Abstract
Motivation: High-throughput screens (HTS) by RNAi or small molecules are among the most promising tools in functional genomics. They enable researchers to observe detailed reactions to experimental perturbations on a genome-wide scale. While there is a core set of computational approaches used in many publications to analyze these data, a specialized software combining them and making them easily accessible has so far been missing.
Results: Here we describe HTSanalyzeR, a flexible software to build integrated analysis pipelines for HTS data that contains over-representation analysis, gene set enrichment analysis, comparative gene set analysis and rich sub-network identification. HTSanalyzeR interfaces with commonly used pre-processing packages for HTS data and presents its results as HTML pages and network plots.
Availability: Our software is written in the R language and freely available via the Bioconductor project at http://www.bioconductor.org.
Contact: florian.markowetz@cancer.org.uk
1 INTRODUCTION
In recent years several technological advances have pushed gene perturbation screens to the forefront of functional genomics. Combining high-throughput screening (HTS) techniques with rich phenotypes enables researchers to observe detailed reactions to experimental perturbations on a genome-wide scale. This makes HTS one of the most promising tools in functional genomics.
Although the phenotypes in HTS data mostly correspond to single genes, it becomes more and more important to analyze them in the context of cellular pathways and networks to understand how genes work together. Network analysis of HTS data depends on the dimensionality of the phenotypic readout (Markowetz, 2010). While specialized analysis approaches exist for high-dimensional phenotyping (e.g. Fröhlich et al., 2008), analysis approaches for low-dimensional screens have so far been spread out over diverse softwares and online tools like DAVID (Huang et al., 2009) or gene set enrichment analysis (GSEA; Subramanian et al., 2005).
Here we provide a software to build integrated analysis pipelines for HTS data that contain gene set and network analysis approaches commonly used in many papers (as reviewed by Markowetz, 2010). HTSanalyzeR is implemented by S4 classes in R (R Development Core Team, 2009) and freely available via the Bioconductor project (Gentleman et al., 2004). The example pipeline provided by HTSanalyzeR interfaces directly with existing HTS pre-processing packages like cellHTS2 (Boutros et al., 2006) or RNAither (Rieber et al., 2009). Additionally, our software will be fully integrated in a web-interface for the analysis of HTS data (Pelz et al., 2010) and thus be easily accessible to non-programmers.
2 AN INTEGRATED ANALYSIS PIPELINE FOR HIGH-THROUGHPUT SCREENING DATA
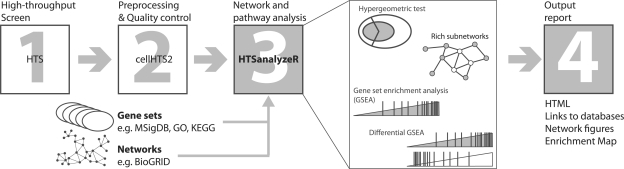
HTSanalyzeR takes as input HTS data that has already undergone pre-processing and quality control (e.g. by using cellHTS2). It then functionally annotates the hits by gene set enrichment and network analysis approaches (see Fig. 1 for an overview).
Fig. 1.
HTSanalyzeR takes as input HTS data that has already been pre-processed, normalized and quality checked, e.g. by cellHTS2. HTSanalyzeR then combines the HTS data with gene sets and networks from freely available sources and performs three types of analysis: (i) hypergeometric tests for overlap between hits and gene sets; (ii) gene set enrichment analysis (GSEA) for concordant trends of a gene set in one phenotype; (iii) differential GSEA to identify gene sets with opposite trends in two phenotypes; and (iv) identification of subnetworks enriched for hits. The results are provided to the user as figures and HTML tables linked to external databases for annotation.
Gene set analysis: HTSanalyzeR implements two approaches: (i) hypergeometric tests for surprising overlap between hits and gene sets, and (ii) gene set enrichment analysis to measure if a gene set shows a concordant trend to stronger phenotypes. HTSanalyzeR uses gene sets from MSigDB (Subramanian et al., 2005), Gene Ontology (Ashburner et al., 2000), KEGG (Kanehisa et al., 2006) and others. The accompanying vignette explains how user-defined gene sets can easily be included. Results are visualized as an enrichment map (Merico et al., 2010).
