Abstract
In order to investigate the cell death-inducing effects of rotenone, a plant extract commonly used as a mitochondrial complex I inhibitor, we studied cancer cell lines with different genetic backgrounds. Rotenone inhibits cell growth through the induction of cell death and cell cycle arrest, associated with the development of mitotic catastrophe. The cell death inducer staurosporine potentiates the inhibition of cell growth by rotenone in a dose-dependent synergistic manner. The tumor suppressor p53 is involved in rotenone-induced cell death, since the drug treatment results in increased expression, phosphorylation and nuclear localization of the protein. The evaluation of the effects of rotenone on a p53-deficient cell line revealed that although not required for the promotion of mitotic catastrophe, functional p53 appears to be essential for the extensive cell death that occurs afterwards. Our results suggest that mitotic slippage also occurs subsequently to the rotenone-induced mitotic arrest and cells treated with the drug for a longer period become senescent. Treatment of mtDNA-depleted cells with rotenone induces cell death and cell cycle arrest as in cells containing wild type mtDNA, but not formation of reactive oxygen species. This suggests that the effects of rotenone are not dependent from the production of reactive oxygen species. This work highlights the multiple effects of rotenone in cancer cells related to its action as an anti-mitotic drug.
Keywords: cell death, rotenone, p53, mitotic catastrophe, cell cycle
1. Introduction
In recent years, the highly sophistication of the mechanisms of programmed cell death which guard the organism against unwanted and potentially harmful cells has become evident. The Nomenclature Committee on Cell Death proposed recently unified criteria for the definition of cell death and of its different morphologies [1]. Nevertheless, one may find several terms to define different (or not so different) ways a cell has to die. The balance between cell division and cell death is of supreme importance for the development and maintenance of multicellular organisms. Deregulation of this balance can lead to pathological conditions, like cancer, neurodegenerative and immune system disorders [2].
One of the cell death processes is mitotic catastrophe. There is not a definitive accepted classification of mitotic catastrophe and its significance is still under debate but, currently, the concept refers to a series of events that result from premature or inadequate entry of cells into mitosis and represent an intermediate stage between a prolonged mitotic arrest and the set off of cell death [3]. It can be triggered either by chemical or physical stresses, namely by drugs that disturb the stability of microtubules [3, 4]. The molecular players involved in mitotic catastrophe are another matter of investigation.
Rotenone is a plant compound derived from the Leguminosae family used primarily as a mitochondrial complex I inhibitor. It induces cell death in several types of cells through the generation of reactive oxygen species (ROS) [5–7]. ROS formation is considered crucial, since treatment with antioxidants could abolish rotenone-induced cell death [6, 7]. Yet, it was recently shown that complex I inhibition is not enough to explain the rotenone effect [8, 9] and earlier studies showed that rotenone has another point of attack within the cells, the mitotic spindle [10]. Later work described that rotenone inhibits the assembly of the mitotic spindle by direct binding to tubulin [11–13], but the molecular mechanisms underlying the effects of rotenone remain elusive. Since compounds that inhibit the spindle assembly are inducers of mitotic catastrophe, it is reasonable to presume that rotenone can be such an inducer. Furthermore, studies in animal models of different forms of cancer demonstrated that rotenone possesses in vivo anticancer properties [14–17].
This prompted us to further investigate the effects of rotenone in cancer cells with different genetic backgrounds. In this work we show that rotenone induces mitotic catastrophe, mitotic slippage, cell death and cellular senescence in cancer cells and acts by two independent pathways. Our results also emphasize the involvement of p53 on the action of rotenone.
2. Materials and methods
2.1. Cell culture, reagents and antibodies
TPC-1 and 8505-C cells were maintained in RPMI-1640 medium supplemented with 10% fetal bovine serum, 100 U/ml penicillin, 100 µg/ml streptomycin and 1.25 µg/ml amphotericin B (all from Invitrogen). 143B ρ0 and CMPBR3 cells were maintained in DMEM (Invitrogen) supplemented with 50 µg/ml uridine (Sigma-Aldrich), 10% fetal bovine serum, 100 U/ml penicillin, 100 µg/ml streptomycin and 1.25 µg/ml amphotericin B. The 143B ρ0 cell line is derived from the parental osteosarcoma cell line 143B after depletion of mitochondrial DNA (mtDNA) using herpes simplex virus [18]. The CMPBR3 cybrid cell line was obtained through the fusion of 143B ρ0 cells with platelets from a healthy individual, as in reference [19]. All cell lines were kept at 37°C and 5% CO2. For the treatments with the drugs, cells were seeded and left to adhere for approximately 24 hours. Afterwards, the medium was replaced by fresh medium supplemented with the desired concentrations of the drugs. Stock solutions of rotenone and staurosporine were prepared in dimethyl sulfoxide (DMSO) at 50 mM and 10 mM, respectively. DMSO was used in all experiments as the vehicle (control).
Rotenone, sulphorhodamine B, propidium iodide, dihydroethidium (DHE) and dihydrorhodamine 123 (DHR123) were purchased from Sigma-Aldrich, DMSO from AppliChem, staurosporine from LC Laboratories, mounting medium with 4',6-diamidino-2-phenylindole (DAPI) from Vector Laboratories, CMXRos (MitoTracker Red) and rhodamine-phalloidin from Molecular Probes, nonyl acridine orange from PromoKine, 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal) from NZYTech and RNase A from Fermentas. The following antibodies were employed: p53 (clone DO-1) (Oncogene Science), phospho-p53 (Serine 15) (Cell Signaling Technology), SSBP1 (Abnova), OPA1 (BD Biosciences), cytochrome c and complex V subunit α (MitoSciences) and actin (Santa Cruz Biotechnology).
2.2. Analysis of cell growth
For the sulphorhodamine B assay, cells were fixed with 10% trichloroacetic acid and stained with 0.1% sulphorhodamine B. The plates were read in a microplate reader (Bio-Rad) at 560 nm. Triplicate wells per condition were evaluated in each experiment.
For cell cycle analysis, cells were fixed with ice-cold 70% ethanol for at least 4 hours and, after washing with PBS, stained with a solution containing 5 µg/ml propidium iodide and 100 µg/ml RNase A. Flow cytometry was performed in a Epics XL-MCL (Beckman-Coulter) and the results analyzed using FlowJo software (Tree Star).
