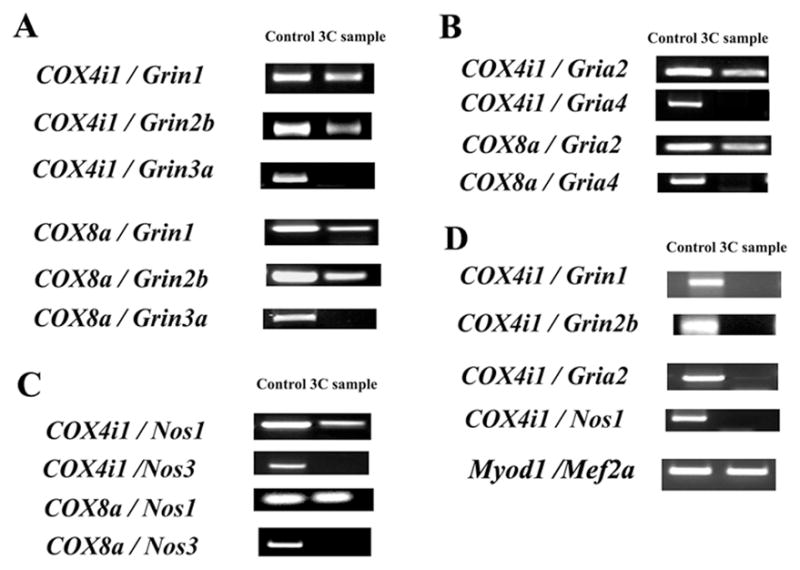
3C interactions in N2a and C2C12 cells between COX subunit genes and glutamatergic neurochemical genes. All interactive pairs between COX4i1 or COX8a and Grin1, Grin2b, Grin3a, Gria2, Gria4, Nos1, or Nos3 were tested by PCR targeting their cross-linking products. A–C. COX4i1 or COX8a consistently interacted with each of the Grin1, Grin2b, Gria2, or Nos1 genes in N2a cells. However, COX4i1 or COX8a did not yield any positive hybrid product when paired with Grin3a, Gria4, or Nos3. D. In C2C12 cells, COX4i1 did not interacted with each of the Grin1, Grin2b, Gria2, or Nos1 genes, whereas Myod1 and Mef2a showed consistent interactions. All experiments were performed in triplicates.
Table 2.
3C primers around Bgl II sites.
| Gene | Sequence | Amplicon length | Tm |
|---|---|---|---|
| COX4i1 | F 5′TTAGGACCTCTGCTCGCTCT 3′ R 5′TCAGCAGGTAAGAGCACTGG 3′ |
204 bps | 60°C |
| COX8a | F 5′CATTATGGTGGGCAACCTTC 3′ R 5′TCCATCTGCCATTCTCACTG 3′ |
261 bps | 60°C |
| Grin1 | F 5′CTCTGTGGCGTAGGAGGAAC 3′ R 5′TTTGTGTGGTGTGGTGTGTG 3′ |
220 bps | 60°C |
| Grin2b | F 5′TTCTTCCTTTCCCCTCTTCC 3′ R 5′TTCCAGCTTGTTGGCTTTCT 3′ |
271 bps | 60°C |
| Grin3a | F 5′CTTGGCGTACCTGAACTCGT 3′ R 5′ AAAGAAGGCCATGGAGAAGC 3′ |
275 bps | 60°C |
| Gria2 | F 5′ACAATGCGCCCAGAGAGA 3′ R 5′CCCAAGAATGGAGTTTAAGTCTTT 3′ |
250 bps | 60°C |
| Gria4 | F 5′TTGGGCATCAGTGAAGAAAA 3′ R 5′CTGTGTTGCCATGTGAGTCC 3′ |
315 bps | 60°C |
| Nos1 | F 5′CCCAAGGTCCTGAGTTCAAA 3′ R 5′TCCCTTCCTTCCTACCTTCG 3′ |
190 bps | 60°C |
| Nos3 | F 5′CTGCCTGTCATCACTCAGGA 3′ R 5′TATCCTTGGGCTCTCCAAAA 3′ |
166 bps | 60°C |
| Myod1 | F 5′TGACCATGTTCTTCCACTGC 3′ R 5′CAGGTGGACTGTATGCAGGA 3′ |
202 bps | 60°C |
| Mef2a | F 5′TCTGGCCCACTGACCTACAT 3′ R 5′CTCACAGCCGCCTAGAACTC 3′ |
206 bps | 60°C |
| CalR (+2.8 kb) | F 5′TTCTTCTACAGGGCGAGTGG 3′ R 5′GACTTGAGGTCTGCCAGGAA 3′ |
197 bps | 60°C |
| CalR (+4.4 kb) | F 5′CTGGCTGCTCCCAATAATGT 3′ R 5′GGGAGGGACAGAAGGAAAAG 3′ |
210 bps | 60°C |
A positive 3C control targeted an “internal” cross-linking product between two Bgl II sites located 1.6 kb apart within the coding region of calreticulin (CalR) gene (+2.8 kb and +4.4 kb relative to CalR transcriptional start point) (Tolhuis et al. 2002) (Fig. 1A).
