Abstract
Summary: We present an R based pipeline, ArrayExpressHTS, for pre-processing, expression estimation and data quality assessment of high-throughput sequencing transcriptional profiling (RNA-seq) datasets. The pipeline starts from raw sequence files and produces standard Bioconductor R objects containing gene or transcript measurements for downstream analysis along with web reports for data quality assessment. It may be run locally on a user's own computer or remotely on a distributed R-cloud farm at the European Bioinformatics Institute. It can be used to analyse user's own datasets or public RNA-seq datasets from the ArrayExpress Archive.
Availability: The R package is available at www.ebi.ac.uk/tools/rcloud with online documentation at www.ebi.ac.uk/Tools/rwiki/, also available as supplementary material.
Contact: angela.goncalves@ebi.ac.uk
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Deep sequencing of cDNA molecules (RNA-seq) is becoming the tool of choice for gene expression studies, often replacing microarrays to estimate gene expression levels, and rapidly superseding exon arrays in alternative splicing analysis (Wang et al., 2008) or tilling arrays in the identification of previously unknown transcribed regions (Guttman et al., 2010; Trapnell et al., 2010). In fact RNA-seq allows researchers to study phenomena that were previously beyond the reach of microarrays, such as allele specific expression (Montgomery et al., 2010). The popularity of the new sequencing methods for gene expression is attested by the numerous recent publications and by the increasing number of submissions to public data repositories such as ArrayExpress (AE) (Parkinson et al., 2008).
Many methods have been developed recently to tackle different aspects of RNA-seq data analysis, but combining them into a robust analysis pipeline is an inherently study-specific task and poses ongoing challenges. The configuration options used for each tool affect the others used downstream, making it necessary for bioinformaticians to have a thorough knowledge of each one of them and its internal workings. Furthermore, RNA-seq methods routinely generate tens of millions of raw sequence reads corresponding to hundreds of gigabytes of data, the analysis of which requires intensive computational processing steps that render the analysis impossible without the use of powerful servers. The gap between experimental throughput and processing speed is widening (Schatz et al., 2010), with the analysis component falling behind. In light of these considerations we have developed ArrayExpressHTS, an automated R/Bioconductor-based pipeline for pre-processing, expression estimation and data quality assessment of RNA-seq datasets. Starting from the raw read files it produces standard R objects containing expression levels for downstream analysis, and graphical HTML reports for data quality assessment. The pipeline has a choice of analysis methods and guides their configuration. Flexibility is provided for power users to adjust all aspects of the pipeline within the well-known and powerful R language.
2 METHODS
2.1 The analysis pipeline
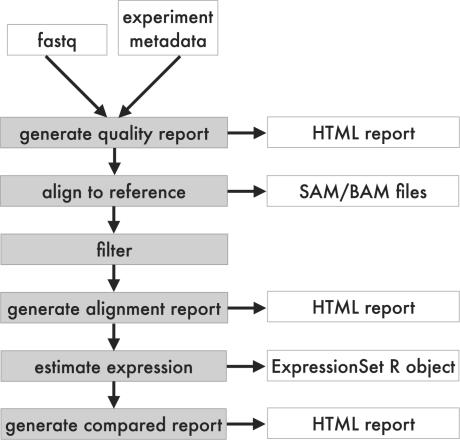
Running ArrayExpressHTS within R with default options is straightforward, with a simple call to function ArrayExpressHTS. Data analysis begins by obtaining the input raw read files and the corresponding experimental metadata. This experimental metadata serves to create a set of options used to configure the analysis and includes experimental protocol information such as the retaining of strand information and the insert size in paired-end reads; experiment design information including the links between files and sample properties (e.g. disease states); and machine-related information, such as the instrument used and quality scale (Fig. 1). Further options passed to the processing methods are documented in the pipeline and a set of reasonable options provided as default.
Fig. 1.
The ArrayExpressHTS analysis pipeline.
Once the necessary data is gathered, an HTML report is created, providing the investigator with diagnostics plots. Plots built upon the ones in the ShortRead (Morgan,M. et al., 2009) package are provided for individual samples, while additional ones are available for between-sample comparisons.
The analysis proceeds by mapping reads to a reference sequence (a genome or a transcriptome), with one of the available aligners: Bowtie (Langmead et al., 2009), TopHat (Trapnell et al., 2009) or BWA (Li and Durbin, 2009). The alignments are saved in the standard SAM format, converted to the BAM format (loadable into R) and sorted seamlessly with SAMtools (Li et al., 2009). References and index files for the aligners can be automatically downloaded from Ensembl (Hubbard et al., 2009) and created upon request, or manually provided in a local directory for alignment to custom references. It is also possible to provide custom alignment files by passing the appropriate options to the main R function (for more details see user documentation online).
Aligned reads can then be submitted to a set of filters including cutoffs on average base call qualities, number of allowed uncalled bases, by the size of runs of the same base at the head and tail of the reads, by read complexity as measured by a complexity score, by number of valid alignments, by number of reads with the same sequence, by genomic regions and other parameters.