The β-galactosidase assay was carried out as previously described [20]. Phase-contrast and β-galactosidase micrographs of cells were taken using a CKX41 (Olympus) inverted microscope.
2.3. Fluorescence microscopy
For observation of nuclei, cells were incubated with 0.075 M KCl (individual chromosomes can be microscopically identified and examined in much greater detail by incubating cells with a hypotonic solution), washed with PBS, fixed (with a 3:1 mixture of methanol and acetic acid at 4°C) and stained with DAPI. For staining of F-actin, cells growing in coverslips were fixed with 4% paraformaldehyde, washed with PBS, permeabilized with 0.2% Triton X-100 and incubated with phalloidin-rhodamine diluted 1:40 in PBS for 20 minutes.
For immunostaining for p53, cells were fixed and permeabilized, incubated 1 hour with blocking buffer (5% normal rabbit serum and 0.3% Triton X-100 in PBS), 1 hour with the primary anti-p53 (DO-1) antibody (1:40 dilution in 10% BSA + 0.3% Triton X-100 in PBS), washed (PBS-0.5% Tween-20-0.05% BSA) and 1 hour with the secondary rabbit anti-mouse antibody conjugated with fluorescein isothiocyanate (FITC) (Dako).
The cytochrome c release assay was performed according to the protocol provided by the manufacturer (MitoSciences). For observation and analysis, the Axio Imager 2 microscope and the AxioVision LE software (Zeiss) were used.
2.4. Western blotting
Whole lysates of cells were obtained first by incubation with RIPA buffer (50 mM Tris-HCl + 1% NP-40 + 150 mM NaCl + 2mM EDTA, ph 7.5), a protease inhibitor cocktail (Roche Applied Science) and a phosphatase inhibitor cocktail (Sigma-Aldrich), followed by centrifugation (14000 rpm, 15’, 4°C) and collection of the supernatant. The lysates were separated by SDS-PAGE and transferred to a nitrocellulose membrane. The membrane was blocked for 1 hour using a 5% non-fat dry milk or bovine serum albumin in PBS-0.5% Tween-20 solution and incubated with the appropriate primary and secondary antibodies. The protein bands were detected by chemiluminescence and X-ray film exposure (GE Healthcare).
2.5. Analysis of mitochondrial parameters
For evaluation of the production of ROS or mitochondrial membrane potential (ΔΨm), 5 µM DHE/10 µM DHR123 or 250 nM CMXRos, respectively, were added to the culture medium for 30 minutes. Then, cells were detached, washed two times and resuspended in PBS. For analysis of the mitochondrial mass, cells were first detached, washed two times and incubated in PBS containing 5 µM nonyl acridine orange. Samples were analyzed by flow cytometry.
For inspection of mitochondrial morphology, cells growing in coverslips were incubated with 250 nM CMXRos for 30 minutes at 37°C, fixed with 4% paraformaldehyde, washed twice in PBS and observed by fluorescence microscopy.
2.6. Statistical analysis
For quantification of synergy two measurements were employed – the combination index (CI) and the dose reduction index (DRI) [21] – using CalcuSyn software (Biosoft). The calculation is based on the median-effect equation derived from the mass-action law. Briefly, the median-effect equation describes dose-effect relationships, allows the construction of the median-effect plot and provides parameters for the calculation of CI and DRI. When two or more drugs are combined and the CI is calculated, CI < 1, = 1, and > 1 indicates synergism, additive effect, and antagonism, respectively. The DRI measures how many folds the dose of each drug in a synergistic combination may be reduced to a given effect level; the greater DRI value indicates a greater dose reduction [21]. The remaining statistical analysis of the data was performed using SPSS 13.0 (SPSS Inc.) by the non-parametric Mann-Whitney test, for comparisons between two groups. P-values < 0.05 were considered to be statistically significant.
3. Results
3.1. Rotenone inhibits the growth of cancer cells and induces cell death, cell cycle arrest, mitotic catastrophe and cellular senescence
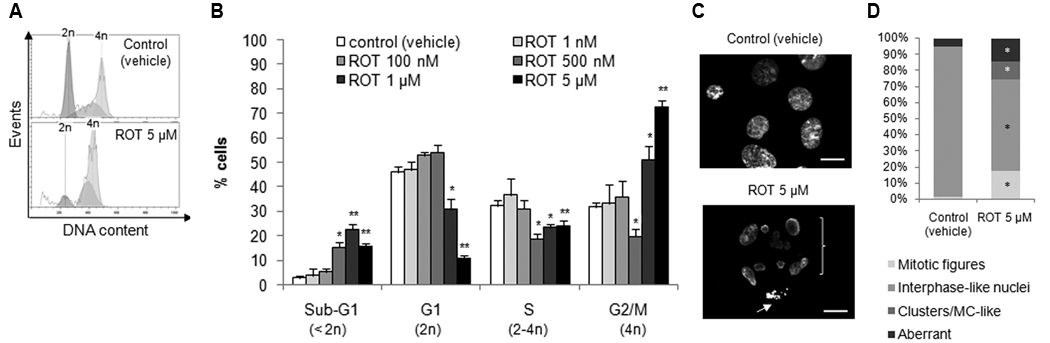
Treatment of TPC-1 cells for 24 hours with rotenone results in growth inhibition, as demonstrated in Supplemental Fig. 1A by phase-contrast micrographs of the cells. This growth inhibition occurred in a dose-dependent manner (Supplemental Fig. 1B). The calculated median-effect dose of the drug or IC50 is 26 µM (data not shown). We analyzed the cell cycle of TPC-1 cells treated for 24 hours with increasing concentrations of rotenone. Fig. 1A shows two representative histograms of the DNA content of TPC-1 cells treated with a control or with 5 µM rotenone and quantification of the data for the various concentrations tested are represented in Fig. 1B. Rotenone induces cell death, represented by the fraction of sub-G1 cells (15.78% for 5 µM rotenone vs 2.86% for the control; P = 0.006), and also cell cycle arrest, showed by the increase of the percentage of cells with 4N DNA content, typical of the G2 and M phases of the cycle (72.38% for 5 µM rotenone vs 31.83% for the control; P = 0.006). Thereby, rotenone-induced growth inhibition of TPC-1 cells can be explained by the induction of death and cell cycle arrest.
Figure 1.