Three 3C negative controls were employed: a) possible cross-linking between the CalR’s 4.4 kb Bgl II site and another Bgl II site on COX 4i1; b) no cross-linking agent (formaldehyde); and c) no ligase (Fig. 1A and B).
PCR assays
The PCR conditions used were very strict in order to detect only specific signals. PCR cycles used were: initial denaturing step for 2 min at 94°C followed by 36 repeats of a cycle with 30 sec at 94°C, 15 sec at 59°C, and 15 sec at 72°C, followed by a final step of 1 min at 72°C. Thirty six PCR cycles were sufficient to detect the specific PCR fragments generated after digestion and ligation that captured rare molecular phenomenon occurring in the nucleus. Cross-linking among COX and NMDAR, AMPAR and nNOS subunit gene loci in 3C samples were detected using semi-quantitative PCR. Specifically, a pair of primers targeting two different genes was used to amplify cross-linked DNA generated from 3C reactions. Each primer targeted a DNA region 100–150 bps from the Bgl II site and generated 200–300 bp amplicons. Primers were designed for all genes as shown in Table 2. The linear range of amplification was determined for 3C samples by serial dilution as described previously (Tolhuis et al. 2002; Lomvardas et al. 2006; Dhar et al. 2009a; Miele and Dekker 2009). An appropriate amount (150 μg) of 3C DNA within the linear range was added to each PCR reaction. PCR product was run on 2% agarose gel stained with ethidium bromide. All PCR reactions were performed in triplicates.
Results
Mouse chromosomal distribution of COX subunit and neurochemical genes
Chromosomal distributions of COX4i1, COX8a, Grin1, Grin2b, Grin3a, Gria2, Gria4, Nos1, and Nos3 genes in the mouse genome are summarized in Table 1. The ubiquitously expressed COX subunit genes 4i1 and 8a are on chromosomes 8 and 19, respectively. Grin1, Grin2b, and Grin3a are located on chromosomes 2, 6, and 4, respectively, whereas Gria2 and Gria4 are on chromosomes 3 and 9, respectively. Both Nos1 and Nos3 isoforms are located on the same chromosome 5 but are 94.42 Mb apart and do not form a gene cluster. Nos1 is located on the 5 F locus, whereas Nos3 is on the 5A3 locus. As all 10 nuclear-encoded COX subunit genes have recently been shown to interact with each other (Dhar et al. 2009a), the present study chose only COX4i1 and COX8a (representing the largest and smallest nuclear-encoded subunits of COX) and their interactions with each of the glutamatergic neurochemical genes to be tested in neurons.
Table 1.
