Abstract
Summary: Dalliance is a new genome viewer which offers a high level of interactivity while running within a web browser. All data is fetched using the established distributed annotation system (DAS) protocol, making it easy to customize the browser and add extra data.
Availability and Implementation: Dalliance runs entirely within your web browser, and relies on existing DAS server infrastructure. Browsers for several mammalian genomes are available at http://www.biodalliance.org/, and the use of DAS means you can add your own data to these browsers. In addition, the source code (Javascript) is available under the BSD license, and is straightforward to install on your own web server and embed within other documents.
Contact: thomas@biodalliance.org
Since the early days of the draft human genome, web-based genome browsers such as Ensembl, GBrowse and the UCSC browser have been popular and important tools for biologists working on datasets large and small (Hubbard et al., 2009; Rhead et al., 2010; Stein et al., 2002). Despite increasing sophistication of data production and analysis methods, the importance of ‘eyeballing’ data to generate hypotheses or simply check the results of new analyses cannot be understated. Perhaps surprisingly, while genome browsing tools remain under very active development, the general approach taken by the major browsers has remained constant: a complex piece of server software reads databases, integrates information and creates bitmap image files that are displayed in the user's web browser window. This approach is reliable and places low demands on the end user's machine. However, it imposes serious limits on the level of interactivity, since any change in the display requires a full reload. There has been some interest in desktop applications, such as IGV (Robinson et al., 2011), which shift more work to the client side and increase the level of interactivity. Examples such as Apollo (Lewis et al., 2002) and Otterlace/Zmap (Searle et al., 2004) have become important tools to support the specialized activity of genome annotation. However, given the availability of reasonably functional tools which run in the web browser, the majority of users have been reluctant to install a heavyweight desktop client.
With that said, the web browser is not a static target, and the possibility of writing rich client applications in Javascript that run in web browsers has increased steadily over the last few years. In particular, making calls back to the server and fetching additional information without fully refreshing the page (‘AJAX’) has become a standard part of many web developers' toolkits. There have already been several attempts to improve genome browsing using a degree of client side interactivity. One approach uses large sets of pre-rendered image tiles, analogous to online mapping applications (Yates et al., 2007) while others have used CSS to perform limited drawing within the user's browser (Skinner et al., 2009). However, the former seriously limits the flexibility of the system—especially when it comes to adding new data—while the latter offers only limited graphical capabilities. We believe that the time is ripe for a new and uncompromising approach to combine the best features of web-based and desktop genome browsers.
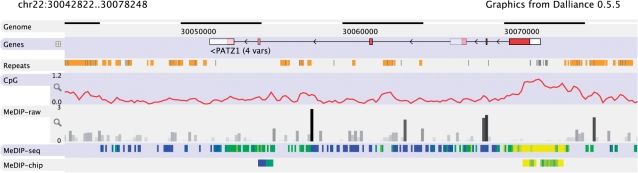
To address the limitations in current browsers, we have developed Dalliance, a new genomics tool which runs within the web browser but uses a number of recent technologies—most importantly, the W3C scalable vector graphic model (SVG)—to offer a level of interactivity which is competitive with desktop applications. Dalliance uses the standard distributed annotation system (DAS) protocol (Jenkinson et al., 2008), already used to add extra tracks to the web-based browsers including Ensembl and Gbrowse, to fetch sequence, annotations and alignments from servers around the network, before integrating the data into a smoothly-scrolling vector graphics display (Fig. 1).
Fig. 1.
Typical display from the Dalliance genome browser. Quantitative data from functional genomics experiments can be displayed in a range of styles, configured using DAS stylesheets.
Taking this approach offers a number of advantages. Following the DAS model means that researchers wanting to show their own data in a browser can easily do so without hosting their own copies of the reference genome and basic annotation databases, and allows data consumers to combine datasets in novel ways. Our choice of SVG gives a rich graphics platform comparable with APIs available on desktop platforms: we currently implement all the glyph types from the DAS stylesheet specification, and it would be straightforward to add more. SVG takes a scene graph approach (i.e. the rendering code builds a tree of objects describing what should be drawn, rather than calling rendering primitives directly), which means that smooth scrolling and export of high-quality vector graphics in SVG or, with some straightfoward server support, PDF format for publication or presentation are both straightforward. Because each ‘track’ of features is fetched using a separate—although usually concurrent—network request, and displayed as soon as the data arrives, one slow data source does not hold up the display of the rest of the data. And by fetching some excess data on each side of what is currently being displayed, the loading time can often be hidden from the user entirely.