Network analysis: In a complementary approach strong hits are mapped to a network and enriched subnetworks are identified. Networks can come from different sources, especially protein interaction networks are often used. In HTSanalyzeR we use networks defined in the BioGRID database (Stark et al., 2006), but other user-defined networks can easily be included in the analysis. To identify rich subnetworks, we use the BioNet package (Beisser et al., 2010), which in its heuristic version is fast and produces close-to-optimal results.
Comparing phenotypes: A goal we expect to become more and more important in the future is to compare phenotypes for the same genes in different cellular conditions. HTSanalyzeR supports comparative analyses for gene sets and networks. Differentially enriched gene sets are computed by comparing GSEA enrichment scores or alternatively by a Wilcoxon test statistic. Subnetworks rich for more than one phenotype can be found with BioNet (Beisser et al., 2010).
3 CORE CLASSES AND METHODS
The two core S4 classes in HTSanalyzeR are ‘GSCA’ (Gene Set Collection Analysis) and ‘NWA’ (NetWork Analysis). S4 methods for both classes cover the following functions:
Preprocessing: The S4 methods ‘preprocess’ reformat the input data, e.g. by removing duplicated genes and converting annotations to Entrez identifiers. This step makes the objects of class ‘GSCA’ and ‘NWA’ ready for the following analyses.
Analyses: The S4 methods ‘analyze’ are provided for gene set and network analyses. Each method depends on several input parameters which can be defined by the user. HTSanalyzeR also implements a standard analysis option using default parameters that we have found to work well in many applications.
Visualization: GSEA random walks, enrichment maps and rich subnetworks can be viewed by S4 methods ‘viewGSEA’, ‘viewEnrichMap’ and ‘viewSubNet’, respectively.
Reporting: The analyses results of class ‘GSCA’ and ‘NWA’ can be reported seperately or together to HTML files using the S4 methods ‘report’ and ‘reportAll’, respectively. The output format was inspired by cellHTS2 and contains network figures as well as tables linked to external databases.
ACKNOWLEDGEMENTS
We thank Oliver Pelz and Michael Boutros for integrating HTSanalyzeR into the web-cellHTS interface. We thank Benilton Carvalho for helping to improve our code.
Funding: The University of Cambridge, Cancer Research UK; Hutchison Whampoa Limited. Fondation Philippe Wiener - Maurice Anspach (to C.T.).
Conflict of Interest: none declared.
REFERENCES
- Ashburner M, et al. Gene ontology: tool for the unification of biology. the gene ontology consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisser D, et al. BioNet: an R-Package for the functional analysis of biological networks. Bioinformatics. 2010;26:1129–1130. doi: 10.1093/bioinformatics/btq089. [DOI] [PubMed] [Google Scholar]
- Boutros M, et al. Analysis of cell-based RNAi screens. Genome Biol. 2006;7:R66. doi: 10.1186/gb-2006-7-7-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröhlich H, et al. Analyzing gene perturbation screens with nested effects models in R and Bioconductor. Bioinformatics. 2008;24:2549–2550. doi: 10.1093/bioinformatics/btn446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, et al. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, et al. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowetz F. How to understand the cell by breaking it: network analysis of gene perturbation screens. PLoS Comput. Biol. 2010;6:e1000655. doi: 10.1371/journal.pcbi.1000655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merico D, et al. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelz O, et al. web-cellHTS2: a web-application for the analysis of high-throughput screening data. BMC Bioinformatics. 2010;11:185. doi: 10.1186/1471-2105-11-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2009. [Google Scholar]
- Rieber N, et al. RNAither, an automated pipeline for the statistical analysis of high-throughput RNAi screens. Bioinformatics. 2009;25:678–679. doi: 10.1093/bioinformatics/btp014. [DOI] [PubMed] [Google Scholar]
- Stark C, et al. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]