Chromosomal location of murine genes arranged by chromosomal number.
| Chromosome | Gene |
|---|---|
| 2 | Grin1 |
| 3 | Gria2 |
| 4 | Grin3a |
| 5 | Nos1 |
| 5 | Nos3 |
| 6 | Grin2b |
| 8 | COX4i1 |
| 9 | Gria4 |
| 19 | COX8a |
Interactions between COX subunit genes and glutamatergic neurochemical genes
To identify interactions between two noncontiguous DNA regions, a positive control PCR was attained using primers targeting an “internal” cross-linkable product between two Bgl II sites located 1.6 kb apart within the coding region of calreticulin (CalR) gene (Fig. 1A). CalR and COX 4i1 are unrelated genes and would co-localize only at a background frequency. As Figure 1A illustrates, the internal cross-linking within the CalR gene was detectable by PCR of our 3C samples. In contrast, any chance cross-linking event between CalR and COX 4i1 was not detectable by our PCRs. In the absence of a cross-linking agent or T4 ligase, PCR products were not detected among CalR, COX4i1, Grin1, Gria2 and Nos1 genes (Fig. 1B).
Possible interactions between COX4i1 or COX8a and neurochemical genes in primary neurons were tested by PCR using primers for COX4i1, COX8a, Grin1, Grin2b, Grin3a, Gria2, Gria4, Nos1 and Nos3 genes (Table 2). COX4i1 and COX8a were individually documented to form a cross-linking pair with Grin1, Grin2b, Gria2, and Nos1 genes, respectively (Fig. 2A–C). All possible interactions among these genes were consistently detected by 3C in our PCR product. However, no cross-linking product between COX4i1 or COX8a and Grin3a, Gria4, or Nos3 was detected in our PCR product (Fig. 2A–C).
To determine whether these interactions are neuron-specific, C2C12 muscle cells were tested for possible interactions between COX4i1 and neurochemical genes. COX4i1 did not form a crosslinking pair with Grin1, Grin2b, Gria2, or Nos1 genes (Fig. 2D). However, muscle-specific transcription factors Myod1 and Mef2a did show positive interactions and were consistently detected by 3C in our PCR product (Fig. 2D).
The effect of KCl stimulation and TTX inactivation on gene interactions in primary neurons
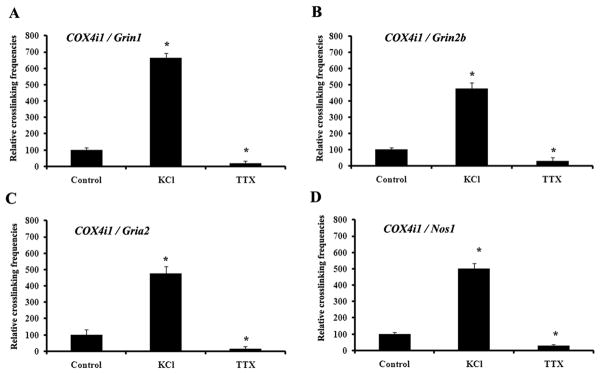
Primary neurons were subjected to depolarizing stimulation with 20 mM KCl for five hours or impulse blockade with TTX for 6 days to verify the effect of functional perturbations on COX4i1 interactions with Grin1, Grin2b, Gria2, and Nos1 genes, respectively. As shown in Fig. 3A–D, KCl treatment induced a 224% to 540% increase in crosslinking frequency (P < 0.05), whereas TTX blockade resulted in a 68 to 83% decrease in crosslinking frequency (P < 0.05) among the genes, as monitored by real-time quantitative PCR.
Fig. 3.
3C interactions in primary neurons exposed to KCl stimulation or TTX impulse blockade. KCl depolarization induced an up-regulation of cross-linking frequencies between COX4i1 and Grin1 (A), COX4i1 and Grin2b (B), COX4i1 and Gria2 (C), and COX4i1 and Nos1 (D), whereas TTX exposure for 6 days down-regulated the interactions of these genes (A–D). *, P < 0.05.
The effect of KCN treatment on gene interactions in neurons
To determine if specific inhibition of COX would disrupt the co-regulation of COX and neurochemical genes, N2a cells were subjected to 300 μM KCN, a specific inhibitor of COX, for 5 hr. As shown in Fig. 4A–D, KCN inhibition resulted in a 60 to 80% decrease in crosslinking frequency (P < 0.05) among the genes, as monitored by real-time quantitative PCR. Other concentrations of KCN and time points were tested. 100 μM and 200 μM KCN for 5 hr yielded no change in interactions, but at 24 hr the frequencies of interactions were reduced but did not reach statistical significance (data not shown). Cells started to die after 24 hr of 300 μM KCN, after 3 days of ≥ 200 μM KCN, or after 5 days of ≥ 100 μM KCN (data not shown). At 300 μM KCN for 5 hours, COX was inhibited but cells all survived.