In order to get the expression of features of interest (genes, transcripts or exons), aligned reads are either counted over those features and given as counts or as estimates calculated by the statistical method Cufflinks using a reference (Trapnell et al., 2010) or MMSEQ (Turro et al., 2011). The type of values returned can be controlled by passing normalization and standardization options to the main R function. The data is stored in a Bioconductor ExpressionSet object, grouping samples by factors, for downstream data analysis. A final HTML report is created providing information on the characteristics of the alignments and data (examples are again provided at the pipeline's website).
For users analysing their own experiments locally, the data must be available on the filesystem while the experiment metadata can be provided in the function call itself as a list of options or through a MAGE-TAB like set of files (Rayner et al., 2006). Optionally, publicly available datasets in AE can be re-analysed by providing the function with the experiment's accession number, upon which all raw data files and relevant metadata will be downloaded from the AE Archive and the European Nucleotide Archive.
The pipeline strives to minimize the amount of computation needed. Each time it is run on the same dataset with the exact same set of options, it will check at each step whether previous results, such as alignment files, reports or expression estimates, already exist and if this is the case the results will be retrieved instead of being re-processed. Runs of the pipeline with different options will have the results saved in different directories so the users can process the data in different ways for comparison.
2.2 Implementation on the R cloud
ArrayExpressHTS can also be run remotely on the R cloud at the European Bioinformatics Institute (EBI). Running it remotely on the cloud has several advantages while retaining the same function interface as the local implementation. In particular, it makes use of the distributed computing power of the EBI cluster. The only difference for the user is that the pipeline must be called from within the R Workbench graphical user interface for R, which provides a pooling framework for dispatching compute-intensive tasks to the server farm. This allows a multi-sample experiment to be automatically distributed among several computing nodes (the steps depicted in Figure 1 are run in parallel for each sample).
A possible way to use the R cloud at EBI to analyse the user's own data is to submit these data to the AE Archive, where the data will remain password protected (simple MAGE-TAB templates can be obtained from www.ebi.ac.uk/arrayexpress, curators will assist in file preparation and validation). When calling the function with an accession number the data already resides in a filesystem accessible to R cloud at EBI. In this way it is possible to use for analysis the data in AE (possibly in combination with users own data) without downloading gigabytes of raw sequences to the local computer. Finally, the R Workbench uses the most up to date package, reference sequences, aligner indexes and annotation for all major organisms, and access to the 3rd party software used, relieving the users from installing these on their own machines.
2.3 Performance and other use cases
We tested ArrayExpressHTS on publicly available human Solexa/Illumina RNA-seq datasets on the EBI R CLOUD. Experiment sizes ranged from ∼660 MB (1 sequencing run) to ∼160 GB (161 runs) with median analysis time (including data gathering) of ∼1.5 h per GB. Running the pipeline with default options gives us expression estimates for known transcripts, but other option settings make different types of analysis possible. We have tested the pipeline for a variety of alternative set-ups including: (i) haplo-isoform-level expression estimation, i.e. obtaining the expression estimates for the different haplotypes of transcripts containing heterozygotes (ii) and strand-specific expression estimation with Cufflinks for anti-sense and non-coding RNA discovery and quantification (possible when the experimental protocol used allows the identification of the read's strand of origin).
3 DISCUSSION
The ArrayExpressHTS package allows the users to obtain a standard Bioconductor ExpressionSet object containing expression levels from raw sequence files with a single R function call. The main benefit of ArrayExpressHTS is the ease of its use, running in the same way either on a local computer or on the R cloud and with private or public data. Written in R and available as open source, it also gives users the opportunity to extend and customize the pipeline for their needs. It can be used for individual data analyses or in routine data production pipelines, and it will be extended in the future to support other sequencing platforms, multiplexed data and the reporting of expression of non-annotated regions.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Wolfgang Huber, Ernest Turro, Anna Farne, Duncan Odom, Stephen Watt, Sarah Leigh-Brown, David Thybert, Johan Rung and John Marioni for their helpful comments.
Funding: The work was partly funded by the European Commission under the Seventh Framework Programme's (FP7) SLING (grant agreement number 226073) and SYBARIS collaborative projects (grant agreement number 242220).
Conflict of Interest: none declared.
REFERENCES
- Guttman M, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat. Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard TJP, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan M, et al. ShortRead: a bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics. 2009;25:2607–2608. doi: 10.1093/bioinformatics/btp450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery S, et al. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature. 2010;464:773–777. doi: 10.1038/nature08903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson H, et al. ArrayExpress update - from an archive of functional genomics experiments to the atlas of gene expression. Nucleic Acids Res. 2008;37:D868–D872. doi: 10.1093/nar/gkn889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner T, et al. A simple spreadsheet-based, MIAME-supportive format for microarray data: MAGE-TAB. BMC Bioinformatics. 2006;7:489. doi: 10.1186/1471-2105-7-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz M, et al. Cloud computing and the DNA data race. Nature Biotechnol. 2010;28:691–693. doi: 10.1038/nbt0710-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, et al. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, et al. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turro E, et al. Haplotype and isoform specific expression estimation using multi-mapping RNA-seq reads. Genome Biol. 2011;12:R13. doi: 10.1186/gb-2011-12-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.