Rotenone induces cell death, cell cycle arrest and mitotic catastrophe. (A) Representative histograms of the cell cycle analysis by propidium iodide staining of DNA of cells treated with a control (vehicle) (upper panel) or with rotenone (lower panel). (B) Quantification of the data for experiments like the one in (A) for different concentrations of the drug. (C) Nuclear morphology of 5 µM rotenone-treated cells was assessed by staining with DAPI; the bracket indicates a mitotic catastrophe-like cluster and the arrow points to a mitotic figure; scale bar: 10 µm. (D) Stained nuclei were counted and classified in four groups (mitotic figures, interphase-like nuclei, clusters/mitotic catastrophe-like and aberrant) based on their morphological features. All treatments in this figure were 24 hours long. Data is representative of at least three independent experiments. Data shown in (B) represents the mean ± standard error of mean (s.e.m.). *, P<0.05; **, P<0.01.
Evaluation of the nuclear morphology of TPC-1 cells by fluorescence microscopy shows that rotenone induces mitotic catastrophe (Fig. 1C). Scoring of the presence of interphase-like nuclei, mitotic figures, aberrant nuclei and clusters of nuclei typical of mitotic catastrophe (Fig. 1D) denotes a significant alteration in all the types of nuclei scored, including an increase in the number of mitotic figures (20.18% for 5 µM rotenone vs 1.22% for the control; P = 0.034 at 24 hours) and in the presence of syncytia of nuclei resembling those characteristic of mitotic catastrophe (12.71% for 5 µM rotenone vs 0% for the control; P = 0.019 at 24 hours). TPC-1 cells treated with rotenone for 5 days display a senescent-like phenotype [22], with a higher size, flattened aspect, numerous vacuoles and foci of heterochromatin (Fig. 2A). Increased expression of β-galactosidase (Fig. 2B and 2C; 14.41% for 5 µM rotenone vs 0.18% for the control; P = 0.004) supports the observation that cells become senescent after prolonged exposure to rotenone. Staining of F-actin with phalloidin revealed overexpression of the filaments and a considerable increase in cell size (Fig. 2D).
Figure 2.
Long rotenone treatments induce cellular senescence in TPC-1 cells. (A) Phase-contrast micrographs of rotenone (ROT)-treated TPC-1 cells; scale bar: 30 µm. The expression of β-galactosidase was evaluated by staining cells treated with rotenone with X-gal (B; scale bar: 50 µm) and by calculating the ratio of positive cells/total number of cells (C). (D) The F-actin filaments of rotenone-treated cells were stained with phalloidin-rhodamine; scale bar: 10 µm. The cells were exposed to rotenone for 5 days (after 3 days of treatment the above alterations were already observable). Data is representative of at least three independent experiments. Data shown in (C) represents the mean ± s.e.m. **, P<0.01.
3.2. Involvement of p53 in rotenone-induced cell death
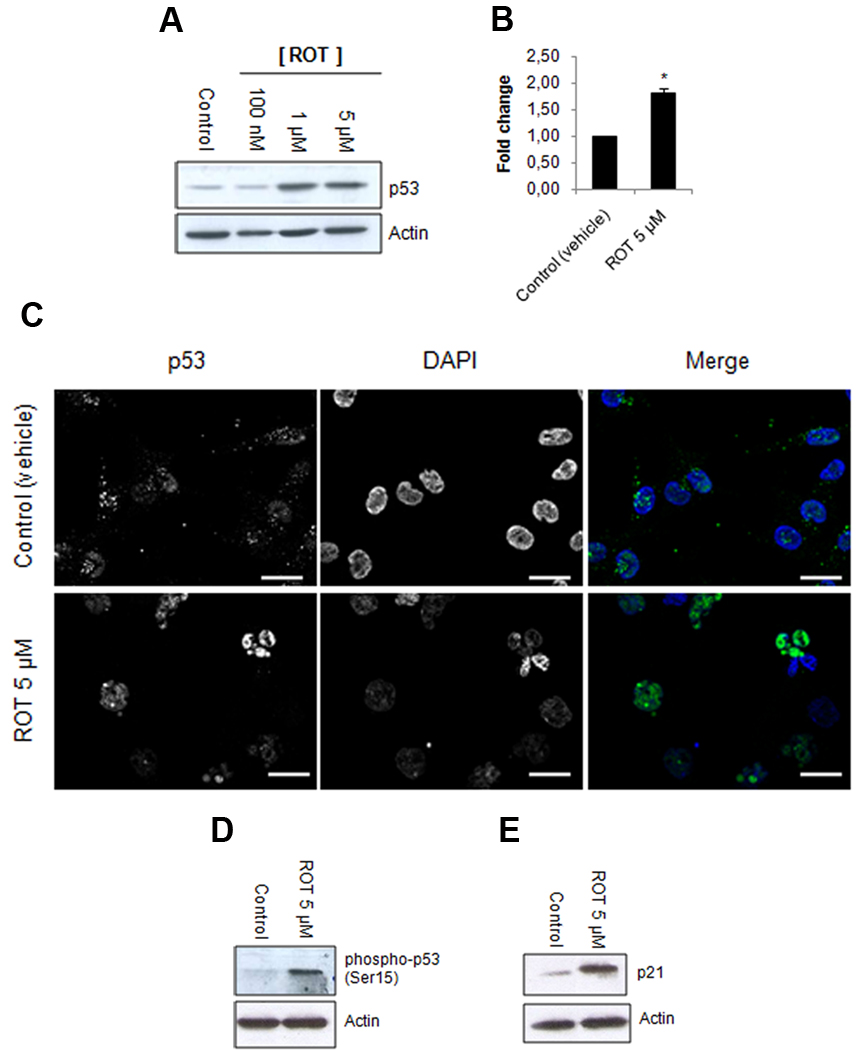
Since p53 is a molecule frequently responsive to cell stress, particularly cell cycle insults, we tested if rotenone was altering its state. Western blot analysis revealed that rotenone induces almost a two-fold increase in the levels of p53 (Fig. 3A and 3B; P = 0.037). In control-treated cells, p53 was located in the nucleus and dispersed in the cytoplasm (Fig. 3C). Conversely, in rotenone-treated cells, p53 was almost exclusively within the nucleus, including mitotic catastrophe-like nuclei (Fig. 3C). Phosphorylation of p53 at its serine 15 residue is a key step during the p53 activation process [23, 24]. Rotenone-treated TPC-1 cells evidenced an increase in the phosporylation of p53 in serine 15 (Fig. 3D). Therefore, rotenone induces a nuclear accumulation of p53 together with an increase in the levels of the protein and possibly its activation, since its phosphorylation at serine 15 was enhanced by rotenone. We also studied the expression of the cyclin-dependent kinase inhibitor p21, which is a p53 target-gene that regulates cell cycle progression at G1 [25, 26]. Treatment with rotenone lead to an increase in the expression of p21 (Fig. 3E). We did not detect alterations in other molecules tested by western blot, specifically Chk2, Bax, XIAP and cyclin D1 (data not shown).