Fig. 4.
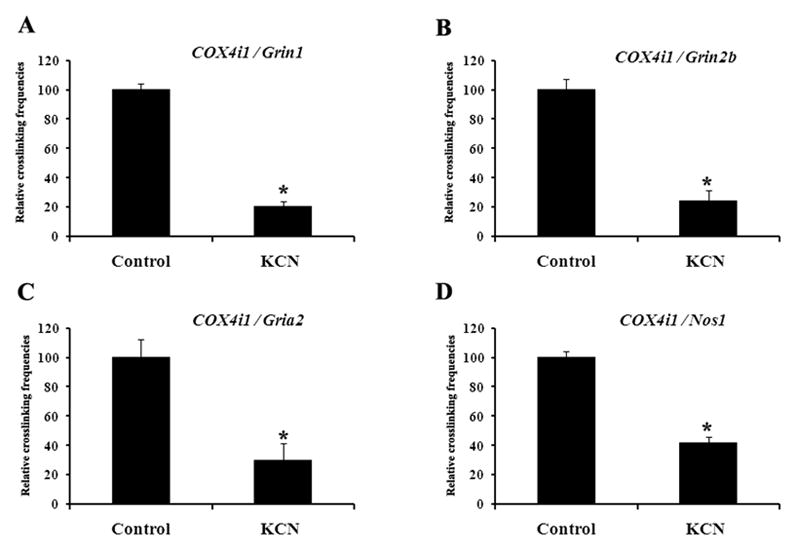
3C interactions in N2a cells exposed to 300 μM KCN treatment for 5 hr. KCN inactivation induced a significant down-regulation of cross-linking frequencies between COX4i1 and Grin1 (A), COX4i1 and Grin2b (B), COX4i1 and Gria2 (C), and COX4i1 and Nos1 (D). *, P < 0.05.
Discussion
The present study documents for the first time that genes responsible for energy metabolism (COX) and those involved in glutamatergic neurotransmission (Grin1, Grin2b, GluR2, and Nos1) are transcribed in presumably the same transcription factory in neurons. Such intermolecular interactions are further enhanced by depolarizing stimulation and suppressed by impulse blockade. The results substantiate our previous findings that these genes are co-regulated by the same transcription factor, nuclear respiratory factor 1 (Dhar and Wong-Riley, 2009; Dhar et al., 2009b,c). The interactions are also neuron-specific, as they were not present in C2C12 muscle cells, where Myod1 and Mef2a genes did interact (present study). Furthermore, specific inhibition of COX by KCN significantly reduced the frequency of interactions among these genes, indicating that, indeed, COX and neurochemical genes are co-regulated.
The 3C technique was developed based on the theory that actively co-expressed or co-regulated genes are physically localized in a “transcription factory” that contains copies of active RNA polymerases, transcription factors, and associated genomic loci undergoing transcription (Xu and Cook 2008; Schoenfelder et al. 2010). Initially, the 3C method was used in studying chromosomal conformation in yeasts (Dekker et al. 2002), but the approach was found to expose folding of complex gene loci and active chromatin hub in mammalian cells as well (Jackson et al. 1998; Osborne et al. 2004; Zhou et al. 2006). As shown in the present study, such interactions are not random but are highly specific. For example, genes that do not share a common transcription factor, in the present case, did not interact, such as the absence of ligation pairs between COX4i1 or COX8a with Grin3a, Gria4, or Nos3. Until recently, 3C has uncovered interactions only between genomic loci of the same chromosome or at most of two different chromosomes (Spilianakis and Flavell 2004; Ling et al. 2006; Lomvardas et al. 2006). However, we found interactions among 13 genomic loci from 11 chromosomes related to the bigenomic enzyme, COX, in neurons (Dhar et al. 2009a). Ten of the loci represent 10 nuclear-encoded COX subunit genes from 9 different chromosomes (Kadenbach et al. 1983; Eppig et al. 2007), whereas the other 3 denote mitochondrial transcription factors Tfam, Tfb1m, and Tfb2m genes important in transcribing the 3 mitochondrial-encoded COX subunit genes (Virbasius and Scarpulla 1994; Gleyzer et al. 2005; Scarpulla 2008a). The present study extended these findings to include interactions of COX subunit genes with genes of glutamatergic neurotransmission that are functionally coupled to energy metabolism.