In recognition that the reference genome sequences of most species are still moving targets, and that data released a few years ago may still be valuable today, even if it isn't actively maintained, we allow DAS sources targeted to one version of a genome (e.g. human NCBI36/hg18) to be remapped on the fly to another (e.g. GRCh37/hg19). The DAS protocol is used even for this. We use the standard DAS alignment command—although in a somewhat novel way—to retrieve the alignment data used for the mapping step, and metadata from the DAS registry tells the client when remapping is necessary.
Dalliance's model of accessing data from multiple sources (via DAS), rather than from a single central server, is also ideally suited to an emerging strategy for the handling of next generation sequencing datasets. The dramatically increased output and decreased costs of sequencing has led to it being used as an assay tool for a wide range of experiment types including genome variation, transcriptional expression, a readout for DNA protein binding. Traditionally, after mapping to a reference genome, sequence reads are processed and stored in a database in order to provide access for users. However, the high overhead in maintaining such databases does not scale to the amounts of data now being generated by such experiments. Kent et al., 2010 have instead proposed processing the output of mapping pipelines for an entire experiment into a single, indexed flat file, made accessible to users by simply placing it on a local web server. This is efficient since browsers can be configured to access portions of these flat files, only downloading data for the region currently being displayed. This indexed file based approach is in the process of being adopted as a submission standard to short read archives at EBI and NCBI using the BAM format implementing Kent's strategy (Li et al., 2009), which will make these files very widely available. As part of the Dalliance project, a lightweight BAMMappingSource has been developed to allow Dalliance to access such indexed files as if they were DAS sources.
Dalliance is written in standard Javascript, using APIs which are being standardized under the HTML5 banner. It places relatively high demands on the web browser, but is tested regularly and runs smoothly on Mozilla Firefox (3.6 or later), Safari (5.0 or later) and Google Chrome (5.0 or later). Microsoft Internet Explorer does not currently include SVG support, but it is promised for version 9, and we are optimistic that at that point it will be possible to support up-to-date versions of all major web browsers. The source code is freely available, and is written as a self contained object which can be inserted into almost any HTML page, so it can be combined with blogs, wikis, etc., or added as a browser component to an existing HTML database interface.
Dalliance is a practical genome browser that provides a smooth, interactive, user experience while handling large volumes of data. Since all data is loaded via DAS, it is straightforward to add additional data, or even a complete new genome dataset. The modern web browser offers a rich platform for data visualization, including complex scientific datasets, and we expect to see similar technological approaches deployed widely in the future.
ACKNOWLEDGEMENTS
Thanks to all the numerous testers who have provided testing and feedback for early versions of the Dalliance software, and to Jonathan Warren for updating the DAS registry to better support Javascript clients.
Funding: Wellcome Trust Research Career Development Fellowship (054523 to T.A.D.).
Conflict of Interest: none declared.
REFERENCES
- Hubbard TJ, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson AM, et al. Integrating biological data–the distributed annotation system. BMC Bioinformatics. 2008;9(Suppl. 8):S3. doi: 10.1186/1471-2105-9-S8-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, et al. Bigwig and bigbed: enabling browsing of large distributed datasets. Bioinformatics. 2010;26:2204–2207. doi: 10.1093/bioinformatics/btq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis SE, et al. Apollo: a sequence annotation editor. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0082. research0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhead B, et al. The UCSC genome browser database: update 2010. Nucleic Acids Res. 2010;38:D613–D619. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, et al. Integrative genomics viewer. Nature Biotech. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle SMJ, et al. The Otter annotation system. Genome Res. 2004;14:936–970. doi: 10.1101/gr.1864804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner ME, et al. Jbrowse: A next-generation genome browser. Genome Res. 2009;19:1630–1638. doi: 10.1101/gr.094607.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LD, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates T, et al. X:Map: annotation and visualization of genome structure for affymetrix exon array analysis. Nucleic Acids Res. 2007;36:D780–D786. doi: 10.1093/nar/gkm779. [DOI] [PMC free article] [PubMed] [Google Scholar]