Figure 3.
Rotenone increases the expression, phosphorylation of serine 15 and nuclear accumulation of p53 in TPC-1 cells. (A) Whole lysates of cells treated with rotenone (ROT) were immunoblotted for p53 and quantification of data (B) was determined by relative intensity (normalized to actin). (C) Cells treated with rotenone were stained for p53 (left column and green in the right column) and with DAPI for nuclei (middle column and blue in the right column); scale bar: 10 µm. (D–G) Whole lysates of cells treated with rotenone were immunoblotted for p53 phospho-serine 15 (D) and p21 (E). The rotenone treatments in this figure correspond to a 24 hours period. Immunoblotting against actin served as a loading control. Data is representative of at least three independent experiments. Data shown in (B) represents the mean ± s.e.m. *, P<0.05.
To further study the involvement of p53, we analyzed the outcome of the treatment with rotenone in 8505-C cells. These cells harbor a point mutation at the DNA-binding domain of TP53 that translates to the substitution Arg248Gly [27], leading to a non-functional protein [28]. The basal cell cycle profile of TPC-1 and 8505-C cells is similar (Supplemental Fig. 2). The cell cycle of 8505-C cells after 24 hours of treatment with 5 µM rotenone is arrested as in TPC-1 cells, as demonstrated by the percentage of cells with a 4N DNA content (80.75% for 5 µM rotenone vs 35.28% for the control; P = 0.05) (Fig. 4A and 4C). Analysis of the nuclear morphology of rotenone-treated 8505-C cells also revealed the presence of mitotic catastrophe-like clusters (Fig. 4B).
Figure 4.
Comparison of the rotenone effects in 8505-C and TPC-1 cells. (A) Quantification of data regarding the cell cycle analysis of 8505-C cells by DNA staining with propidium iodide after a 24 hours treatment with 5 µM rotenone. (B) Nuclear morphology of control (vehicle)-treated (upper panel) or 5 µM rotenone-treated 8505-C cells was assessed by staining with DAPI; scale bar: 10 µm. (C–D) Comparison of the cell cycle of TPC-1 and 8505-C cell lines upon treatment with 5 µM rotenone for 24 hours (C) or 48 hours (D). Data is representative of at least three independent experiments. Data shown in (A), (C) and (D) represents the mean ± s.e.m. *, P<0.05; **, P<0.01.
It is currently accepted that mitotic catastrophe is a biological state that precedes cell death [3]. Thereby, we tested if there is induction of cell death after the mitotic catastrophe-state induced by the drug. As depicted in Fig. 4C and 4D, in TPC-1 cells, there was a dramatic increase in the percentage of hypodiploid cells from 24 hours to 48 hours of treatment (15.78% at 24 hours vs 57.70% at 48 hours; P = 0.009) and a decrease in the percentage of cells with a DNA content typical of G2/M (72.38% at 24 hours vs 18.48% at 48 hours; P = 0.009). In contrast, p53-mutant 8505-C cells remain arrested in the cell cycle after 48 hours of treatment and undergo death to a lesser extent than TPC-1 cells (Fig. 4D; TPC-1 vs 8505-C cells, P = 0.009 for sub-G1 cells and P = 0.008 for G2/M cells). Taken together, these results suggest that a functional p53, although not required for the rotenone-induced cell cycle arrest and mitotic catastrophe, plays a role in the extensive cell death that occurs afterwards.
There are other differences between these two cell lines. In contrast to TPC-1, in 8505-C cells p53 is already located in the nucleus even without rotenone treatment (Supplemental Fig. 3). 8505-C also undergo cellular senescence after prolonged treatment with rotenone, as evidenced by typical morphological alterations (Supplemental Fig. 4A) and by a significant increase in the number of β-galactosidase-positive cells (2.21% for 5 µM rotenone vs 0.13% for the control; P = 0.001; Supplemental Fig. 4B and 4C). However, the percentage of positive cells is significantly higher in TPC-1 than in the 8505-C cell line (14.41% for TPC-1 cells vs 2.21% for 8505-C cells; P = 0.021). These results indicate that p53 is also important for the senescence effect of rotenone.
3.3. Rotenone-induced cell death and cell cycle arrest is independent from the mitochondrial production of ROS
Since oxidative stress induced by rotenone was advanced to be required to trigger cell death [6, 7], we evaluated the formation of ROS in rotenone-treated TPC-1 cells. Using DHE (for superoxide detection), the production of ROS showed a significant increase both at 6 hours (Fig. 5A and 5C; 1.25 folds with 5 µM rotenone; P = 0.037), and at 24 hours (Fig. 5B and 5C; 2.12 folds with 5 µM rotenone; P = 0.037), in a dose-dependent manner. This effect was not cell type-specific, since 8505-C and HEK293 cells were also stimulated to produce ROS (Supplemental Fig. 5A and 5B). Detection of ROS with DHR 123 (detects hydrogen peroxide, peroxonitrite and hypoclorous acid) provided similar results (Supplemental Fig. 5C–5E).
Figure 5.
Rotenone-induced cell death and cell cycle arrest is independent from the mitochondrial production of ROS. (A–F) The production of superoxide was measured using dihydroethidium as a probe; (A) and (B) are representative flow cytometry histograms of TPC-1 cells after treatment with rotenone (ROT) and quantification of the data is presented in (C); (D) and (E) are representative flow cytometry histograms of CMPBR3 (D) and 143B ρ0 (E) cells after treatment with rotenone for 24 hours and quantification of the data is presented in (F). In the histograms presented in (A), (B), (D) and (E), the gray areas correspond to control (vehicle) and the white areas represent treatment with 5 µM rotenone. (G–H) Cell cycle analysis of CMPBR3 (G) and 143B ρ0 (H) cells by staining with propidium iodide after treatment with control (vehicle) or 5 µM rotenone for 24 hours. Data is representative of at least three independent experiments. Data shown in (C), (F), (G) and (H) represents the mean ± s.e.m. *, P<0.05.