COX is a critical oxidative enzyme without which oxidative metabolism cannot be carried to completion and mitochondrial ATP cannot be produced (Wikström et al. 1981; Kadenbach et al. 1983; Wong-Riley 1989). Recently, we found that all 13 subunits of cytochrome c oxidase derived from both the nuclear and mitochondrial genomes are regulated by the transcription factor NRF-1 (Dhar et al. 2008). NRF-1 is also known to be a key transcriptional activator of nuclear genes encoding a number of mitochondrial respiratory enzymes, including subunits of the five respiratory chain complexes (Scarpulla 2008a,b). Neuronal activity and energy metabolism are tightly coupled processes, and the bulk of energy in neurons is used to repolarize their membranes subsequent to glutamate-induced depolarization (Wong-Riley 1989). When neuronal activity is altered, such as with TTX-induced impulse blockade or KCl-stimulated depolarization, neuronal COX activity is down- or up-regulated to match the new energy demand (Wong-Riley 1989), and the expressions of glutamatergic NMDA receptors, AMPA receptor GluR2 subunits, and neuronal NOS are adjusted in parallel (Wong-Riley et al. 1998a,b; Wong-Riley and Jacobs 2002; Bai and Wong-Riley 2003; Dhar et al. 2009b). Remarkably, NRF-1 also directly regulates the expression of critical components of glutamatergic synapses, including NR1 and NR2B of the NMDA receptors, subunit GluR2 of the AMPA receptors, and neuronal nitric oxide synthase (Dhar et al., 2009b,c; Dhar and Wong-Riley 2009). Thus, the tight coupling of neuronal activity and energy metabolism is extended to the molecular level of regulation. The present study further documents that these functionally coupled metabolic and neurochemical genes that are regulated by the same transcription factor interact within the same presumed transcription factory in neurons. These interactions are specific, as genes not regulated by NRF-1 (Grin3a, Gria4, or Nos3) did not interact with COX subunit genes. Comparable associations between co-regulated mouse globin genes and their regulatory factors have recently been described in erythroid cells as a transcriptional interactome (Schoenfelder et al. 2010). Such transcription factories or interactomes are likely to be dynamic depending on the functional demands of the cell. As shown in the present study, interactions among functionally associated genes are up-regulated by KCl stimulation and down regulated by TTX. This presumably leads to the concurrent up- or down-regulation of the respective transcripts, including those for COX, Grin1, Grin2, Gria2, and Nos1 (Dhar et al. 2009a).
Besides NRF-1, nuclear respiratory factor 2 (NRF-2) also regulates all 13 COX subunit genes from the two genomes (Ongwijitwat and Wong-Riley 2005; Ongwijitwat et al. 2006). Moreover, an important coactivator of NRF-1 and NRF-2, known as peroxisome proliferator activated receptor γ coactivator 1α (PGC-1α) (Wu et al. 1999; Scarpulla 2008a,b), is also involved in the transcriptional regulation of COX in neurons (Liang and Wong-Riley 2006; Meng et al. 2007). It is possible that these factors participate in the regulation of the same and/or comparable neurochemicals of glutamatergic neurotransmission. Future studies will be directed at uncovering interactions among these critical factors of transcription in neurons.
Acknowledgments
Supported by NIH grant R01 EY018441.