On the other hand, we checked ROS production in 143B ρ0 cells (depleted of mtDNA) and in CMPBR3 cells (obtained by fusing the previous cell line with mitochondria from platelets harboring wild type mtDNA). The results showed a remarkable increase in ROS formation in CMPBR3 cells upon rotenone treatment (Fig. 5D and 5F; about 2.5 folds with 5 µM rotenone; P = 0.028), but not in 143B ρ0 cells (Fig. 5E and 5F; 1,16 folds with 5 µM rotenone; P = 0.487).
Analysis of the cell cycle in 143B ρ0 and CMPBR3 cells treated with 5 µM rotenone for 24 hours show that both cell lines displayed a significant increase in the percentage of dead cells (Fig. 5G and 5H; 23.16% for 5 µM rotenone vs 3.01% for the control in CMPBR3 cells, P = 0.018; 17.53% for 5 µM rotenone vs 1.70% for the control in 143 B ρ0 cells, P = 0.028) and in the percentage of cells in G2/M (64.80% for 5 µM rotenone vs 34.73% for the control in CMPBR3 cells, P = 0.021; 53.49% for 5 µM rotenone vs 27.37% for the control in 143B ρ0 cells, P = 0.034), similarly to the results obtained with TPC-1 and 8505-C cells. Thus, it appears that cell death and cell cycle arrest induced by rotenone is independent from the mitochondrial production of ROS, at least in this model. In addition, the fact that 143B cells harbor a point mutation at the DNA-binding domain of TP53 that translates to the substitution Arg156Pro [29], leading to a non-functional protein [28], further corroborates that a functional p53 is not required for the rotenone-induced cell cycle arrest.
3.4. Treatment with rotenone leads to alterations in ΔΨm, biogenesis and morphology
We assessed mitochondrial function in TPC-1 cells treated with rotenone by estimation of different parameters. A dual effect on the ΔΨm was observed (Fig. 6A and 6B). There is a fraction of cells with loss of ΔΨm (21.25% for 5 µM rotenone vs control; P = 0.014) and a second population of cells with gain of ΔΨm (7.82% for 5 µM rotenone vs control; P = 0.014) after treatment with rotenone for 24 hours. The population of cells with loss of ΔΨm likely corresponds to those that are in the process of cell death. The observed hyperpolarization may result from an increase in mitochondrial mass [30]. In fact, there is a significant 1.5 fold increase in the number of mitochondria (Fig. 6C and 6D; P = 0.037), suggesting that the drug is stimulating mitochondrial biogenesis. Concordantly, we observed an increase in the expression of SSBP1 (Fig. 6E), a protein that is upregulated during mitochondrial biogenesis [31]. Thus, the gain of ΔΨm may be a consequence of the increase in the number of mitochondria. Rotenone treatment also leads to the presence of cells with a giant nucleus surrounded by a massive network of fused mitochondria (Fig. 6G). This correlates with an increase in the expression of the pro-fusion protein OPA1 (Fig. 6F). Integrity of mitochondria seems to be maintained since we did not observe release of cytochrome c to the cytoplasm (Supplemental Fig. 6).
Figure 6.

Rotenone leads to alterations in mitochondrial transmembrane potential, biogenesis and morphology of TPC-1 cells. The mitochondrial membrane potential was determined by incubating cells with CMXRos followed by analysis through flow cytometry histograms (A) and quantification of the data (B). The mitochondrial mass was assessed by staining cells with nonyl acridine orange followed by analysis through flow cytometry histograms (C) and quantification of the data (D). In the histograms presented in (A) and (C), the gray areas correspond to control (vehicle) and the white areas represent treatment with 5 µM rotenone. Whole lysates of cells treated with the indicated concentrations of rotenone (ROT) were immunoblotted against SSBP1 (E) and OPA1 (F); in (F), the arrow head points to the mature form of OPA1. (G) The morphology of mitochondria was evaluated by microscopy after treatment of cells with 5 µM rotenone and incubation with CMXRos (left column and red in the right column) and staining with DAPI for nuclei (middle column and blue in the right column); scale bar: 10 µm. Data is representative of at least three independent experiments. Data shown in (B) and (D) represents the mean ± s.e.m. *, P<0.05.
The growth of HEK293 cells, which are derived from non-neoplastic embryonic kidney tissue, is also inhibited by rotenone and the other effects described above for TPC-1 cells, such as cell cycle arrest followed by massive cell death induction, increased expression of p53 and p21 and production of ROS are also observed (data not shown).
3.5. Rotenone and staurosporine act synergistically in inhibiting the growth of thyroid cancer cells in a dose-dependent manner
Synergistic combinations of drugs represent a good strategy to treat diseases, namely cancer, because they offer several advantages such as increasing the efficacy of the treatment, decreasing the dose of the compounds and slowing down the development of drug resistance [21]. Hence, we decided to evaluate whether rotenone could interact synergistically with staurosporine, a potent cell death inducer, as observed recently in the filamentous fungus Neurospora crassa [9]. In fact, there is an increase in the percentage of inhibition of cell growth when the drugs were combined vs either drug used alone (Fig. 7A). The median-effect dose of the 1:1000 (staurosporine:rotenone) combination was 2 nM/µM, meaning a more powerful effect when compared with the median-effect doses of 23 nM and 26 µM, for staurosporine or rotenone alone, respectively) (data not shown). To quantify the synergistic interaction between the drugs we used the combination index (CI), which is based on the median-effect equation and according which there is synergy when CI < 1 [21]. As depicted in Fig. 7B, despite the presence of 4 points with CI > 1, there is a dose-dependent tendency to have experimental points falling into the CI < 1 region, as illustrated by the trend line presented in the graph. Another index calculated using the same data, the dose reduction index (DRI), further supports our conclusions. DRI is a measure of how many folds the dose of each drug in a synergistic combination may be reduced at a given effect level compared with the doses of each drug alone [21]. In this case the greater DRI (DRI > 1) value indicates a greater dose reduction for a given therapeutic effect [21]. The DRI calculations (Fig. 7C) for almost all doses of the drugs are greater than 1.
Figure 7.
Staurosporine and rotenone have a synergistic effect against the growth of TPC-1 cells. (A) Growth inhibition was determined by the sulphorhodamine B assay, after 24 hours of treatment, for the indicated concentrations of staurosporine, rotenone or the combination of both drugs. Synergy was evaluated according to the combination index (B) and the dose reduction index (C) using the software CalcuSyn; in (B) the crosses correspond to experimental values and the curve is a trend line; in (C) the dashed line corresponds to DRI = 1. Data is representative of at least three independent experiments.
4. Discussion
A deregulation in the normal processes of cell death and cell cycle is a common feature of cancer cells [32]. Rotenone was previously shown to possess anti-tumoral properties by its capacity to induce cell death, both in vitro and in vivo [5–7, 14–17, 33]. Here we show that rotenone-induced inhibition of the growth of cancer cells is potentiated by the death-inducer staurosporine in a dose-dependent manner. A similar synergistic effect of this drug combination was observed in cells of the filamentous fungus Neurospora crassa [9]. Because the administration of drugs to cells usually elicit collateral toxic effects, a strategy to minimize them could be decreasing the dose of the drug applied and this may be achieved by using drug combinations as we report. Future experiments are required in order to evaluate the possible translation of the results to in vivo models.
Since rotenone is an inhibitor of respiratory chain complex I [6], we evaluated its effect on mitochondria. The mitochondrial network morphology was modified but the organelles seemed intact and we did not observe release of cytochrome c. As expected, a significant percentage of the cells lost their ΔΨm. Yet, other cells exhibited an increase of ΔΨm, which may be explained by some stimulation of mitochondrial biogenesis by rotenone. The observed higher number of mitochondria may account for the increase in the potential per cell, as already proposed [30]. It was proposed recently that p53 regulates mitochondrial biogenesis and mtDNA copy number [34, 35], in line with our findings that the effects of rotenone involve p53. Rotenone treatment also increased ROS production. Nevertheless, ROS do not appear to be essential for rotenone-induced cell death in our model. Cells with depleted mtDNA are not stimulated to produce ROS, but become arrested in the cell cycle and undergo death to a similar extent as cells containing wild type mtDNA. This is in agreement with reports that rotenone-induced apoptosis does not depend directly on inhibition of the mitochondrial respiratory chain [8, 9].
We found that treatment with rotenone induces cell cycle arrest in the G2/M phases, as previously shown [5, 7]. Further, we could observe morphological features of mitotic catastrophe, in particular the appearance of typical giant clusters of nuclei along with micronucleation and multinucleation [3, 4]. Other observations support the idea that rotenone induces mitotic catastrophe. Mitotic catastrophe can be triggered upon microtubule instability leading to an abnormal mitotic checkpoint [3, 4], and rotenone binding to tubulin results in the inhibition of the spindle assembly and thus in an anomalous spindle checkpoint [10–13]. In addition, rotenone can induce endoreduplication [36], a process through which successive rounds of DNA replication take place without mitosis, which may facilitate the polyploidization of cells [37].
Treatment of TPC-1 cells with rotenone increased the levels of p53 together with its phosphorylation at serine 15 and its nuclear accumulation. These three consequences are common responses to DNA or spindle damage [24], suggesting the involvement of the protein in rotenone-induced cell death. Cells arrested in response to spindle assembly inhibition eventually bypass the block and exit mitosis without an appropriate division, in a phenomenon known as mitotic slippage [3, 4, 38]. Consequently, cells enter in G1 with a 4N DNA content and this abnormality leads to the activation of the G1 checkpoint [3, 4, 38]. This halt is mediated by p53 and p21 appears to be a crucial p53-target gene in this postmitotic checkpoint [25, 26]. Moreover, serine 15 phosphorylation stimulates p53 transactivation and is required for this postmitotic checkpoint [23, 39]. Similarly, we observed p53 increase and phosphorylation and upregulation of p21 following rotenone exposure. An increase in the levels of p53 after treatment with microtubule disruptors occurring only after tetraploid cells exit mitotic arrest and progress to interphase was also demonstrated [26].
On the other hand, functional p53 is not an absolute requirement for the rotenone-induced cell cycle arrest and mitotic catastrophe because cells harboring a mutation in the DNA-binding domain of the molecule still become arrested and display polyploidy after treatment with the drug. This is consistent with evidence that in the absence of p53, cells have the ability to activate alternative pathways to halt the cell cycle [3]. However, our results stress that the extensive cell death occurring after the mitotic catastrophe state is p53-dependent. The majority of the cells with wild type p53 overrides the cell cycle blockage and dies after 48 hours of rotenone treatment whereas cells with mutant p53 remain arrested in the cell cycle in the same period and do not show extensive cell death. Kato et al showed that the mutation present in the 8505-C cell line (Arg248Gly) leads to a total absence of the capacity of p53 to transactivate key players of cell death execution such as Bax, NOXA and Apoptosis-inducing Protein 1 [40]. This may explain why 8505-C cells are less prone to die upon treatment with rotenone.
Our results also suggest involvement of p53 in rotenone-induced cellular senescence, a phenomenon that can occur as a consequence of mitotic catastrophe [41–43]. We observed that when treated for a longer period with rotenone, cells exhibit a senescent-like phenotype. Cells possessing non-functional p53 display significantly less senescence than cells harboring the wild type protein, suggesting that the rotenone-induced cellular senescence occurs through a p53-pathway. In fact, p53 and p16 are the initiators of two independent senescence pathways [22] and we did not observe increase in the expression of p16 (data not shown).
In summary, we show that rotenone induces a panoply of effects, including mitotic catastrophe, mitotic slippage, cell death and cellular senescence. Our results are in agreement with findings that rotenone may act by at least two different pathways [5, 7, 8, 12, 13]: acting directly on mitochondria and in the nucleus. The tumor suppressor protein p53 is involved in these processes, specially regarding the extensive cell death that occurs following a polyploidy state.
Supplementary Material
Acknowledgements
We thank Joana Nunes (IPATIMUP) for the help with the ρ0 and cybrid cell lines. A.P.G. was recipient of a fellowship from Fundação Calouste Gulbenkian (reference 104210). K.K.S. was supported by a NIH grant (RO1-121904). This work was supported by Fundação para a Ciência e Tecnologia.