Abbreviations
- 3C
chromosome conformation capture
- AMPA
alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid
- COX
cytochrome c oxidase
- Nos1
neuronal nitric oxide synthase
- Nos3
endothelial nitric oxide synthase
- NRF-1
nuclear respiratory factor-1
- NRF-2
nuclear respiratory factor 2
- PGC-1α
peroxisome proliferator-activated receptor-γ coactivator 1α
- Tfam
transcription factor A of mitochondria
- Tfb1m
transcription factor B1 of mitochondria
- Tfb2m
transcription factor B2 of mitochondria
References
- Bai X, Wong-Riley MT. Neuronal activity regulates protein and gene expressions of GluR2 in postnatal rat visual cortical neurons in culture. J Neurocytol. 2003;32:71–78. doi: 10.1023/a:1027380315902. [DOI] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Dhar SS, Wong-Riley MT. Coupling of energy metabolism and synaptic transmission at the transcriptional level: role of nuclear respiratory factor 1 in regulating both cytochrome c oxidase and NMDA glutamate receptor subunit genes. J Neurosci. 2009;29:483–492. doi: 10.1523/JNEUROSCI.3704-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar SS, Ongwijitwat S, Wong-Riley MT. Nuclear respiratory factor 1 regulates all ten nuclear-encoded subunits of cytochrome c oxidase in neurons. J Biol Chem. 2008;283:3120–3129. doi: 10.1074/jbc.M707587200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar SS, Ongwijitwat S, Wong-Riley MT. Chromosome conformation capture of all 13 genomic Loci in the transcriptional regulation of the multisubunit bigenomic cytochrome C oxidase in neurons. J Biol Chem. 2009a;284:18644–18650. doi: 10.1074/jbc.M109.019976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar SS, Liang HL, Wong-Riley MT. Nuclear respiratory factor 1 co-regulates AMPA glutamate receptor subunit 2 and cytochrome c oxidase: tight coupling of glutamatergic transmission and energy metabolism in neurons. J Neurochem. 2009b;108:1595–1606. doi: 10.1111/j.1471-4159.2009.05929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar SS, Liang HL, Wong-Riley MT. Transcriptional coupling of synaptic transmission and energy metabolism: role of nuclear respiratory factor 1 in co-regulating neuronal nitric oxide synthase and cytochrome c oxidase genes in neurons. Biochim Biophys Acta. 2009c;1793:1604–1613. doi: 10.1016/j.bbamcr.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppig JT, Blake JA, Bult CJ, Richardson JE, Kadin JA, Ringwald M. Mouse genome informatics (MGI) resources for pathology and toxicology. Toxicol Pathol. 2007;35:456–457. doi: 10.1080/01926230701310536. [DOI] [PubMed] [Google Scholar]
- Fonnum F. Glutamate: a neurotransmitter in mammalian brain. J Neurochem. 1984;42:1–11. doi: 10.1111/j.1471-4159.1984.tb09689.x. [DOI] [PubMed] [Google Scholar]
- Gleyzer N, Vercauteren K, Scarpulla RC. Control of mitochondrial transcription specificity factors (TFB1M and TFB2M) by nuclear respiratory factors (NRF-1 and NRF-2) and PGC-1 family coactivators. Mol Cell Biol. 2005;25:1354–1366. doi: 10.1128/MCB.25.4.1354-1366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Iborra FJ, Manders EM, Cook PR. Numbers and organization of RNA polymerases, nascent transcripts, and transcription units in HeLa nuclei. Mol Biol Cell. 1998;9:1523–1536. doi: 10.1091/mbc.9.6.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadenbach B, Jarausch J, Hartmann R, Merle P. Separation of mammalian cytochrome c oxidase into 13 polypeptides by a sodium dodecyl sulfate-gel electrophoretic procedure. Anal Biochem. 1983;129:51. doi: 10.1016/0003-2697(83)90586-9. [DOI] [PubMed] [Google Scholar]
- Liang HL, Wong-Riley MTT. Activity-dependent regulation of nuclear respiratory factor-1, nuclear respiratory factor-2, and peroxisome proliferator-activated receptor gamma coactivator-1 in neurons. Neuroreport. 2006;17:401–405. doi: 10.1097/01.wnr.0000204980.98876.11. [DOI] [PubMed] [Google Scholar]
- Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW, Cherry AM, Hoffman AR. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- Meng H, Liang HL, Wong-Riley M. Quantitative immuno-electron microscopic analysis of depolarization-induced expression of PGC-1alpha in cultured rat visual cortical neurons. Brain Res. 2007;1175:10–16. doi: 10.1016/j.brainres.2007.07.063. [DOI] [PubMed] [Google Scholar]