Abbreviations
- ROS
reactive oxygen species
- mtDNA
mitochondrial DNA
- DMSO
dimethyl sulfoxide
- DHE
dihydroethidium
- DHR 123
dihydrorhodamine 123
- X-gal
5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside
- DAPI
4',6-diamidino-2-phenylindole
- FITC
fluorescein isothiocyanate
- ΔΨm
mitochondrial membrane potential
- CI
combination index
- DRI
dose reduction index
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kroemer G, Galluzzi L, Vandenabeele P, Abrams J, Alnemri ES, Baehrecke EH, Blagosklonny MV, El-Deiry WS, Golstein P, Green DR, Hengartner M, Knight RA, Kumar S, Lipton SA, Malorni W, Nunez G, Peter ME, Tschopp J, Yuan J, Piacentini M, Zhivotovsky B, Melino G. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 2009;16:3–11. doi: 10.1038/cdd.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hotchkiss RS, Strasser A, McDunn JE, Swanson PE. Cell death. N Engl J Med. 2009;361:1570–1583. doi: 10.1056/NEJMra0901217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vakifahmetoglu H, Olsson M, Zhivotovsky B. Death through a tragedy: mitotic catastrophe. Cell Death Differ. 2008;15:1153–1162. doi: 10.1038/cdd.2008.47. [DOI] [PubMed] [Google Scholar]
- 4.Castedo M, Perfettini JL, Roumier T, Andreau K, Medema R, Kroemer G. Cell death by mitotic catastrophe: a molecular definition. Oncogene. 2004;23:2825–2837. doi: 10.1038/sj.onc.1207528. [DOI] [PubMed] [Google Scholar]
- 5.Armstrong JS, Hornung B, Lecane P, Jones DP, Knox SJ. Rotenone-induced G2/M cell cycle arrest and apoptosis in a human B lymphoma cell line PW. Biochem Biophys Res Commun. 2001;289:973–978. doi: 10.1006/bbrc.2001.6054. [DOI] [PubMed] [Google Scholar]
- 6.Li N, Ragheb K, Lawler G, Sturgis J, Rajwa B, Melendez JA, Robinson JP. Mitochondrial complex I inhibitor rotenone induces apoptosis through enhancing mitochondrial reactive oxygen species production. J Biol Chem. 2003;278:8516–8525. doi: 10.1074/jbc.M210432200. [DOI] [PubMed] [Google Scholar]
- 7.Lee J, Huang MS, Yang IC, Lai TC, Wang JL, Pang VF, Hsiao M, Kuo MY. Essential roles of caspases and their upstream regulators in rotenone-induced apoptosis. Biochem Biophys Res Commun. 2008;371:33–38. doi: 10.1016/j.bbrc.2008.03.149. [DOI] [PubMed] [Google Scholar]
- 8.Choi WS, Kruse SE, Palmiter RD, Xia Z. Mitochondrial complex I inhibition is not required for dopaminergic neuron death induced by rotenone, MPP+, or paraquat. Proc Natl Acad Sci U S A. 2008;105:15136–15141. doi: 10.1073/pnas.0807581105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castro A, Lemos C, Falcao A, Fernandes AS, Glass NL, Videira A. Rotenone enhances the antifungal properties of staurosporine. Eukaryot Cell. 2010;9:906–914. doi: 10.1128/EC.00003-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meisner HM, Sorensen L. Metaphase arrest of Chinese hamster cells with rotenone. Exp Cell Res. 1966;42:291–295. doi: 10.1016/0014-4827(66)90292-8. [DOI] [PubMed] [Google Scholar]
- 11.Brinkley BR, Barham SS, Barranco SC, Fuller GM. Rotenone inhibition of spindle microtubule assembly in mammalian cells. Exp Cell Res. 1974;85:41–46. doi: 10.1016/0014-4827(74)90210-9. [DOI] [PubMed] [Google Scholar]
- 12.Ren Y, Liu W, Jiang H, Jiang Q, Feng J. Selective vulnerability of dopaminergic neurons to microtubule depolymerization. J Biol Chem. 2005;280:34105–34112. doi: 10.1074/jbc.M503483200. [DOI] [PubMed] [Google Scholar]
- 13.Srivastava P, Panda D. Rotenone inhibits mammalian cell proliferation by inhibiting microtubule assembly through tubulin binding. FEBS J. 2007;274:4788–4801. doi: 10.1111/j.1742-4658.2007.06004.x. [DOI] [PubMed] [Google Scholar]
- 14.Cunningham ML, Soliman MS, Badr MZ, Matthews HB. Rotenone, an anticarcinogen, inhibits cellular proliferation but not peroxisome proliferation in mouse liver. Cancer Lett. 1995;95:93–97. doi: 10.1016/0304-3835(95)03869-x. [DOI] [PubMed] [Google Scholar]
- 15.Gerhauser C, Mar W, Lee SK, Suh N, Luo Y, Kosmeder J, Luyengi L, Fong HH, Kinghorn AD, Moriarty RM, et al. Rotenoids mediate potent cancer chemopreventive activity through transcriptional regulation of ornithine decarboxylase. Nat Med. 1995;1:260–266. doi: 10.1038/nm0395-260. [DOI] [PubMed] [Google Scholar]
- 16.Yoshitani SI, Tanaka T, Kohno H, Takashima S. Chemoprevention of azoxymethane-induced rat colon carcinogenesis by dietary capsaicin and rotenone. Int J Oncol. 2001;19:929–939. doi: 10.3892/ijo.19.5.929. [DOI] [PubMed] [Google Scholar]
- 17.Tanaka T, Kohno H, Sakata K, Yamada Y, Hirose Y, Sugie S, Mori H. Modifying effects of dietary capsaicin and rotenone on 4-nitroquinoline 1-oxide-induced rat tongue carcinogenesis. Carcinogenesis. 2002;23:1361–1367. doi: 10.1093/carcin/23.8.1361. [DOI] [PubMed] [Google Scholar]
- 18.Saffran HA, Pare JM, Corcoran JA, Weller SK, Smiley JR. Herpes simplex virus eliminates host mitochondrial DNA. EMBO Rep. 2007;8:188–193. doi: 10.1038/sj.embor.7400878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kulawiec M, Owens KM, Singh KK. mtDNA G10398A variant in African-American women with breast cancer provides resistance to apoptosis and promotes metastasis in mice. J Hum Genet. 2009;54:647–654. doi: 10.1038/jhg.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 2006;58:621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 22.Campisi J, d'Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 23.Dumaz N, Meek DW. Serine15 phosphorylation stimulates p53 transactivation but does not directly influence interaction with HDM2. EMBO J. 1999;18:7002–7010. doi: 10.1093/emboj/18.24.7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jimenez GS, Bryntesson F, Torres-Arzayus MI, Priestley A, Beeche M, Saito S, Sakaguchi K, Appella E, Jeggo PA, Taccioli GE, Wahl GM, Hubank M. DNA-dependent protein kinase is not required for the p53-dependent response to DNA damage. Nature. 1999;400:81–83. doi: 10.1038/21913. [DOI] [PubMed] [Google Scholar]
- 25.Lanni JS, Jacks T. Characterization of the p53-dependent postmitotic checkpoint following spindle disruption. Mol Cell Biol. 1998;18:1055–1064. doi: 10.1128/mcb.18.2.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andreassen PR, Lohez OD, Lacroix FB, Margolis RL. Tetraploid state induces p53-dependent arrest of nontransformed mammalian cells in G1. Mol Biol Cell. 2001;12:1315–1328. doi: 10.1091/mbc.12.5.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meireles AM, Preto A, Rocha AS, Rebocho AP, Maximo V, Pereira-Castro I, Moreira S, Feijao T, Botelho T, Marques R, Trovisco V, Cirnes L, Alves C, Velho S, Soares P, Sobrinho-Simoes M. Molecular and genotypic characterization of human thyroid follicular cell carcinoma-derived cell lines. Thyroid. 2007;17:707–715. doi: 10.1089/thy.2007.0097. [DOI] [PubMed] [Google Scholar]
- 28.Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, Olivier M. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007;28:622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 29.Ottaviano L, Schaefer KL, Gajewski M, Huckenbeck W, Baldus S, Rogel U, Mackintosh C, de Alava E, Myklebost O, Kresse SH, Meza-Zepeda LA, Serra M, Cleton-Jansen AM, Hogendoorn PC, Buerger H, Aigner T, Gabbert HE, Poremba C. Molecular characterization of commonly used cell lines for bone tumor research: a trans-European EuroBoNet effort. Genes Chromosomes Cancer. 2010;49:40–51. doi: 10.1002/gcc.20717. [DOI] [PubMed] [Google Scholar]
- 30.Reed JC, Jurgensmeier JM, Matsuyama S. Bcl-2 family proteins and mitochondria. Biochim Biophys Acta. 1998;1366:127–137. doi: 10.1016/s0005-2728(98)00108-x. [DOI] [PubMed] [Google Scholar]
- 31.Schultz RA, Swoap SJ, McDaniel LD, Zhang B, Koon EC, Garry DJ, Li K, Williams RS. Differential expression of mitochondrial DNA replication factors in mammalian tissues. J Biol Chem. 1998;273:3447–3451. doi: 10.1074/jbc.273.6.3447. [DOI] [PubMed] [Google Scholar]
- 32.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 33.Chen Y, McMillan-Ward E, Kong J, Israels SJ, Gibson SB. Mitochondrial electron-transport-chain inhibitors of complexes I and II induce autophagic cell death mediated by reactive oxygen species. J Cell Sci. 2007;120:4155–4166. doi: 10.1242/jcs.011163. [DOI] [PubMed] [Google Scholar]
- 34.Kulawiec M, Ayyasamy V, Singh KK. p53 regulates mtDNA copy number and mitocheckpoint pathway. J Carcinog. 2009;8:8. doi: 10.4103/1477-3163.50893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lebedeva MA, Eaton JS, Shadel GS. Loss of p53 causes mitochondrial DNA depletion and altered mitochondrial reactive oxygen species homeostasis. Biochim Biophys Acta. 2009;1787:328–334. doi: 10.1016/j.bbabio.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsumoto K, Ohta T. Rotenone induces aneuploidy, polyploidy and endoreduplication in cultured Chinese hamster cells. Mutat Res. 1991;263:173–177. doi: 10.1016/0165-7992(91)90058-c. [DOI] [PubMed] [Google Scholar]
- 37.Lee HO, Davidson JM, Duronio RJ. Endoreplication: polyploidy with purpose. Genes Dev. 2009;23:2461–2477. doi: 10.1101/gad.1829209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gascoigne KE, Taylor SS. How do anti-mitotic drugs kill cancer cells? J Cell Sci. 2009;122:2579–2585. doi: 10.1242/jcs.039719. [DOI] [PubMed] [Google Scholar]
- 39.Tritarelli A, Oricchio E, Ciciarello M, Mangiacasale R, Palena A, Lavia P, Soddu S, Cundari E. p53 localization at centrosomes during mitosis and postmitotic checkpoint are ATM-dependent and require serine 15 phosphorylation. Mol Biol Cell. 2004;15:3751–3757. doi: 10.1091/mbc.E03-12-0900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kato S, Han SY, Liu W, Otsuka K, Shibata H, Kanamaru R, Ishioka C. Understanding the function-structure and function-mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc Natl Acad Sci U S A. 2003;100:8424–8429. doi: 10.1073/pnas.1431692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eom YW, Kim MA, Park SS, Goo MJ, Kwon HJ, Sohn S, Kim WH, Yoon G, Choi KS. Two distinct modes of cell death induced by doxorubicin: apoptosis and cell death through mitotic catastrophe accompanied by senescence-like phenotype. Oncogene. 2005;24:4765–4777. doi: 10.1038/sj.onc.1208627. [DOI] [PubMed] [Google Scholar]
- 42.Huang X, Tran T, Zhang L, Hatcher R, Zhang P. DNA damage-induced mitotic catastrophe is mediated by the Chk1-dependent mitotic exit DNA damage checkpoint. Proc Natl Acad Sci U S A. 2005;102:1065–1070. doi: 10.1073/pnas.0409130102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li S, Szymborski A, Miron MJ, Marcellus R, Binda O, Lavoie JN, Branton PE. The adenovirus E4orf4 protein induces growth arrest and mitotic catastrophe in H1299 human lung carcinoma cells. Oncogene. 2009;28:390–400. doi: 10.1038/onc.2008.393. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.