- Miele A, Dekker J. Long-range chromosomal interactions and gene regulation. Mol Biosyst. 2008;4:1046–1057. doi: 10.1039/b803580f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele A, Dekker J. Mapping Cis- and Trans- Chromatin Interaction Networks Using Chromosome Conformation Capture (3C) Methods Mol Biol. 2009;464:105–121. doi: 10.1007/978-1-60327-461-6_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongwijitwat S, Wong-Riley MTT. Is nuclear respiratory factor 2 a master transcriptional coordinator for all ten nuclear-encoded cytochrome c oxidase subunits in neurons? Gene. 2005;360:65–77. doi: 10.1016/j.gene.2005.06.015. [DOI] [PubMed] [Google Scholar]
- Ongwijitwat S, Liang HL, Graboyes EM, Wong-Riley MTT. Nuclear respiratory factor 2 senses changing cellular energy demands and its silencing down-regulates cytochrome oxidase and other target gene mRNAs. Gene. 2006;374:39–49. doi: 10.1016/j.gene.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, Fraser P. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev. 2008a;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- Scarpulla RC. Nuclear control of respiratory chain expression by nuclear respiratory factors and PGC-1-related coactivator. Ann N Y Acad Sci. 2008b;1147:321–334. doi: 10.1196/annals.1427.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, Eskiw CH, Luo Y, Wei CL, Ruan Y, Bieker JJ, Fraser P. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet. 2010;42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spilianakis CG, Flavell RA. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat Immunol. 2004;5:1017–1027. doi: 10.1038/ni1115. [DOI] [PubMed] [Google Scholar]
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- Virbasius JV, Scarpulla RC. Activation of the human mitochondrial transcription factor A gene by nuclear respiratory factors: a potential regulatory link between nuclear and mitochondrial gene expression in organelle biogenesis. Proc Natl Acad Sci U S A. 1994;91:1309–1313. doi: 10.1073/pnas.91.4.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikström M, Krab K, Saraste M. A Synthesis. Academic Press; New York: 1981. Cytochrome Oxidase. [Google Scholar]
- Wong-Riley MTT. Cytochrome oxidase: an endogenous metabolic marker for neuronal activity. Trends Neurosci. 1989;3:94–101. doi: 10.1016/0166-2236(89)90165-3. [DOI] [PubMed] [Google Scholar]
- Wong-Riley MTT, Jacobs P. AMPA glutamate receptor subunit 2 in normal and visually deprived macaque visual cortex. Vis Neurosci. 2002;19:563–573. doi: 10.1017/s0952523802195022. [DOI] [PubMed] [Google Scholar]
- Wong-Riley M, Anderson B, Liebl W, Huang Z. Neurochemical organization of the macaque striate cortex: correlation of cytochrome oxidase with Na+K+ATPase, NADPH-diaphorase, nitric oxide synthase, and N-methyl-D-aspartate receptor subunit 1. Neuroscience. 1998a;83:1025–1045. doi: 10.1016/s0306-4522(97)00432-6. [DOI] [PubMed] [Google Scholar]
- Wong-Riley MTT, Huang Z, Liebl W, Nie F, Xu H, Zhang C. Neurochemical organization of the macaque retina: effect of TTX on levels and gene expression of cytochrome oxidase and nitric oxide synthase and on the immunoreactivity of Na+K+ATPase and NMDA receptor subunit 1. Vision Res. 1998b;38:1455–1477. doi: 10.1016/s0042-6989(98)00001-7. [DOI] [PubMed] [Google Scholar]
- Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, Spiegelman BM. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- Xu M, Cook PR. Similar active genes cluster in specialized transcription factories. J Cell Biol. 2008;181:615–623. doi: 10.1083/jcb.200710053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou GL, Xin L, Song W, Di LJ, Liu G, Wu XS, Liu DP, Liang CC. Active chromatin hub of the mouse alpha-globin locus forms in a transcription factory of clustered housekeeping genes. Mol Cell Biol. 2006;26:5096–5105. doi: 10.1128/MCB.02454-05. [DOI] [PMC free article] [PubMed] [Google Scholar]